Transcriptional Signatures of Immune, Neural, and Endocrine Functions in the Brain and Kidney of Rainbow Trout (Oncorhynchus mykiss) in Response to Aeromonas salmonicida Infection
Abstract
:1. Introduction
2. Results
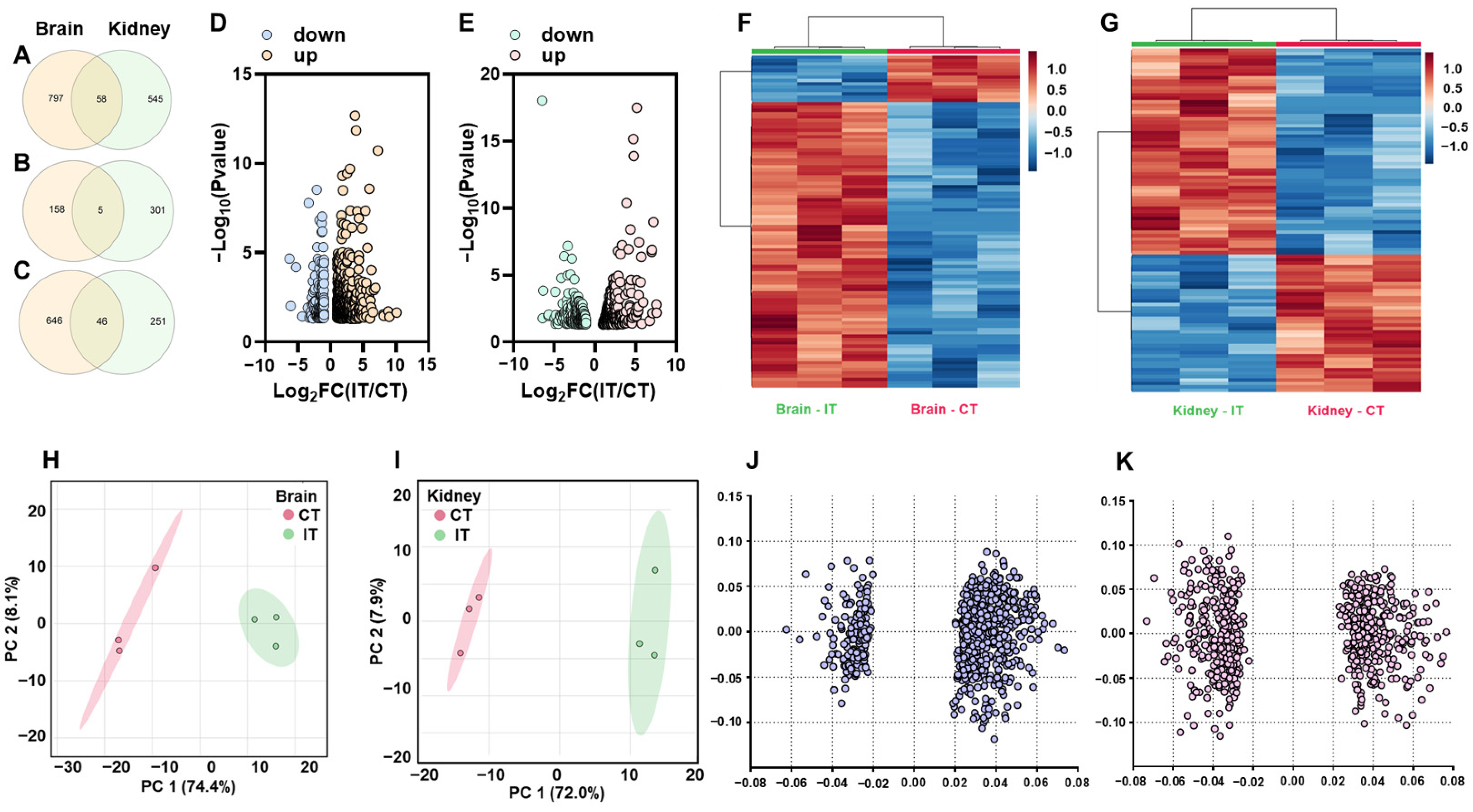
2.1. General Transcriptomic Profiles of the Brain and Kidney in Response to A. salmonicida Infection
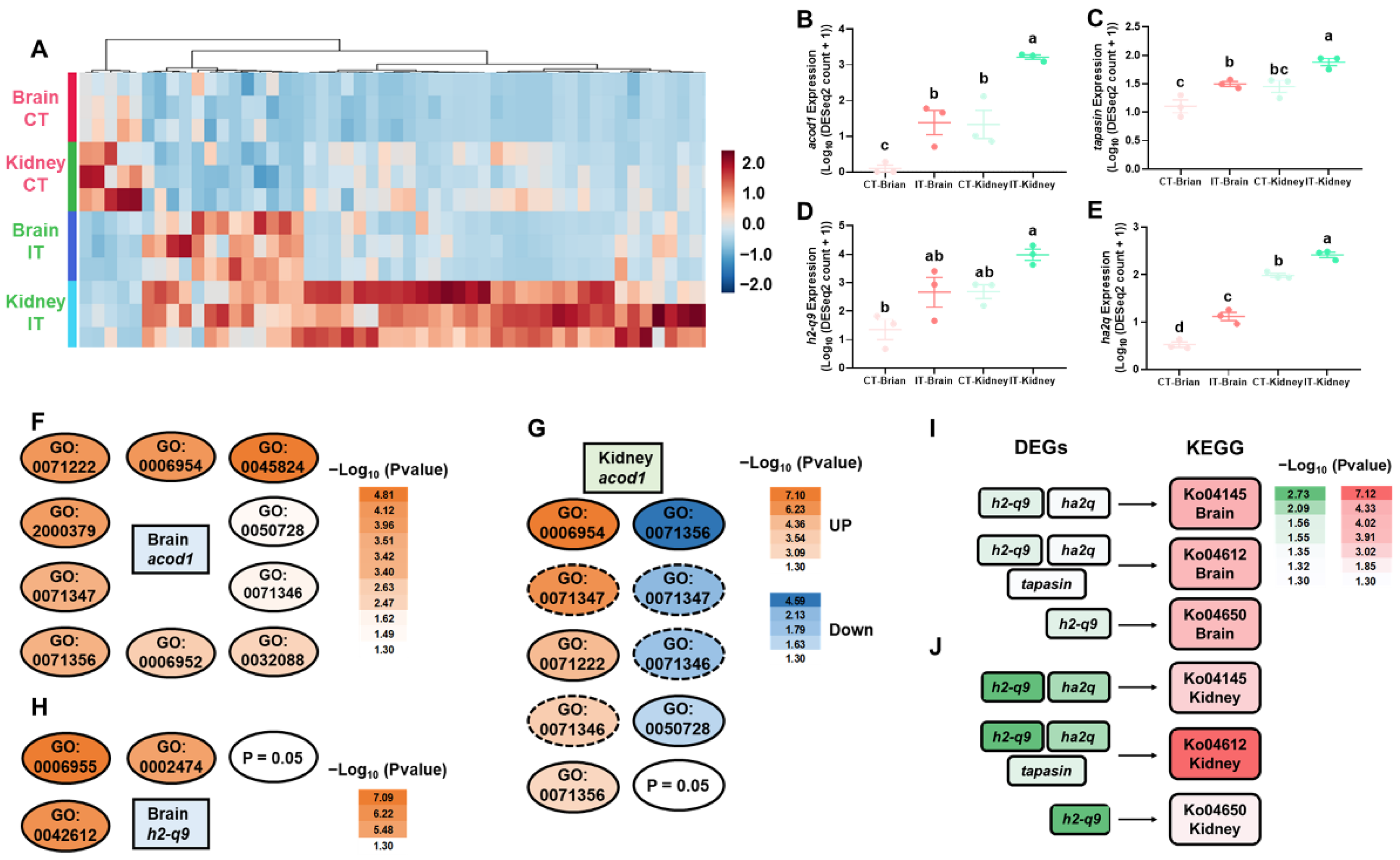
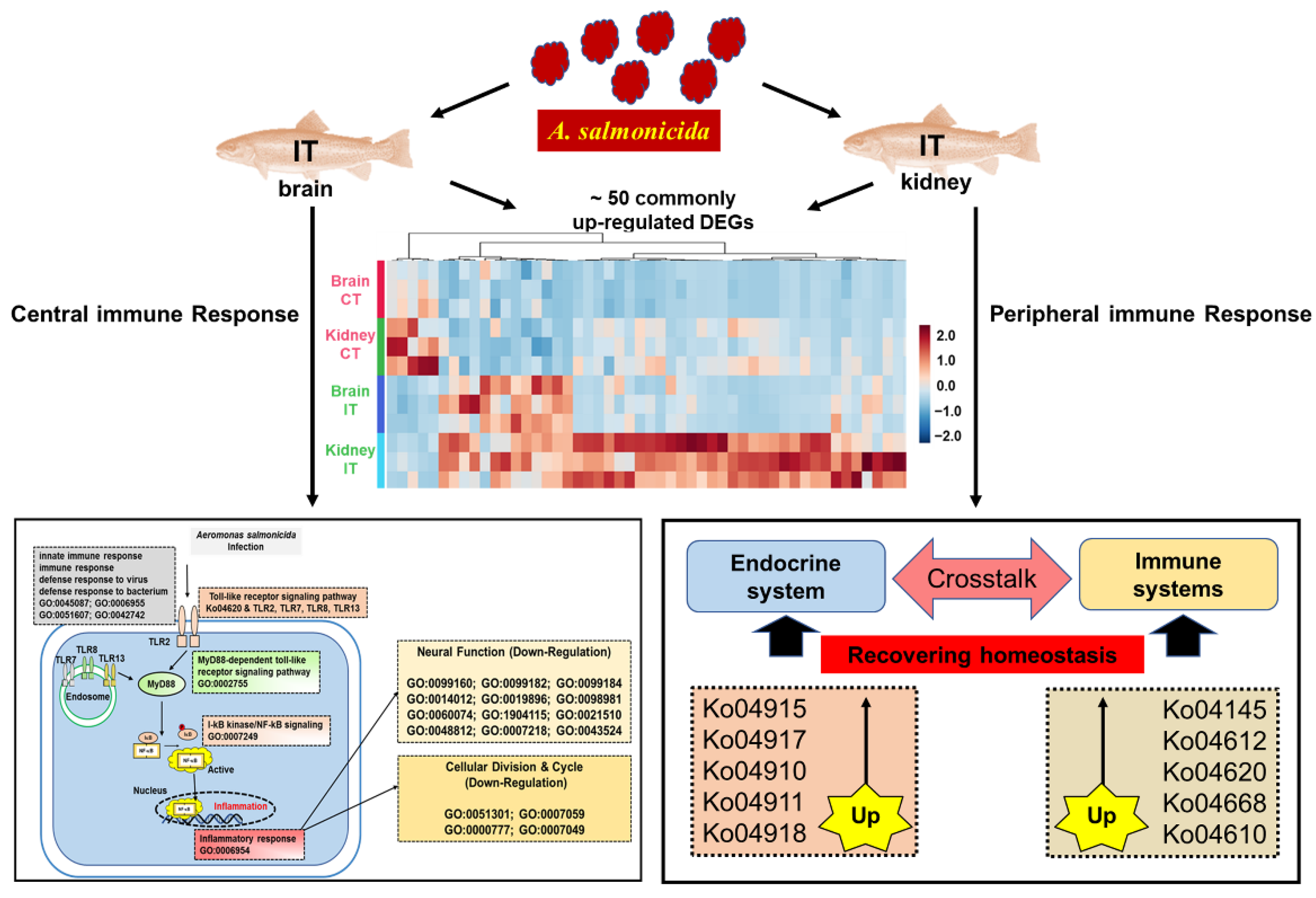
2.2. DEGs Commonly Identified in Brain and Kidney
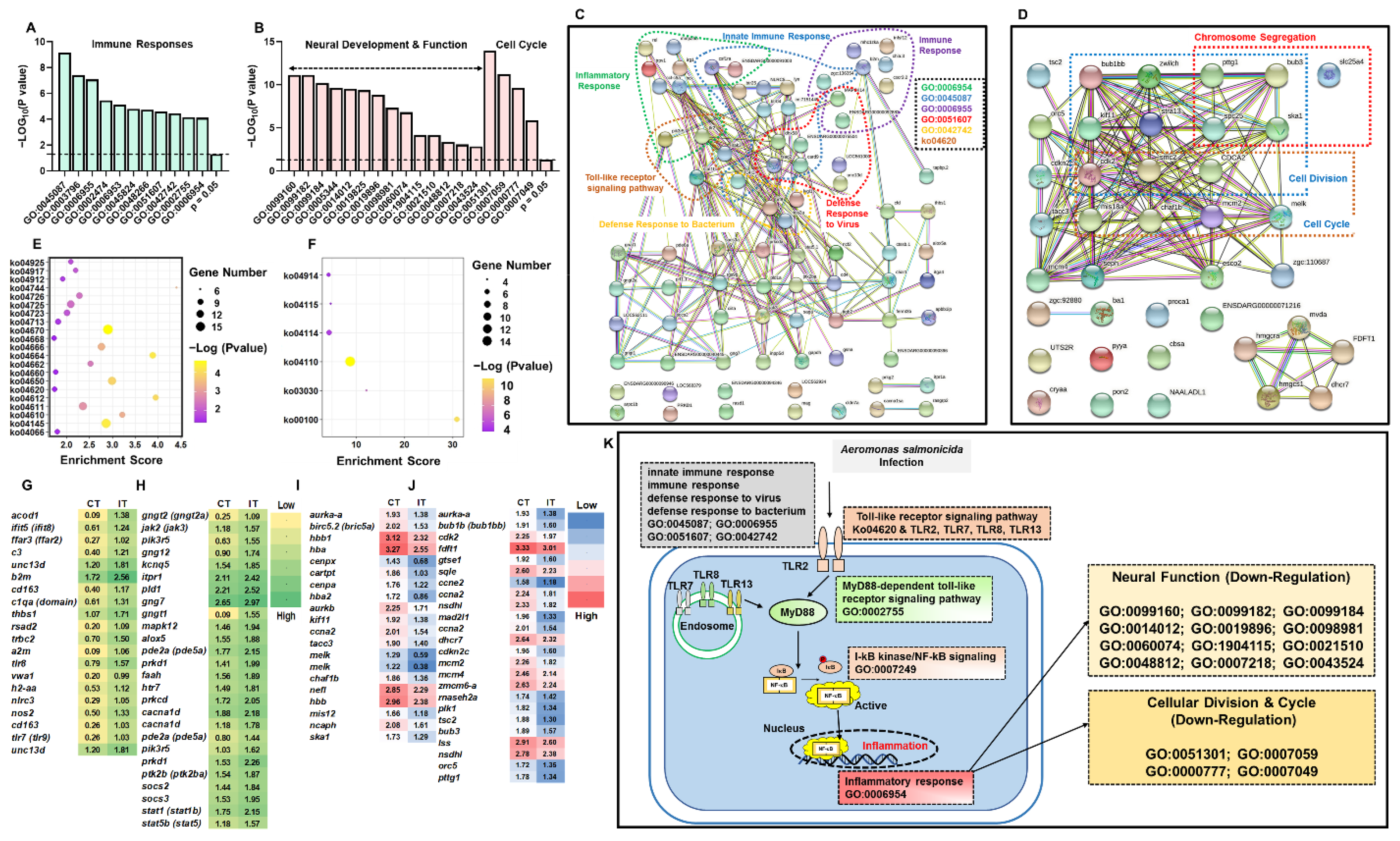
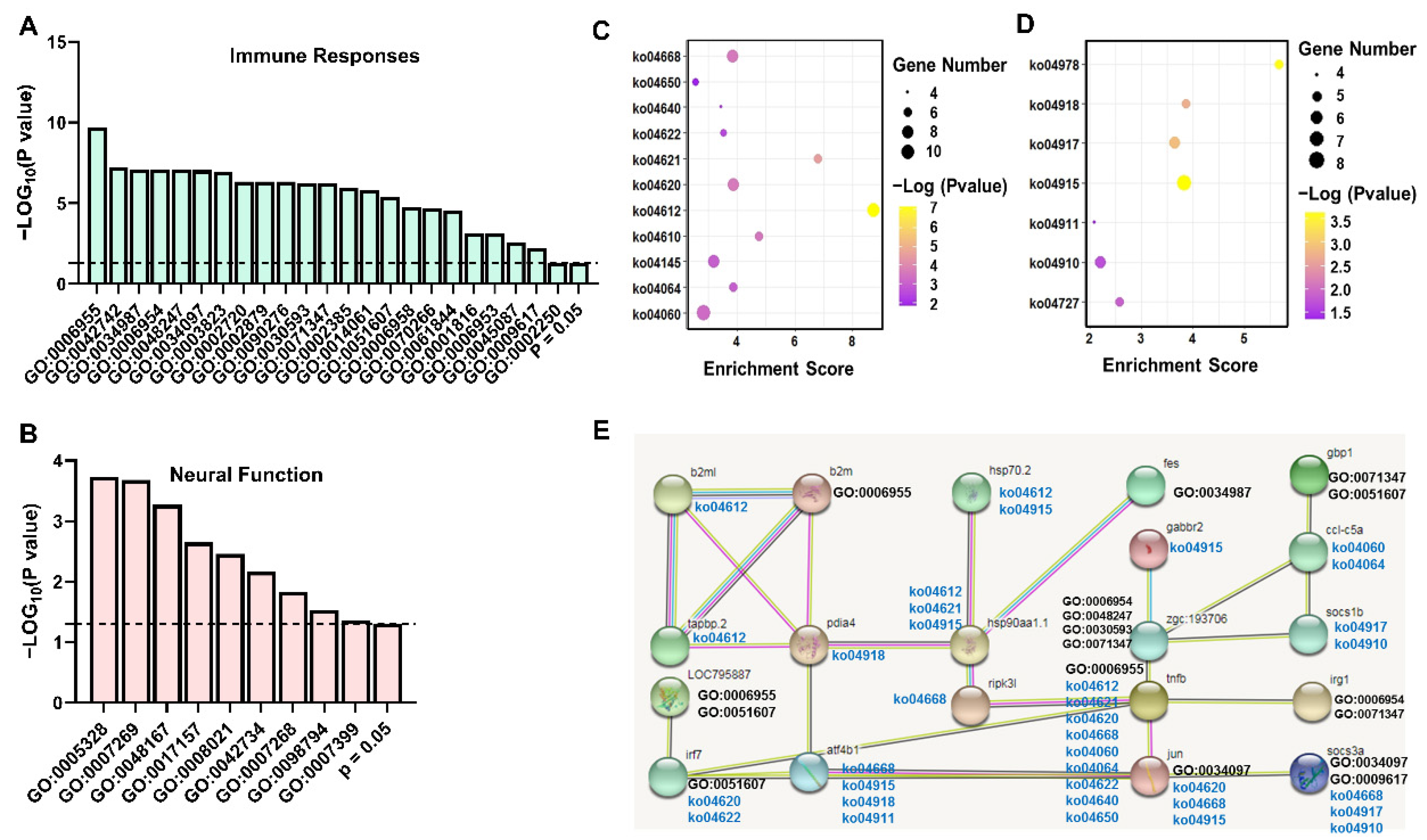
2.3. Enrichment Analysis of DEGs in Brain and Kidney
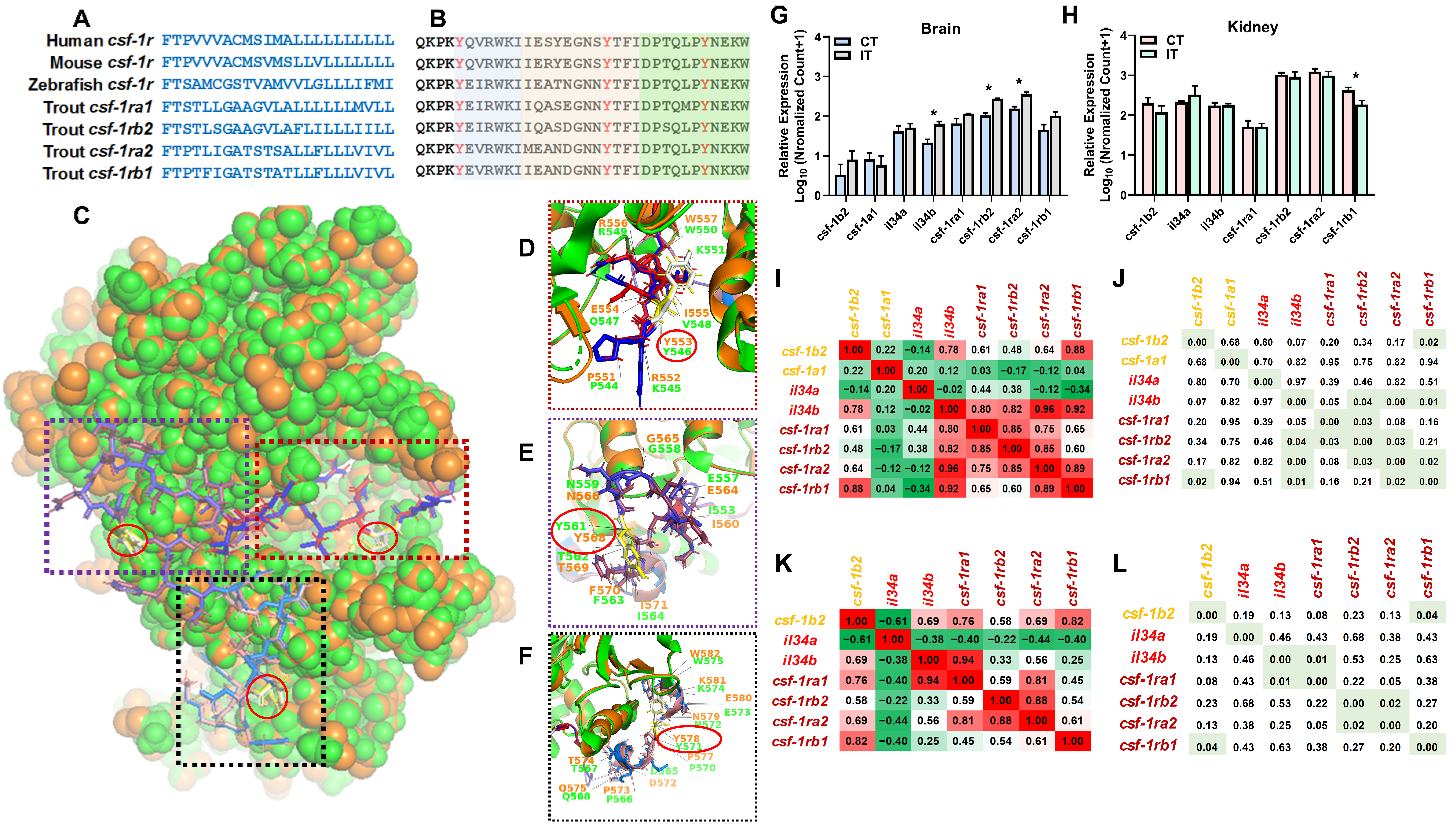
2.4. Identification of the Novel Macrophage csf-1r
3. Discussion
3.1. Key Immune-Related Genes Commonly Identified in the Brain and Kidney
3.2. The Pathway Analysis of Neural and Peripheral Immune Modulation
3.3. Identification of Novel Genes Associated with Immunomodulation
4. Materials and Methods
4.1. Ethics Statement
4.2. Experiment Design and Sample Collection
4.3. RNA-Seq Analysis
4.4. Identification of Novel Immune Genes Based on RNA-Seq Data
4.5. Validation of RNA-seq Data by qPCR
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tidwell, J.H.; Allan, G.L. Fish as food: Aquaculture’s contribution. EMBO Rep. 2001, 2, 958–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naylor, R.L.; Goldburg, R.J.; Primavera, J.H.; Kautsky, N.; Beveridge, M.C.; Clay, J.; Folke, C.; Lubchenco, J.; Mooney, H.; Troell, M. Effect of aquaculture on world fish supplies. Nature 2000, 405, 1017–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naylor, R.L.; Hardy, R.W.; Buschmann, A.H.; Bush, S.R.; Cao, L.; Klinger, D.H.; Little, D.C.; Lubchenco, J.; Shumway, S.E.; Troell, M. A 20-year retrospective review of global aquaculture. Nature 2021, 591, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Belton, B.; Thilsted, S.H. Fisheries in transition: Food and nutrition security implications for the global South. Glob. Food Sec. 2014, 3, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Béné, C.; Arthur, R.; Norbury, H.; Allison, E.H.; Beveridge, M.; Bush, S.; Campling, L.; Leschen, W.; Little, D.; Squires, D.; et al. Contribution of Fisheries and Aquaculture to Food Security and Poverty Reduction: Assessing the Current Evidence. World Dev. 2016, 79, 177–196. [Google Scholar] [CrossRef]
- Thilsted, S.H.; Thorne-Lyman, A.; Webb, P.; Bogard, J.R.; Subasinghe, R.; Phillips, M.J.; Allison, E.H. Sustaining healthy diets: The role of capture fisheries and aquaculture for improving nutrition in the post-2015 era. Food Policy 2016, 61, 126–131. [Google Scholar] [CrossRef] [Green Version]
- Belton, B.; Little, D.C.; Zhang, W.; Edwards, P.; Skladany, M.; Thilsted, S.H. Farming fish in the sea will not nourish the world. Nat. Commun. 2020, 11, 5804. [Google Scholar] [CrossRef]
- Khansari, A.R.; Balasch, J.C.; Vallejos-Vidal, E.; Parra, D.; Reyes-López, F.E.; Tort, L. Comparative immune- and stress-related transcript response induced by air exposure and Vibrio anguillarum bacterin in rainbow trout (Oncorhynchus mykiss) and gilthead seabream (Sparus aurata) mucosal surfaces. Front. Immunol. 2018, 9, 856. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Wang, Q.; Huang, Z.; Ding, L.; Xu, Z. Immunoglobulins, Mucosal Immunity and Vaccination in Teleost Fish. Front. Immunol. 2020, 11, 2597. [Google Scholar] [CrossRef]
- Ndong, D.; Chen, Y.Y.; Lin, Y.H.; Vaseeharan, B.; Chen, J.C. The immune response of tilapia Oreochromis mossambicus and its susceptibility to Streptococcus iniae under stress in low and high temperatures. Fish Shellfish Immunol. 2007, 22, 686–694. [Google Scholar] [CrossRef]
- Yada, T.; Tort, L. 10-Stress and Disease Resistance: Immune System and Immunoendocrine Interactions. In Biology of Stress in Fish; Schreck, C.B., Tort, L., Farrell, A.P., Brauner, C.J., Eds.; Fish Physiology: Amsterdam, The Netherlands, 2016; Volume 35, pp. 365–403. [Google Scholar]
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalsgaard, I.; Gudmundsdóttir, B.K.; Helgason, S.; Høie, S.; Thoresen, O.F.; Wichardt, U.P.; Wiklund, T. Identification of atypical Aeromonas salmonicida: Inter-laboratory evaluation and harmonization of methods. J. Appl. Microbiol. 1998, 84, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Han, J.E.; Kwon, H.; Park, S.C.; Kim, J.H. Recent insights into Aeromonas salmonicida and its bacteriophages in aquaculture: A comprehensive review. J. Microbiol. Biotechnol. 2020, 30, 1443–1457. [Google Scholar] [CrossRef] [PubMed]
- Austin, B.; Austin, D. Bacterial Fish Pathogens: Diseases of Farmed and Wild Fish; Springer: Dordrecht, The Netherlands, 2007; ISBN 978-1-4020-6068-7. [Google Scholar] [CrossRef]
- Farto, R.; Milton, D.L.; Bermúdez, M.B.; Nieto, T.P. Colonization of turbot tissues by virulent and avirulent Aeromonas salmonicida subsp. salmonicida strains during infection. Dis. Aquat. Organ. 2011, 95, 167–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Ringø, E.; Hoseinifar, S.H.; Lauzon, H.L.; Birkbeck, H.; Yang, D. The adherence and colonization of microorganisms in fish gastrointestinal tract. Rev. Aquac. 2019, 11, 603–618. [Google Scholar] [CrossRef]
- Shaalan, M.; El-Mahdy, M.; Theiner, S.; Dinhopl, N.; El-Matbouli, M.; Saleh, M. Silver Nanoparticles: Their Role as Antibacterial Agent against Aeromonas salmonicida Subsp. salmonicida in Rainbow Trout (Oncorhynchus mykiss). Res. Vet. Sci. 2018, 119, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Bernoth, E.-M. Furunculosis: The history of the disease and of disease research. In Furunculosis; Academic Press: Cambridge, MA, USA, 1997; pp. 1–20. [Google Scholar] [CrossRef]
- Dallaire-Dufresne, S.; Tanaka, K.H.; Trudel, M.V.; Lafaille, A.; Charette, S.J. Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet. Microbiol. 2014, 169, 1–7. [Google Scholar] [CrossRef]
- Altmann, S.; Korytář, T.; Kaczmarzyk, D.; Nipkow, M.; Kühn, C.; Goldammer, T.; Rebl, A. Toll-like receptors in maraena whitefish: Evolutionary relationship among salmonid fishes and patterns of response to Aeromonas salmonicida. Fish Shellfish Immunol. 2016, 54, 391–401. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, D.; Li, S.; Zhao, J.; Xu, L.; Liu, H.; Lu, T.; Mou, Z. A transcriptome analysis focusing on splenic immune-related mciroRNAs of rainbow trout upon Aeromonas salmonicida subsp. salmonicida infection. Fish Shellfish Immunol. 2019, 91, 350–357. [Google Scholar] [CrossRef]
- Miest, J.J.; Falco, A.; Pionnier, N.P.M.; Frost, P.; Irnazarow, I.; Williams, G.T.; Hoole, D. The influence of dietary β-glucan, PAMP exposure and Aeromonas salmonicida on apoptosis modulation in common carp (Cyprinus carpio). Fish Shellfish Immunol. 2012, 33, 846–856. [Google Scholar] [CrossRef]
- Soto-Dávila, M.; Hossain, A.; Chakraborty, S.; Rise, M.L.; Santander, J. Aeromonas salmonicida subsp. Salmonicida Early infection and immune response of atlantic cod (Gadus morhua L.) primary macrophages. Front. Immunol. 2019, 10, 1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyd, J.; Williams, J.; Curtis, B.; Kozera, C.; Singh, R.; Reith, M. Three small, cryptic plasmids from Aeromonas salmonicida subsp. salmonicida A449. Plasmid 2003, 50, 131–144. [Google Scholar] [CrossRef]
- Burr, S.E.; Pugovkin, D.; Wahli, T.; Segner, H.; Frey, J. Attenuated virulence of an Aeromonas salmonicida subsp. salmonicida type III secretion mutant in a rainbow trout model. Microbiology 2005, 151, 2111–2118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulder, I.E.; Wadsworth, S.; Secombes, C.J. Cytokine expression in the intestine of rainbow trout (Oncorhynchus mykiss) during infection with Aeromonas salmonicida. Fish Shellfish Immunol. 2007, 23, 747–759. [Google Scholar] [CrossRef]
- Ji, L.; Fu, S.; Sun, G.; Li, X.; Liu, Y. Dietary β-glucan modulate haematological parameters, cytokines and gene expression in TLR and ERK pathways of rainbow trout (Oncorhynchus mykiss) during infection by Aeromonas salmonicida. Aquac. Res. 2020, 51, 906–917. [Google Scholar] [CrossRef]
- Komatsu, K.; Tsutsui, S.; Hino, K.; Araki, K.; Yoshiura, Y.; Yamamoto, A.; Nakamura, O.; Watanabe, T. Expression profiles of cytokines released in intestinal epithelial cells of the rainbow trout, Oncorhynchus mykiss, in response to bacterial infection. Dev. Comp. Immunol. 2009, 33, 499–506. [Google Scholar] [CrossRef]
- Soleto, I.; Morel, E.; Muñoz-Atienza, E.; Díaz-Rosales, P.; Tafalla, C. Aeromonas salmonicida activates rainbow trout IgM+ B cells signalling through Toll like receptors. Sci. Rep. 2020, 10, 16810. [Google Scholar] [CrossRef]
- Korytář, T.; Jaros, J.; Verleih, M.; Rebl, A.; Kotterba, G.; Kühn, C.; Goldammer, T.; Köllner, B. Novel insights into the peritoneal inflammation of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2013, 35, 1192–1199. [Google Scholar] [CrossRef]
- Origgi, F.C.; Benedicenti, O.; Segner, H.; Sattler, U.; Wahli, T.; Frey, J. Aeromonas salmonicida type III secretion system-effectors-mediated immune suppression in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2017, 60, 334–345. [Google Scholar] [CrossRef]
- Liu, P.F.; Du, Y.; Meng, L.; Li, X.; Yang, D.; Liu, Y. Phosphoproteomic analyses of kidneys of Atlantic salmon infected with Aeromonas salmonicida. Sci. Rep. 2019, 9, 2101. [Google Scholar] [CrossRef]
- Liu, P.F.; Du, Y.; Meng, L.; Li, X.; Liu, Y. Metabolic profiling in kidneys of Atlantic salmon infected with Aeromonas salmonicida based on 1H NMR. Fish Shellfish Immunol. 2016, 58, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; Zhao, J.; Li, T.; Tafalla, C.; Zhang, Q.; Wang, X.; Gong, X.; Shen, Z.; Li, A. Transcriptomic and proteomic analyses of splenic immune mechanisms of rainbow trout (Oncorhynchus mykiss) infected by Aeromonas salmonicida subsp. salmonicida. J. Proteom. 2015, 122, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Curtis, L.R.; Bravo, C.F.; Bayne, C.J.; Tilton, F.; Arkoosh, M.R.; Lambertini, E.; Loge, F.J.; Collier, T.K.; Meador, J.P.; Tilton, S.C. Transcriptional changes in innate immunity genes in head kidneys from Aeromonas salmonicida-challenged rainbow trout fed a mixture of polycyclic aromatic hydrocarbons. Ecotoxicol. Environ. Saf. 2017, 142, 157–163. [Google Scholar] [CrossRef]
- Liu, P.F.; Du, Y.; Meng, L.; Li, X.; Liu, Y. Proteomic analysis in kidneys of Atlantic salmon infected with Aeromonas salmonicida by iTRAQ. Dev. Comp. Immunol. 2017, 72, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Marana, M.H.; Karami, A.M.; Ødegård, J.; Zuo, S.; Jaafar, R.M.; Mathiessen, H.; von Gersdorff Jørgensen, L.; Kania, P.W.; Dalsgaard, I.; Nielsen, T.; et al. Whole-genome association study searching for QTL for Aeromonas salmonicida resistance in rainbow trout. Sci. Rep. 2021, 11, 17857. [Google Scholar] [CrossRef] [PubMed]
- Panula, P.; Sallinen, V.; Sundvik, M.; Kolehmainen, J.; Torkko, V.; Tiittula, A.; Moshnyakov, M.; Podlasz, P. Modulatory neurotransmitter systems and behavior: Towards zebrafish models of neurodegenerative diseases. Zebrafish 2006, 3, 235–247. [Google Scholar] [CrossRef]
- Quan, N.; Banks, W.A. Brain-immune communication pathways. Brain. Behav. Immun. 2007, 21, 727–735. [Google Scholar] [CrossRef]
- Kipnis, J.; Filiano, A.J. Neuroimmunology in 2017: The central nervous system: Privileged by immune connections. Nat. Rev. Immunol. 2018, 18, 83–84. [Google Scholar] [CrossRef]
- Rummel, C. Inflammatory transcription factors as activation markers and functional readouts in immune-to-brain communication. Brain. Behav. Immun. 2016, 54, 1–14. [Google Scholar] [CrossRef]
- D’Mello, C.; Le, T.; Swain, M.G. Cerebral microglia recruit monocytes into the brain in response to tumor necrosis factora signaling during peripheral organ inflammation. J. Neurosci. 2009, 29, 2089–2102. [Google Scholar] [CrossRef] [Green Version]
- Al-Obaidi, M.M.J.; Desa, M.N.M. Mechanisms of Blood Brain Barrier Disruption by Different Types of Bacteria, and Bacterial–Host Interactions Facilitate the Bacterial Pathogen Invading the Brain. Cell. Mol. Neurobiol. 2018, 38, 1349–1368. [Google Scholar] [CrossRef] [PubMed]
- Baldissera, M.D.; Souza, C.F.; Santos, R.C.V.; Baldisserotto, B. Blood–brain barrier breakdown and myeloperoxidase activity in silver catfish experimentally infected with Pseudomonas aeruginosa. J. Fish Dis. 2018, 41, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Alzaid, A.; Castro, R.; Wang, T.; Secombes, C.J.; Boudinot, P.; Macqueen, D.J.; Martin, S.A.M. Cross talk between growth and immunity: Coupling of the igf axis to conserved cytokine pathways in rainbow trout. Endocrinology 2016, 157, 1942–1955. [Google Scholar] [CrossRef] [PubMed]
- MacQueen, D.J.; Garcia De La Serrana, D.; Johnston, I.A. Evolution of ancient functions in the vertebrate insulin-like growth factor system uncovered by study of duplicated salmonid fish genomes. Mol. Biol. Evol. 2013, 30, 1060–1076. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Lee, E.; Hestir, K.; Leo, C.; Huang, M.; Bosch, E.; Halenbeck, R.; Wu, G.; Zhou, A.; Behrens, D.; et al. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science 2008, 320, 807–811. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Lin, W.Y.; Chen, Y.; Stawicki, S.; Mukhyala, K.; Wu, Y.; Martin, F.; Bazan, J.F.; Starovasnik, M.A. Structural basis for the dual recognition of helical cytokines IL-34 and CSF-1 by CSF-1R. Structure 2012, 20, 676–687. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Lu, X.J.; Li, M.Y.; Chen, J. Molecular cloning, pathologically-correlated expression and functional characterization of the colony stimulating factor 1 receptor (CSF-1R) gene from a teleost, Plecoglossus altivelis. Dong Wu Xue Yan Jiu Zool Res. 2016, 37, 96–102. [Google Scholar] [CrossRef]
- Dai, X.M.; Ryan, G.R.; Hapel, A.J.; Dominguez, M.G.; Russell, R.G.; Kapp, S.; Sylvestre, V.; Stanley, E.R. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 2002, 99, 111–120. [Google Scholar] [CrossRef]
- Droin, N.; Solary, E. Editorial: CSF1R, CSF-1, and IL-34, a “ménage à trois” conserved across vertebrates. J. Leukoc. Biol. 2010, 87, 745–747. [Google Scholar] [CrossRef]
- Lee, C.G.L.; Jenkins, N.A.; Gilbert, D.J.; Copeland, N.G.; O’Brien, W.E. Cloning and analysis of gene regulation of a novel LPS-inducible cDNA. Immunogenetics 1995, 41, 263–270. [Google Scholar] [CrossRef]
- Michelucci, A.; Cordes, T.; Ghelfi, J.; Pailot, A.; Reiling, N.; Goldmann, O.; Binz, T.; Wegner, A.; Tallam, A.; Rausell, A.; et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. USA 2013, 110, 7820–7825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, R.; Chen, F.; Wang, N.; Tang, D.; Kang, R. ACOD1 in immunometabolism and disease. Cell. Mol. Immunol. 2020, 17, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Cordes, T.; Wallace, M.; Michelucci, A.; Divakaruni, A.S.; Sapcariu, S.C.; Sousa, C.; Koseki, H.; Cabrales, P.; Murphy, A.N.; Hiller, K.; et al. Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J. Biol. Chem. 2016, 291, 14274–14284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wentzel, A.S.; Janssen, J.J.E.; de Boer, V.C.J.; van Veen, W.G.; Forlenza, M.; Wiegertjes, G.F. Fish Macrophages Show Distinct Metabolic Signatures Upon Polarization. Front. Immunol. 2020, 11, 152. [Google Scholar] [CrossRef]
- Ryan, S.O.; Cobb, B.A. Roles for major histocompatibility complex glycosylation in immune function. Semin. Immunopathol. 2012, 34, 425–441. [Google Scholar] [CrossRef] [Green Version]
- Janeway, C.A., Jr.; Travers, P.; Walport, M. Immunobiology: The Immune System in Health and Disease: The major histocompatibility complex and its functions. Immunobiol. Immune Syst. Health Dis. 2001. Available online: https://www.ncbi.nlm.nih.gov/books/NBK27156/ (accessed on 1 December 2021).
- Hewitt, E.W. The MHC class I antigen presentation pathway: Strategies for viral immune evasion. Immunology 2003, 110, 163–169. [Google Scholar] [CrossRef]
- Delamarre, L.; Holcombe, H.; Mellman, I. Presentation of exogenous antigens on major histocompatibility complex (MHC) class I and MHC class II molecules is differentially regulated during dendritic cell maturation. J. Exp. Med. 2003, 198, 111–122. [Google Scholar] [CrossRef] [Green Version]
- Rock, K.L.; York, I.A.; Saric, T.; Goldberg, A.L. Protein degradation and the generation of MHC class I-presented peptides. Adv. Immunol. 2002, 80, 1–70. [Google Scholar] [CrossRef]
- Wieczorek, M.; Abualrous, E.T.; Sticht, J.; Álvaro-Benito, M.; Stolzenberg, S.; Noé, F.; Freund, C. Major histocompatibility complex (MHC) class I and MHC class II proteins: Conformational plasticity in antigen presentation. Front. Immunol. 2017, 8, 292. [Google Scholar] [CrossRef] [Green Version]
- Kjøglum, S.; Larsen, S.; Bakke, H.G.; Grimholt, U. The effect of specific MHC class I and class II combinations on resistance to furunculosis in Atlantic salmon (Salmo salar). Scand. J. Immunol. 2008, 67, 160–168. [Google Scholar] [CrossRef]
- Kjøglum, S.; Larsen, S.; Bakke, H.G.; Grimholt, U. How specific MHC class I and class II combinations affect disease resistance against infectious salmon anaemia in Atlantic salmon (Salmo salar). Fish Shellfish Immunol. 2006, 21, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Yewdell, J.W.; Norbury, C.C.; Bennink, J.R. Mechanisms of Exogenous Antigen Presentation by MHC Class I Molecules In Vitro and In Vivo: Implications for Generating CD8+ T Cell Responses to Infectious Agents, Tumors, Transplants, and Vaccines. Adv. Immunol. 1999, 73, 1–77, ISBN 0065-2776. [Google Scholar] [PubMed]
- Croisetière, S.; Tarte, P.D.; Bernatchez, L.; Belhumeur, P. Identification of MHC class IIβ resistance/susceptibility alleles to Aeromonas salmonicida in brook charr (Salvelinus fontinalis). Mol. Immunol. 2008, 45, 3107–3116. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.S.; Son, Y.J.; Ryou, C.; Sung, G.H.; Kim, J.H.; Cho, J.Y. Functional roles of Syk in macrophage-mediated inflammatory responses. Mediators Inflamm. 2014, 2014, 270302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shastri, A.; Bonifati, D.M.; Kishore, U. Innate immunity and neuroinflammation. Mediators Inflamm. 2013, 2013, 342931. [Google Scholar] [CrossRef] [PubMed]
- Galea, I.; Bechmann, I.; Perry, V.H. What is immune privilege (not)? Trends Immunol. 2007, 28, 12–18. [Google Scholar] [CrossRef]
- Huang, C.; Irwin, M.G.; Wong, G.T.C.; Chang, R.C.C. Evidence of the impact of systemic inflammation on neuroinflammation from a non-bacterial endotoxin animal model. J. Neuroinflamm. 2018, 15, 147. [Google Scholar] [CrossRef] [Green Version]
- Hoogland, I.C.M.; Houbolt, C.; van Westerloo, D.J.; van Gool, W.A.; van de Beek, D. Systemic inflammation and microglial activation: Systematic review of animal experiments. J. Neuroinflamm. 2015, 12, 114. [Google Scholar] [CrossRef] [Green Version]
- Zotova, E.; Bharambe, V.; Cheaveau, M.; Morgan, W.; Holmes, C.; Harris, S.; Neal, J.W.; Love, S.; Nicoll, J.A.R.; Boche, D. Inflammatory components in human Alzheimer’s disease and after active amyloid-β42 immunization. Brain 2013, 136, 2677–2696. [Google Scholar] [CrossRef] [Green Version]
- Ransohoff, R.M. How neuroinflammation contributes to neurodegeneration. Science 2016, 353, 777–783. [Google Scholar] [CrossRef]
- Capuron, L.; Miller, A.H. Immune system to brain signaling: Neuropsychopharmacological implications. Pharmacol. Ther. 2011, 130, 226–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowlati, Y.; Herrmann, N.; Swardfager, W.; Liu, H.; Sham, L.; Reim, E.K.; Lanctôt, K.L. A Meta-Analysis of Cytokines in Major Depression. Biol. Psychiatry 2010, 67, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Howren, M.B.; Lamkin, D.M.; Suls, J. Associations of depression with c-reactive protein, IL-1, and IL-6: A meta-analysis. Psychosom. Med. 2009, 71, 171–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, V. Toll-like receptors in the pathogenesis of neuroinflammation. J. Neuroimmunol. 2019, 332, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.H.; Staudt, L.M. Toll-Like receptor signaling. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, L.; Sun, G.; Li, X.; Liu, Y. Comparative transcriptome analysis reveals the mechanism of β-glucan in protecting rainbow trout (Oncorhynchus mykiss) from Aeromonas salmonicida infection. Fish Shellfish Immunol. 2020, 98, 87–99. [Google Scholar] [CrossRef]
- Rebl, A.; Siegl, E.; Köllner, B.; Fischer, U.; Seyfert, H.M. Characterization of twin toll-like receptors from rainbow trout (Oncorhynchus mykiss): Evolutionary relationship and induced expression by Aeromonas salmonicida salmonicida. Dev. Comp. Immunol. 2007, 31, 499–510. [Google Scholar] [CrossRef]
- Besedovsky, H.O.; del Rey, A. Immune-neuro-endocrine interactions: Facts and hypotheses. Endocr. Rev. 1996, 17, 64–102. [Google Scholar] [CrossRef]
- Harris, J.; Bird, D.J. Modulation of the fish immune system by hormones. Vet. Immunol. Immunopathol. 2000, 77, 163–176. [Google Scholar] [CrossRef]
- Tort, L. Stress and immune modulation in fish. Dev. Comp. Immunol. 2011, 35, 1366–1375. [Google Scholar] [CrossRef] [PubMed]
- Del Rey, A.; Besedovsky, H.O. Immune-Neuro-Endocrine Reflexes, Circuits, and Networks: Physiologic and Evolutionary Implications. Front. Horm. Res. 2017, 48, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Wensveen, F.M.; Šestan, M.; Turk Wensveen, T.; Polić, B. ‘Beauty and the beast’ in infection: How immune–endocrine interactions regulate systemic metabolism in the context of infection. Eur. J. Immunol. 2019, 49, 982–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milla, S.; Depiereux, S.; Kestemont, P. The effects of estrogenic and androgenic endocrine disruptors on the immune system of fish: A review. Ecotoxicology 2011, 20, 305–319. [Google Scholar] [CrossRef] [PubMed]
- Cabas, I.; Chaves-Pozo, E.; Mulero, V.; García-Ayala, A. Role of estrogens in fish immunity with special emphasis on GPER1. Dev. Comp. Immunol. 2018, 89, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Geissmann, F.; Manz, M.G.; Jung, S.; Sieweke, M.H.; Merad, M.; Ley, K. Development of monocytes, macrophages, and dendritic cells. Science 2010, 327, 656–661. [Google Scholar] [CrossRef] [Green Version]
- Hume, D.A.; Ross, I.L.; Himes, S.R.; Sasmono, R.T.; Wells, C.A.; Ravasi, T. The mononuclear phagocyte system revisited. J. Leukoc. Biol. 2002, 72, 621–627. [Google Scholar] [CrossRef]
- Rettenmier, C.W.; Roussel, M.F. Differential processing of colony-stimulating factor 1 precursors encoded by two human cDNAs. Mol. Cell. Biol. 1988, 8, 5026–5034. [Google Scholar] [CrossRef]
- Wu, Z.; Harne, R.; Chintoan-Uta, C.; Hu, T.J.; Wallace, R.; MacCallum, A.; Stevens, M.P.; Kaiser, P.; Balic, A.; Hume, D.A. Regulation and function of macrophage colony-stimulating factor (CSF1) in the chicken immune system. Dev. Comp. Immunol. 2020, 105, 103586. [Google Scholar] [CrossRef]
- Hanington, P.C.; Wang, T.; Secombes, C.J.; Belosevic, M. Growth factors of lower vertebrates: Characterization of goldfish (Carassius auratus L.) macrophage colony-stimulating factor-1. J. Biol. Chem. 2007, 282, 31865–31872. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Kono, T.; Monte, M.M.; Kuse, H.; Costa, M.M.; Korenaga, H.; Maehr, T.; Husain, M.; Sakai, M.; Secombes, C.J. Identification of IL-34 in teleost fish: Differential expression of rainbow trout IL-34, MCSF1 and MCSF2, ligands of the MCSF receptor. Mol. Immunol. 2013, 53, 398–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.Y.; Zhou, Y.; Zhou, Q.J.; Li, M.Y.; Chen, J. Mudskipper interleukin-34 modulates the functions of monocytes/macrophages via the colony-stimulating factor-1 receptor 1. Zool. Res. 2020, 41, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noël, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A.; et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014, 5, 3657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, H.; Wang, B.; He, J.; Hu, Y. Macrophage colony stimulating factor (MCSF) of Japanese flounder (Paralichthys olivaceus): Immunoregulatory property, anti-infectious function, and interaction with MCSF receptor. Dev. Comp. Immunol. 2021, 116, 103920. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.Q.; Li, Y.W.; Zhou, L.; Li, A.X.; Luo, X.C.; Dan, X.M. Grouper (Epinephelus coioides) IL-34/MCSF2 and MCSFR1/MCSFR2 were involved in mononuclear phagocytes activation against Cryptocaryon irritans infection. Fish Shellfish Immunol. 2015, 43, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Hou, Z.S.; Zhao, H.K.; Xin, Y.R.; Liu, M.Q.; Yang, X.D.; Wen, H.S.; Li, J.F. Identification and characterization of caspases genes in rainbow trout (Oncorhynchus mykiss) and their expression profiles after Aeromonas salmonicida and Vibrio anguillarum infection. Dev. Comp. Immunol. 2021, 118, 103987. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression of RNA-Seq data at the gene level—The DESeq package. Heidelberg, Ger. Eur. Mol. Biol. Lab. 2012, 10, f1000research. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Soufan, O.; Xia, J.; Tang, R.; Li, L.; Li, D. Transcriptome and physiological analysis reveal alterations in muscle metabolisms and immune responses of grass carp (Ctenopharyngodon idellus) cultured at different stocking densities. Aquaculture 2019, 503, 186–197. [Google Scholar] [CrossRef]
- Mouton, A.J.; Ma, Y.; Rivera Gonzalez, O.J.; Daseke, M.J.; Flynn, E.R.; Freeman, T.C.; Garrett, M.R.; DeLeon-Pennell, K.Y.; Lindsey, M.L. Fibroblast polarization over the myocardial infarction time continuum shifts roles from inflammation to angiogenesis. Basic Res. Cardiol. 2019, 114, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, 1–128. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Yang, X.; Zeng, C.; Zhao, H.; Li, J.; Hou, Z.; Wen, H. Transcriptional Signatures of Immune, Neural, and Endocrine Functions in the Brain and Kidney of Rainbow Trout (Oncorhynchus mykiss) in Response to Aeromonas salmonicida Infection. Int. J. Mol. Sci. 2022, 23, 1340. https://doi.org/10.3390/ijms23031340
Liu M, Yang X, Zeng C, Zhao H, Li J, Hou Z, Wen H. Transcriptional Signatures of Immune, Neural, and Endocrine Functions in the Brain and Kidney of Rainbow Trout (Oncorhynchus mykiss) in Response to Aeromonas salmonicida Infection. International Journal of Molecular Sciences. 2022; 23(3):1340. https://doi.org/10.3390/ijms23031340
Chicago/Turabian StyleLiu, Mengqun, Xiaodong Yang, Chu Zeng, Hongkui Zhao, Jifang Li, Zhishuai Hou, and Haishen Wen. 2022. "Transcriptional Signatures of Immune, Neural, and Endocrine Functions in the Brain and Kidney of Rainbow Trout (Oncorhynchus mykiss) in Response to Aeromonas salmonicida Infection" International Journal of Molecular Sciences 23, no. 3: 1340. https://doi.org/10.3390/ijms23031340
APA StyleLiu, M., Yang, X., Zeng, C., Zhao, H., Li, J., Hou, Z., & Wen, H. (2022). Transcriptional Signatures of Immune, Neural, and Endocrine Functions in the Brain and Kidney of Rainbow Trout (Oncorhynchus mykiss) in Response to Aeromonas salmonicida Infection. International Journal of Molecular Sciences, 23(3), 1340. https://doi.org/10.3390/ijms23031340




