Salicylic Acid Is Required for Broad-Spectrum Disease Resistance in Rice
Abstract
:1. Introduction
2. Results
2.1. OsSAH Proteins Have SA 3- and/or 5-Hydroxylase Activity In Vitro
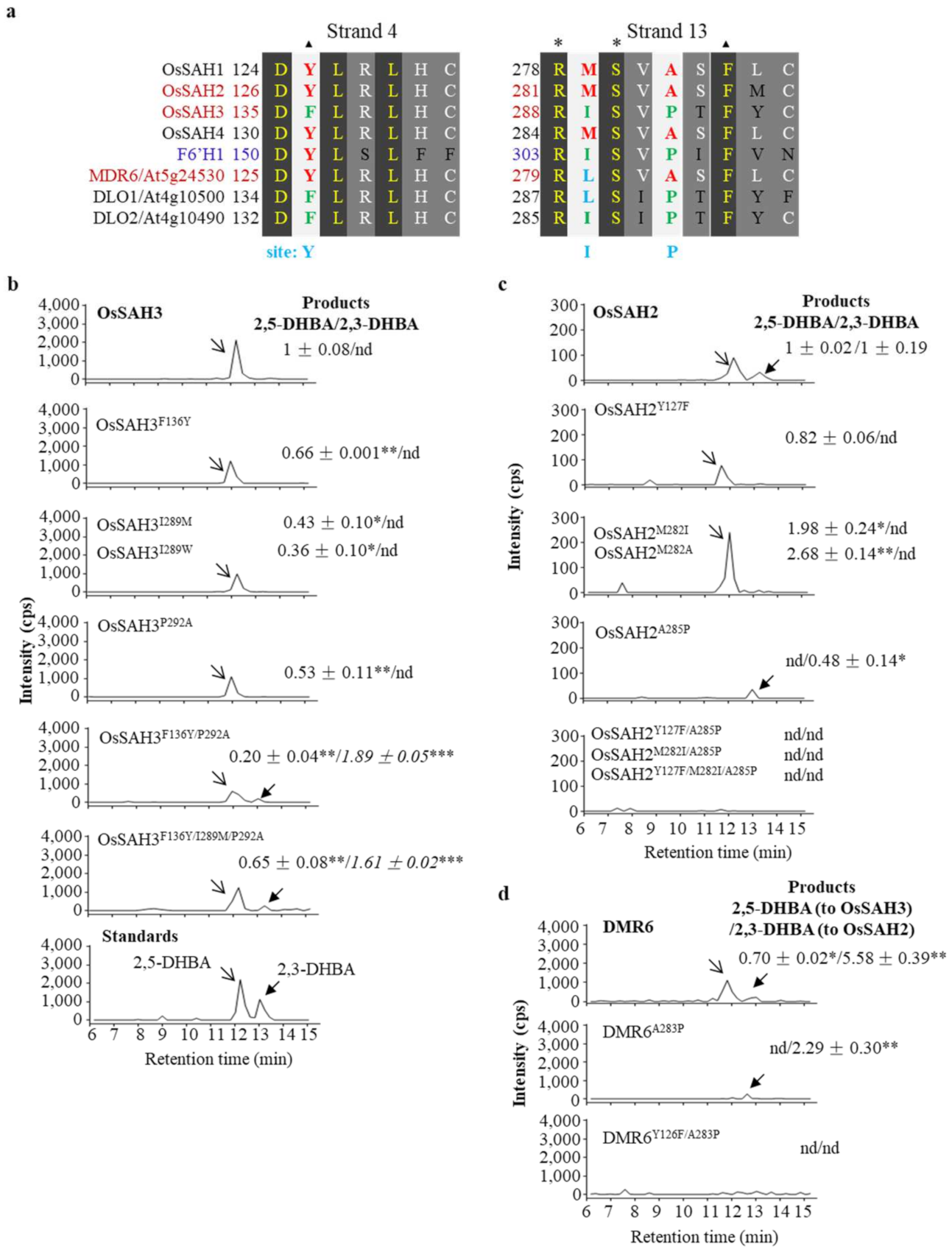
2.2. Key Amino Acids of SAHs Affect Enzymatic Activity
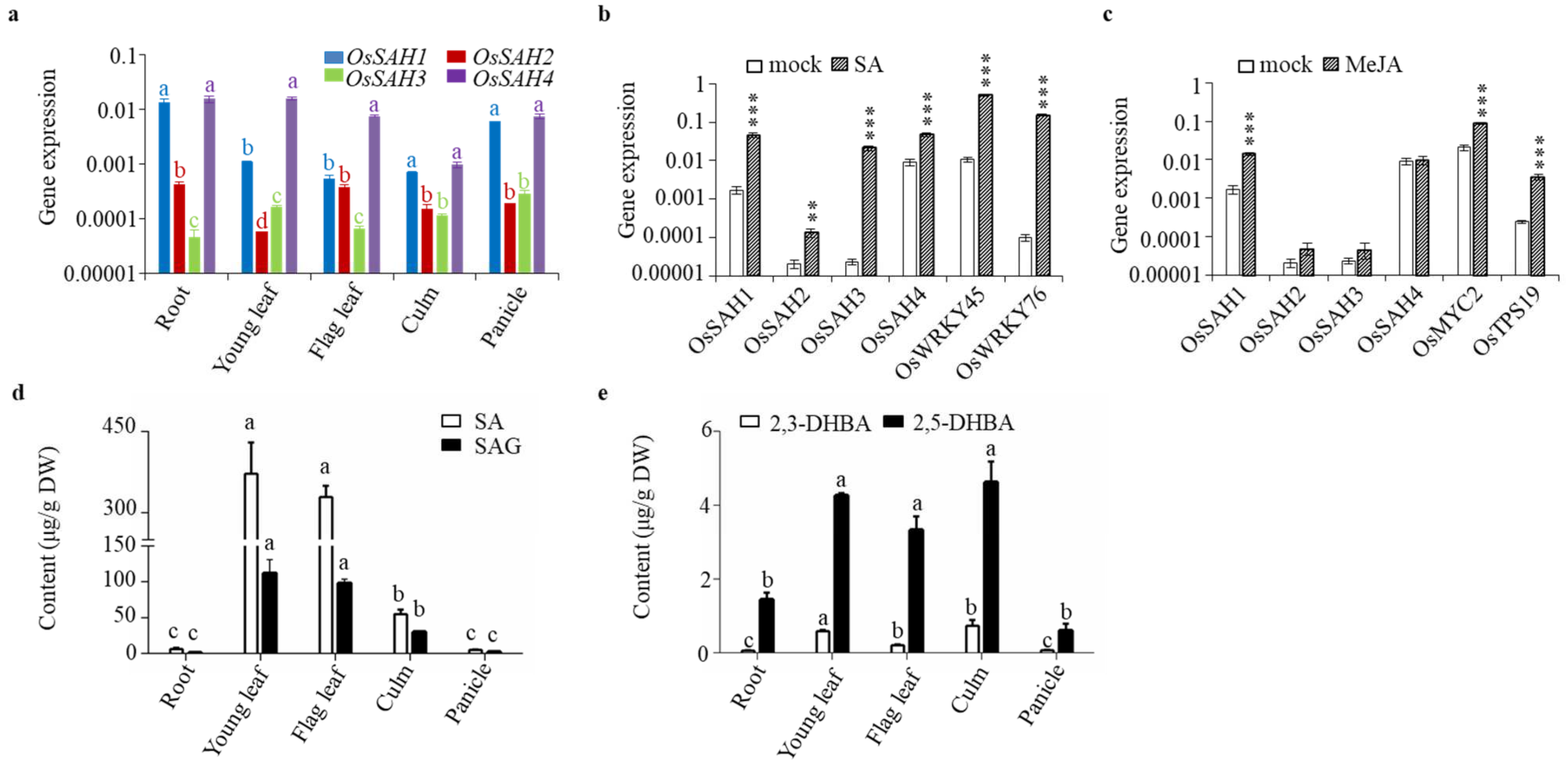
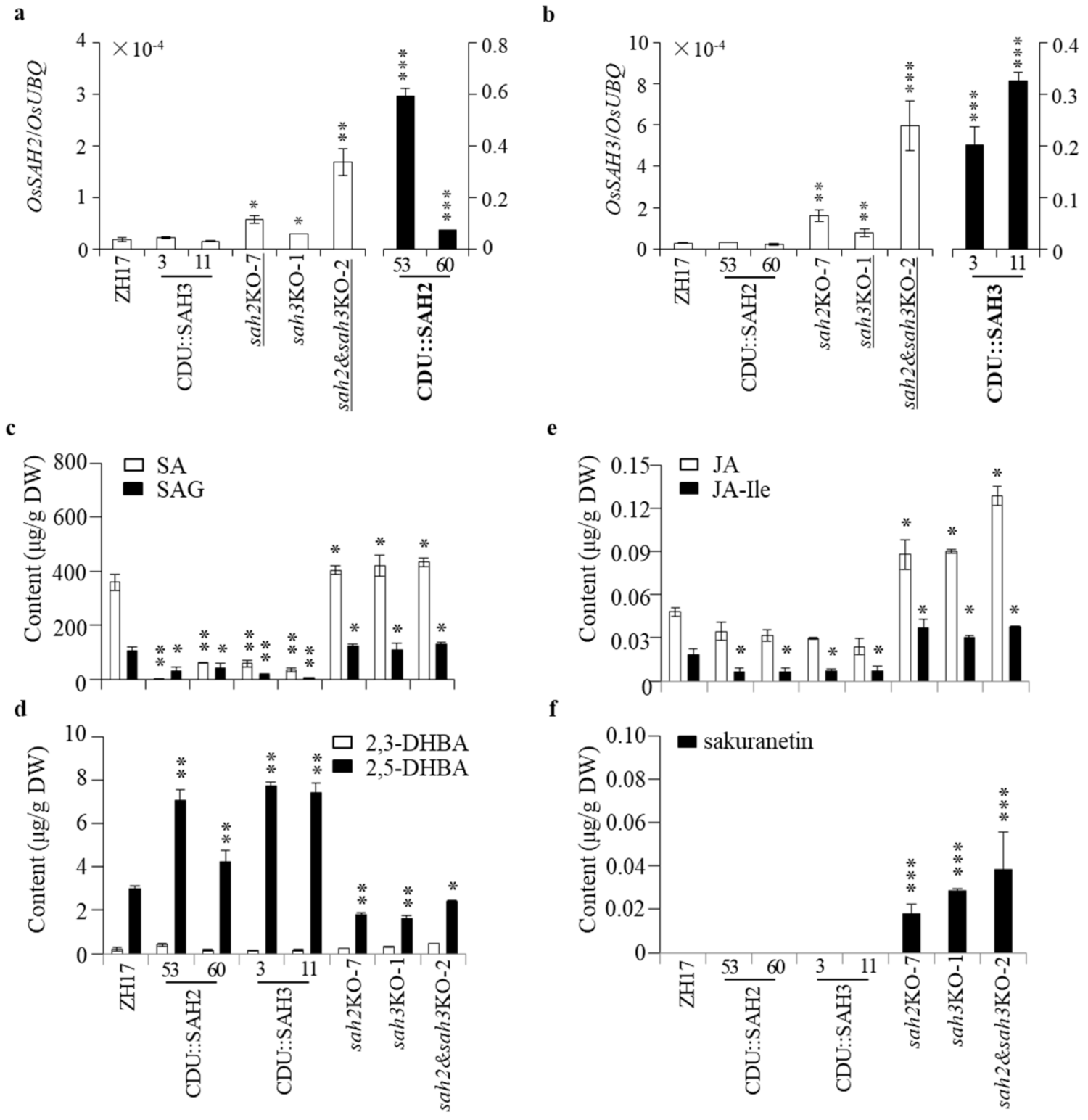
2.3. Patterns of OsSAH Expression and Phenolic Accumulation
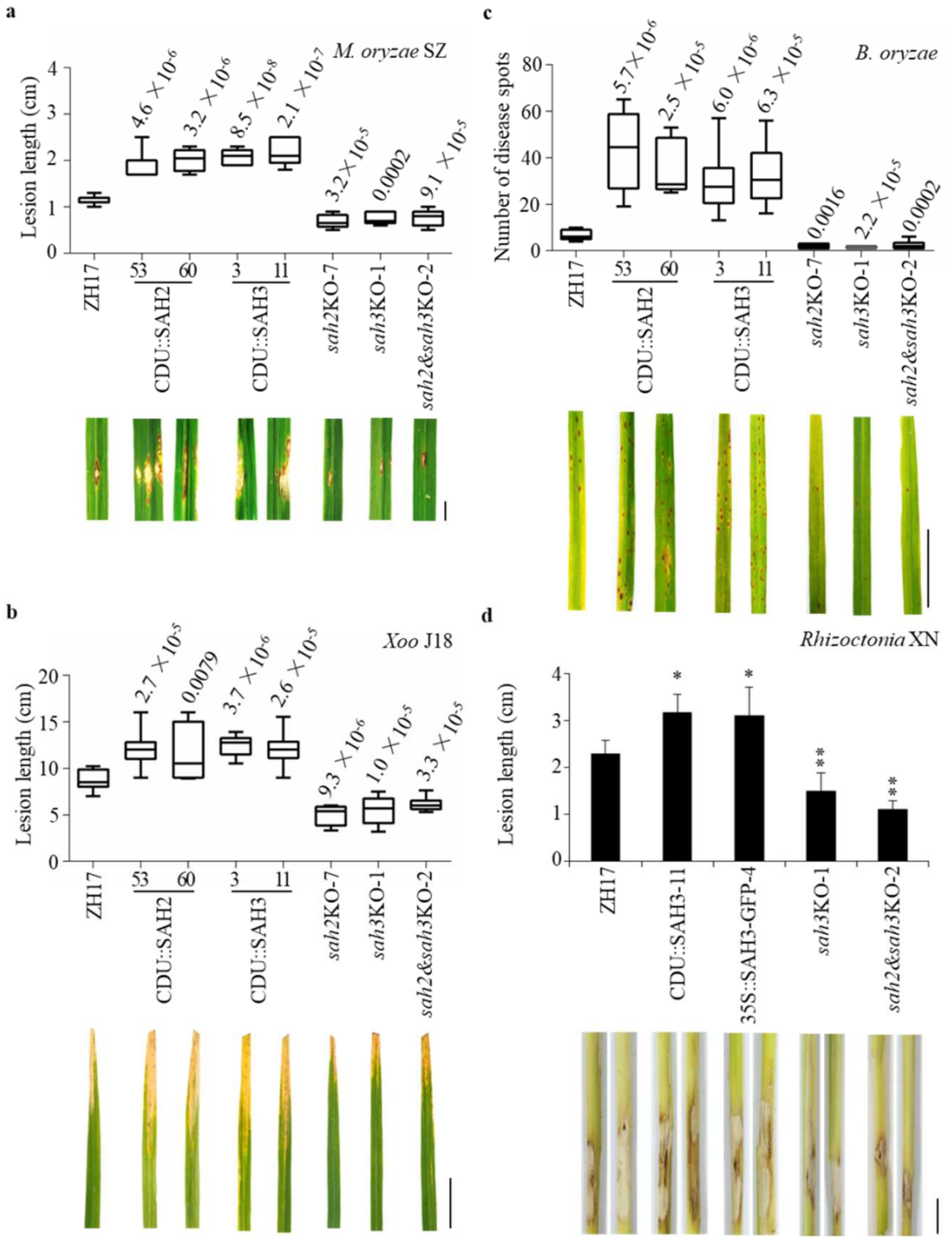
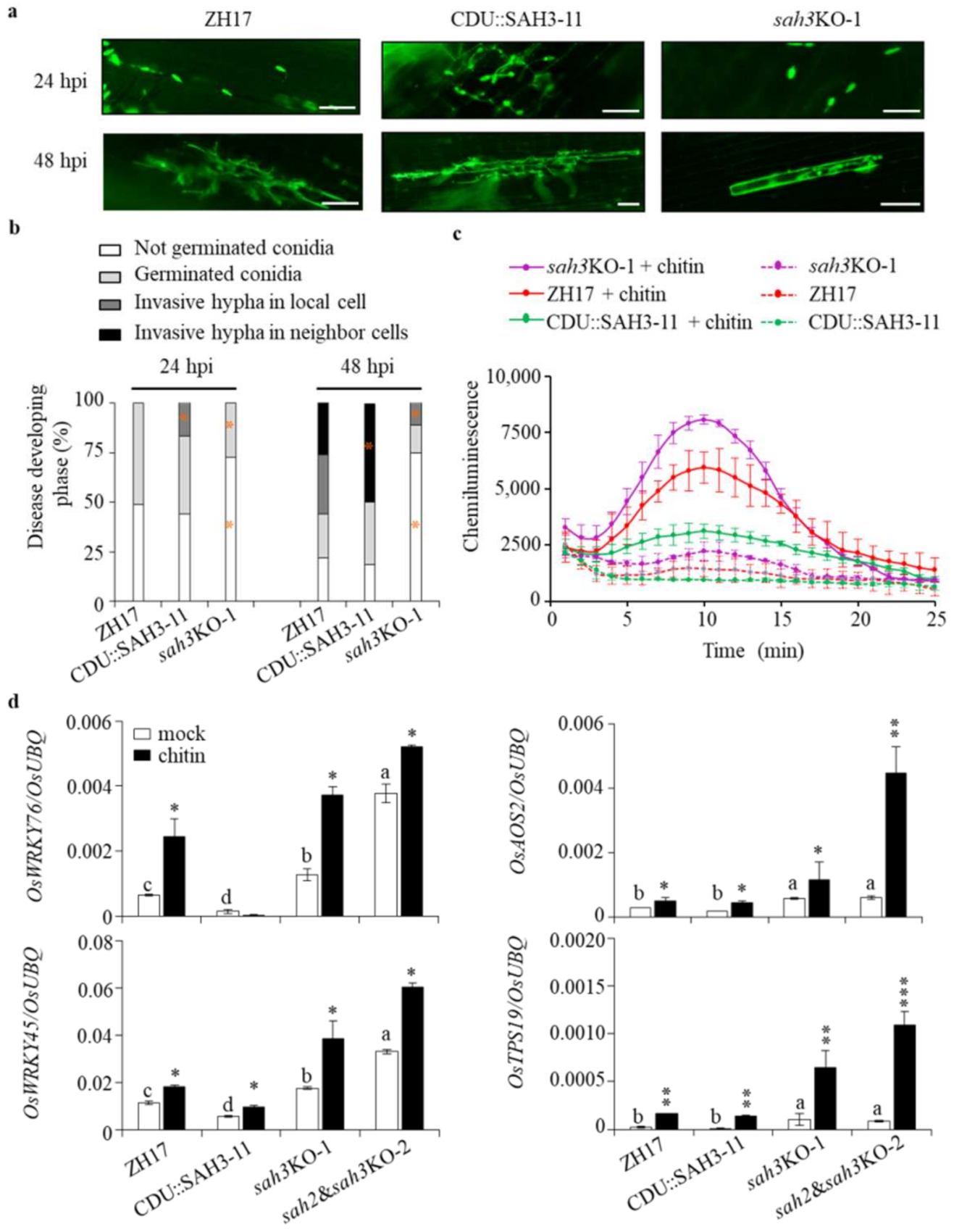
2.4. Knockout OsSAH2–3 Genes Enhance Resistance against Hemibiotrophic and Necrotrophic Pathogens
2.5. Changes of OsSAHs Expression Alter SA Homeostasis
2.6. Restricted Induction of OsSAH3 Promoter by M. oryzae
2.7. Knockout OsSAH Genes Elevate Basal Resistance
3. Discussion
4. Materials and Methods
4.1. Generation of Transgenic Plants
4.2. Plant Growth and Treatments
4.3. Pathogen Inoculation
4.4. ROS Detection
4.5. Protein Expression and Enzyme Assays
4.6. Determination of Metabolites
4.7. Structural Modeling and Substrate Docking
4.8. GUS Staining
4.9. Reverse Transcribed Quantitative (qRT-PCR) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, X. Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 2019, 50, 29–36. [Google Scholar] [CrossRef] [PubMed]
- LefeverE, H.; Bauters, L.; Gheysen, G. Salicylic Acid Biosynthesis in Plants. Front. Plant Sci. 2020, 11, 338. [Google Scholar] [CrossRef] [PubMed]
- Garcion, C.; Lohmann, A.; Lamodière, E.; Catinot, J.; Buchala, A.; Doermann, P.; Métraux, J.-P. Characterization and Biological Function of the ISOCHORISMATE SYNTHASE2 Gene of Arabidopsis. Plant Physiol. 2008, 147, 1279–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rekhter, D.; Lüdke, D.; Ding, Y.; Feussner, K.; Zienkiewicz, K.; Lipka, V.; Wiermer, M.; Zhang, Y.; Feussner, I. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 2019, 365, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Torrens-Spence, M.P.; Bobokalonova, A.; Carballo, V.; Glinkerman, C.M.; Pluskal, T.; Shen, A.; Weng, J.-K. PBS3 and EPS1 Complete Salicylic Acid Biosynthesis from Isochorismate in Arabidopsis. Mol. Plant 2019, 12, 1577–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dean, J.V.; Delaney, S.P. Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant. 2008, 132, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Noutoshi, Y.; Okazaki, M.; Kida, T.; Nishina, Y.; Morishita, Y.; Ogawa, T.; Suzuki, H.; Shibata, D.; Jikumaru, Y.; Hanada, A.; et al. Novel Plant Immune-Priming Compounds Identified via High-Throughput Chemical Screening Target Salicylic Acid Glucosyltransferases in Arabidopsis. Plant Cell 2012, 24, 3795–3804. [Google Scholar] [CrossRef] [Green Version]
- Van Damme, M.; Huibers, R.P.; Elberse, J.; Van den Ackerveken, G. Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J. 2008, 54, 785–793. [Google Scholar] [CrossRef]
- Zeilmaker, T.; Ludwig, N.R.; Elberse, J.; Seidl, M.F.; Berke, L.; Van Doorn, A.; Schuurink, R.C.; Snel, B.; Van den Ackerveken, G. DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 2014, 81, 210–222. [Google Scholar] [CrossRef]
- Zhang, K.W.; Halitschke, R.; Yin, C.X.; Liu, C.J.; Gan, S.S. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. USA 2013, 110, 14807–14812. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhao, L.; Zhao, J.; Li, Y.; Wang, J.; Guo, R.; Gan, S.; Liu, C.-J.; Zhang, K. S5H/DMR6 Encodes a Salicylic Acid 5-Hydroxylase That Fine-Tunes Salicylic Acid Homeostasis. Plant Physiol. 2017, 175, 1082–1093. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Svedin, E.; Mo, H.; Atwell, S.; Dilkes, B.P.; Chapple, C. Exploiting Natural Variation of Secondary Metabolism Identifies a Gene Controlling the Glycosylation Diversity of Dihydroxybenzoic Acids in Arabidopsis thaliana. Genetics 2014, 198, 1267–1276. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.X.; Zhu, G.Q.; Liu, Q.; Chen, L.; Li, Y.J.; Hou, B.K. Modulation of Plant Salicylic Acid-Associated Immune Responses via Glycosylation of Dihydroxybenzoic Acids. Plant Physiol. 2018, 176, 3103–3119. [Google Scholar] [CrossRef]
- Zhou, F.; Last, R.L.; Pichersky, E. Degradation of salicylic acid to catechol in Solanaceae by SA 1-hydroxylase. Plant Physiol. 2021, 185, 876–891. [Google Scholar] [CrossRef]
- Martinez, S.; Hausinger, R.P. Catalytic Mechanisms of Fe(II)- and 2-Oxoglutarate-dependent Oxygenases. J. Biol. Chem. 2015, 290, 20702–20711. [Google Scholar] [CrossRef] [Green Version]
- Hegg, E.L.; Que, L. The 2-His-1-carboxylate facial triad: An emerging structural motif in mononuclear non-heme iron (II) enzymes. Eur. J. Biochem. 1997, 250, 625–629. [Google Scholar] [CrossRef]
- Sun, X.X.; Zhou, D.Y.; Kandavelu, P.; Zhang, H.; Yuan, Q.P.; Wang, B.C.; Rose, J.; Yan, Y. Structural insights into substrate specificity of feruloyl-CoA 6’-hydroxylase from Arabidopsis thaliana. Sci. Rep. 2015, 5, 10355. [Google Scholar] [CrossRef] [Green Version]
- Kawai, Y.; Ono, E.; Mizutani, M. Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 2014, 78, 328–343. [Google Scholar] [CrossRef]
- Herr, C.Q.; Hausinger, R.P. Amazing diversity in biochemical roles of Fe(II)/2-oxoglutarate oxygenases. Trends Biochem. Sci. 2018, 43, 517–532. [Google Scholar] [CrossRef]
- Nadi, R.; Mateo-Bonmati, E.; Juan-Vicente, L.; Micol, J.L. The 2OGD Superfamily: Emerging Functions in Plant Epigenetics and Hormone Metabolism. Mol. Plant 2018, 11, 1222–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.G.; Zhang, Y.H.; Liu, X.; Zhang, X.; Liu, S.C.; Yu, X.W.; Ren, Y.L.; Zheng, X.M.; Zhou, K.N.; Jiang, L.; et al. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev. Cell 2013, 27, 113–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caarls, L.; Elberse, J.; Awwanah, M.; Ludwig, N.R.; De Vries, M.; Zeilmaker, T.; Van Wees, S.C.; Schuurink, R.C.; Ackerveken, G.V.D. Arabidopsis JASMONATE-INDUCED OXYGENASES down-regulate plant immunity by hydroxylation and inactivation of the hormone jasmonic acid. Proc. Natl. Acad. Sci. USA 2017, 114, 6388–6393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Zhao, H.Y.; Ruan, W.Y.; Deng, M.J.; Wang, F.; Peng, J.R.; Luo, J.; Chen, Z.X.; Yi, K. ABNORMAL INFLORESCENCE MERISTEM1 Functions in Salicylic Acid Biosynthesis to Maintain Proper Reactive Oxygen Species Levels for Root Meristem Activity in Rice. Plant Cell 2017, 29, 560–574. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.Q.; Chen, X.J.; Liang, X.X.; Zhou, X.G.; Yang, F.; Liu, J.; He, S.Y.; Guo, Z.J. Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in pathogen defense. Plant Physiol. 2016, 171, 1427–1442. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.N.; Qi, M.; Mei, C.S. Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 2004, 40, 909–919. [Google Scholar] [CrossRef]
- Chen, X.J.; Chen, H.; Yuan, J.S.; Köllner, T.G.; Chen, Y.Y.; Guo, Y.F.; Zhuang, X.F.; Chen, X.L.; Zhang, Y.J.; Fu, J.Y. The rice terpene synthase gene OsTPS19 functions as an (S)-limonene synthase in planta, and its overexpression leads to enhanced resistance to the blast fungus Magnaporthe oryzae. Plant Biotechnol. J. 2018, 16, 1778–1787. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.J. Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: Do We Understand What They Are Whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Guo, Z.J.; Zhou, S.Y.; Han, Q.Y.; Zhang, M.M.; Peng, Y.L.; Hsiang, T.; Chen, X. Evolutionary and genomic comparisons of hybrid uninucleate and nonhybrid Rhizoctonia fungi. Commun. Biol. 2021, 4, 201. [Google Scholar] [CrossRef]
- Yamada, S.; Kano, A.; Tamaoki, D.; Miyamoto, A.; Shishido, H.; Miyoshi, S.; Taniguchi, S.; Akimitsu, K.; Gomi, K. Involvement of OsJAZ8 in Jasmonate-Induced Resistance to Bacterial Blight in Rice. Plant Cell Physiol. 2012, 53, 2060–2072. [Google Scholar] [CrossRef] [Green Version]
- Riemann, M.; Haga, K.; Shimizu, T.; Okada, K.; Ando, S.; Mochizuki, S.; Nishizawa, Y.; Yamanouchi, U.; Nick, P.; Yano, M.; et al. Identification of rice allene oxide cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 2013, 74, 226–238. [Google Scholar] [CrossRef]
- Conrath, U.; Beckers, G.J.; Langenbach, C.J.; Jaskiewicz, M.R. Priming for Enhanced Defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic Acid: Biosynthesis and Signaling. Annu Rev Plant Biol. 2021, 72, 761–791. [Google Scholar] [CrossRef]
- Greenberg, J. Programmed Cell Death in Plant-Pathogen Interactions. Annu. Rev. Plant Biol. 1997, 48, 525–545. [Google Scholar] [CrossRef] [Green Version]
- Radojičić, A.; Li, X.; Zhang, Y. Salicylic Acid: A Double-Edged Sword for Programed Cell Death in Plants. Front. Plant Sci. 2018, 9, 1133. [Google Scholar] [CrossRef] [Green Version]
- Shirasu, K.; Nakajima, H.; Rajasekhar, V.K.; Dixon, R.A.; Lamb, C.J. Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 1997, 9, 261–270. [Google Scholar]
- Rudrappa, T.; Quinn, W.J.; Stanley-Wall, N.R.; Bais, H.P. A degradation product of the salicylic acid pathway triggers oxidative stress resulting in down-regulation of Bacillus subtilis biofilm formation on Arabidopsis thaliana roots. Planta 2007, 226, 283–297. [Google Scholar] [CrossRef]
- Leon-Reyes, A.; Van der Does, D.; De Lange, E.S.; Delker, C.; Wasternack, C.; Van Wees, S.C.; Ritsema, T. Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 2010, 232, 1423–1432. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.Y.; Spivey, N.W.; Zeng, W.; Liu, P.P.; Fu, Z.Q.; Klessig, D.F.; He, S.Y. Coronatine Promotes Pseudomonas syringae Virulence in Plants by Activating a Signaling Cascade that Inhibits Salicylic Acid Accumulation. Cell Host Microbe 2012, 11, 587–596. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.Q.; Li, F.; Tang, J.Y.; Wang, W.H.; Zhang, F.X.; Wang, G.D.; Chu, J.F.; Yan, C.Y.; Wang, T.Q.; Chu, C.; et al. Activation of the Jasmonic Acid Pathway by Depletion of the Hydroperoxide Lyase OsHPL3 Reveals Crosstalk between the HPL and AOS Branches of the Oxylipin Pathway in Rice. PLoS ONE 2012, 7, e50089. [Google Scholar] [CrossRef]
- Tong, X.H.; Qi, J.F.; Zhu, X.D.; Mao, B.Z.; Zeng, L.J.; Wang, B.H.; Li, Q.; Zhou, G.X.; Xu, X.J.; Lou, Y.G.; et al. The rice hydroperoxide lyase OsHPL3 functions in defense responses by modulating the oxylipin pathway. Plant J. 2012, 71, 763–775. [Google Scholar] [CrossRef]
- Gao, M.J.; He, Y.; Yin, X.; Zhong, X.B.; Yan, B.X.; Wu, Y.; Chen, J.; Li, X.Y.; Zhai, K.R.; Huang, Y.F.; et al. Ca2+ sensor-mediated ROS scavenging suppresses rice immunity and is exploited by a fungal effector. Cell 2021, 184, 5391–5404.e17. [Google Scholar] [CrossRef] [PubMed]
- Tamaoki, D.; Seo, S.; Yamada, S.; Kano, A.; Miyamoto, A.; Shishido, H.; Miyoshi, S.; Taniguchi, S.; Akimitsu, K.; Gomi, K. Jasmonic acid and salicylic acid activate a common defense system in rice. Plant Signal. Behav. 2013, 8, e24260. [Google Scholar] [CrossRef]
- Wang, Y.; Duan, G.; Li, C.; Ma, X.; Yang, J. Application of jasmonic acid at the stage of visible brown necrotic spots in Magnaporthe oryzae infection as a novel and environment-friendly control strategy for rice blast disease. Protoplasma 2021, 258, 743–752. [Google Scholar] [CrossRef]
- Navarro, L.; Bari, R.; Achard, P.; Lisón, P.; Nemri, A.; Harberd, N.P.; Jones, J.D. DELLAs Control Plant Immune Responses by Modulating the Balance of Jasmonic Acid and Salicylic Acid Signaling. Curr. Biol. 2008, 18, 650–655. [Google Scholar] [CrossRef]
- De Vleesschauwer, D.; Seifi, H.S.; Filipe, O.; Haeck, A.; Huu, S.N.; Demeestere, K.; Höfte, M. The DELLA Protein SLR1 Integrates and Amplifies Salicylic Acid- and Jasmonic Acid-Dependent Innate Immunity in Rice. Plant Physiol. 2016, 170, 1831–1847. [Google Scholar] [CrossRef] [Green Version]
- Tsuda, K.; Sato, M.; Stoddard, T.; Glazebrook, J.; Katagiri, F. Network Properties of Robust Immunity in Plants. PLoS Genet. 2009, 5, e1000772. [Google Scholar] [CrossRef] [Green Version]
- Betsuyaku, S.; Katou, S.; Takebayashi, Y.; Sakakibara, H.; Nomura, N.; Fukuda, H. Salicylic Acid and Jasmonic Acid Pathways are Activated in Spatially Different Domains Around the Infection Site During Effector-Triggered Immunity in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 8–16. [Google Scholar] [CrossRef]
- Mishra, A.K.; Murli, C.; Pandey, K.K.; Sakuntala, T.; Poswal, H.K.; Verma, A.K. Competing Interactions: Evolution of Inter and Intramolecular Hydrogen Bonds in Salicylic Acid at High Pressures. J. Phys. Chem. B 2020, 124, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yao, J.; Withers, J.; Xin, X.-F.; Banerjee, R.; Fariduddin, Q.; Nakamura, Y.; Nomura, K.; Howe, G.A.; Boland, W.; et al. Host target modification as a strategy to counter pathogen hijacking of the jasmonate hormone receptor. Proc. Natl. Acad. Sci. USA 2015, 112, 14354–14359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kankanala, P.; Czymmek, K.; Valent, B. Roles for Rice Membrane Dynamics and Plasmodesmata during Biotrophic Invasion by the Blast Fungus. Plant Cell 2007, 19, 706–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Júnior, L.A.Z.; Rodrigues, F.Á.; Fontes, R.L.F.; Korndörfer, G.H.; Neves, J.C. Rice Resistance to Brown Spot Mediated by Silicon and its Interaction with Manganese. J. Phytopathol. 2009, 157, 73–78. [Google Scholar] [CrossRef]
- Shi, X.; Long, Y.; He, F.; Zhang, C.; Wang, R.; Zhang, T.; Wu, W.; Hao, Z.; Wang, Y.; Wang, G.-L.; et al. The fungal pathogen Magnaporthe oryzae suppresses innate immunity by modulating a host potassium channel. PLoS Pathog. 2018, 14, e1006878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutsuno, Y.; Sumida, K.; Itoh, T.; Tukey, R.H.; Fujiwara, R. Glucuronidation of drugs in humanized UDP -glucuronosyltransferase 1 mice: Similarity with glucuronidation in human liver microsomes. Pharmacol. Res. Perspect. 2013, 1, e00002. [Google Scholar] [CrossRef]
- Liang, X.; Chen, X.; Li, C.; Fan, J.; Guo, Z. Metabolic and transcriptional alternations for defense by interfering OsWRKY62 and OsWRKY76 transcriptions in rice. Sci. Rep. 2017, 7, 2474. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef]
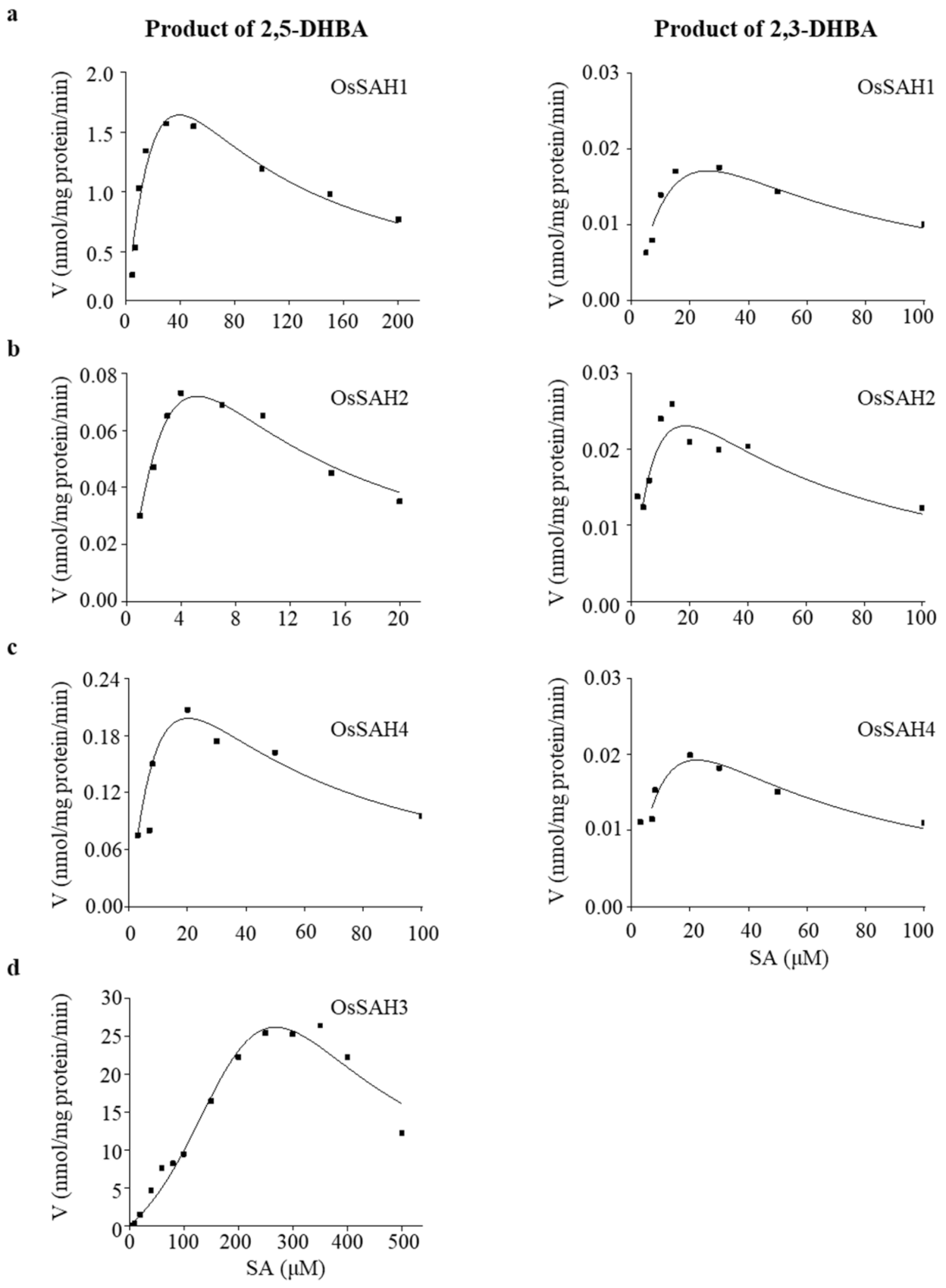

| Products | Protein | Specific Activity * | Km (μM) | Vmax * | Ksi (μM) | Kcat (min−1) | Kcat/Km (s−1·M−1) | R2 |
|---|---|---|---|---|---|---|---|---|
| 2,3-DHBA | SAH1 | 0.02 | 41.07 ± 39.93 | 0.07 ± 0.06 | 16.49 ± 16.49 | 0.05 ± 0.04 | 20.21 | 0.85 |
| SAH2 | 0.03 | 5.87 ± 3.97 | 0.05 ± 0.02 | 14.52 ± 10.36 | 0.03 ± 0.01 | 85.18 | 0.80 | |
| SAH3 | nd | |||||||
| SAH4 | 0.02 | 20.44 ± 13.88 | 0.05 ± 0.02 | 24.26 ± 16.79 | 0.03 ± 0.01 | 24.51 | 0.85 | |
| 2,5-DHBA | SAH1 | 1.57 | 57.41 ± 44.01 | 6.42 ± 3.94 | 27.13 ± 21.11 | 4.40 ± 0.65 | 1277.30 | 0.91 |
| SAH2 | 0.07 | 12.48 ± 10.96 | 0.42 ± 0.31 | 2.14 ± 1.86 | 0.29 ± 0.21 | 387.26 | 0.95 | |
| SAH3 | 26.37 | 197.08 ± 19.42 | 22.28 ± 2.89 | 365.12 ± 30.58 | 15.36 ± 1.99 | 1287.71 | 0.96 | |
| SAH4 | 0.21 | 18.32 ± 6.97 | 0.52 ± 0.14 | 26.52 ± 10.71 | 1.52 ± 0.41 | 363.90 | 0.98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, B.; Wang, H.; Yang, C.; Wang, L.; Qi, L.; Guo, Z.; Chen, X. Salicylic Acid Is Required for Broad-Spectrum Disease Resistance in Rice. Int. J. Mol. Sci. 2022, 23, 1354. https://doi.org/10.3390/ijms23031354
Liang B, Wang H, Yang C, Wang L, Qi L, Guo Z, Chen X. Salicylic Acid Is Required for Broad-Spectrum Disease Resistance in Rice. International Journal of Molecular Sciences. 2022; 23(3):1354. https://doi.org/10.3390/ijms23031354
Chicago/Turabian StyleLiang, Bingbing, Han Wang, Ce Yang, Luyao Wang, Linlu Qi, Zejian Guo, and Xujun Chen. 2022. "Salicylic Acid Is Required for Broad-Spectrum Disease Resistance in Rice" International Journal of Molecular Sciences 23, no. 3: 1354. https://doi.org/10.3390/ijms23031354
APA StyleLiang, B., Wang, H., Yang, C., Wang, L., Qi, L., Guo, Z., & Chen, X. (2022). Salicylic Acid Is Required for Broad-Spectrum Disease Resistance in Rice. International Journal of Molecular Sciences, 23(3), 1354. https://doi.org/10.3390/ijms23031354






