Metabolome and Transcriptome Analyses Unravel the Molecular Regulatory Mechanisms Involved in Photosynthesis of Cyclocarya paliurus under Salt Stress
Abstract
:1. Introduction
2. Results
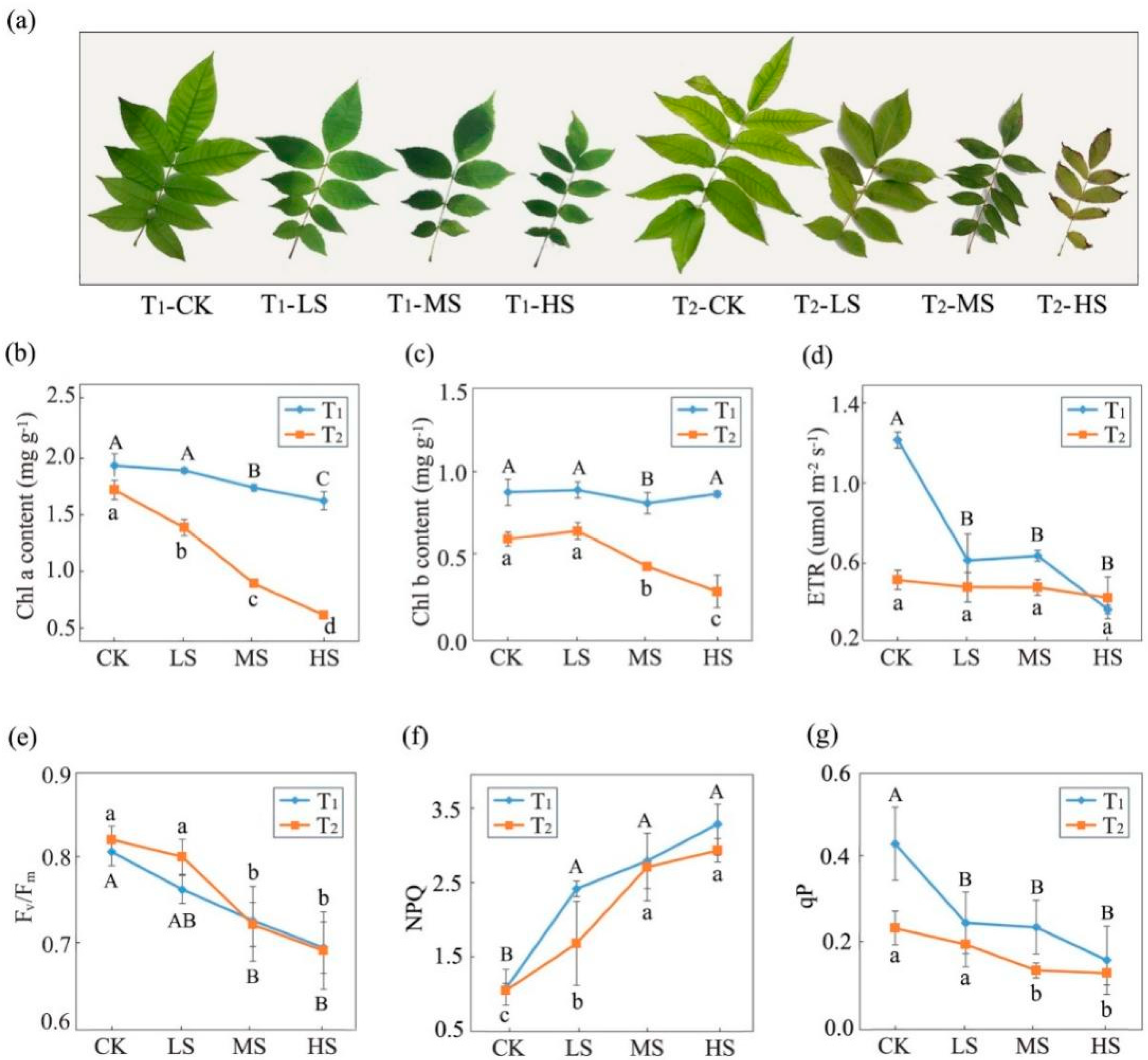
2.1. Variations in Photosynthetic and Fluorescence Characteristics
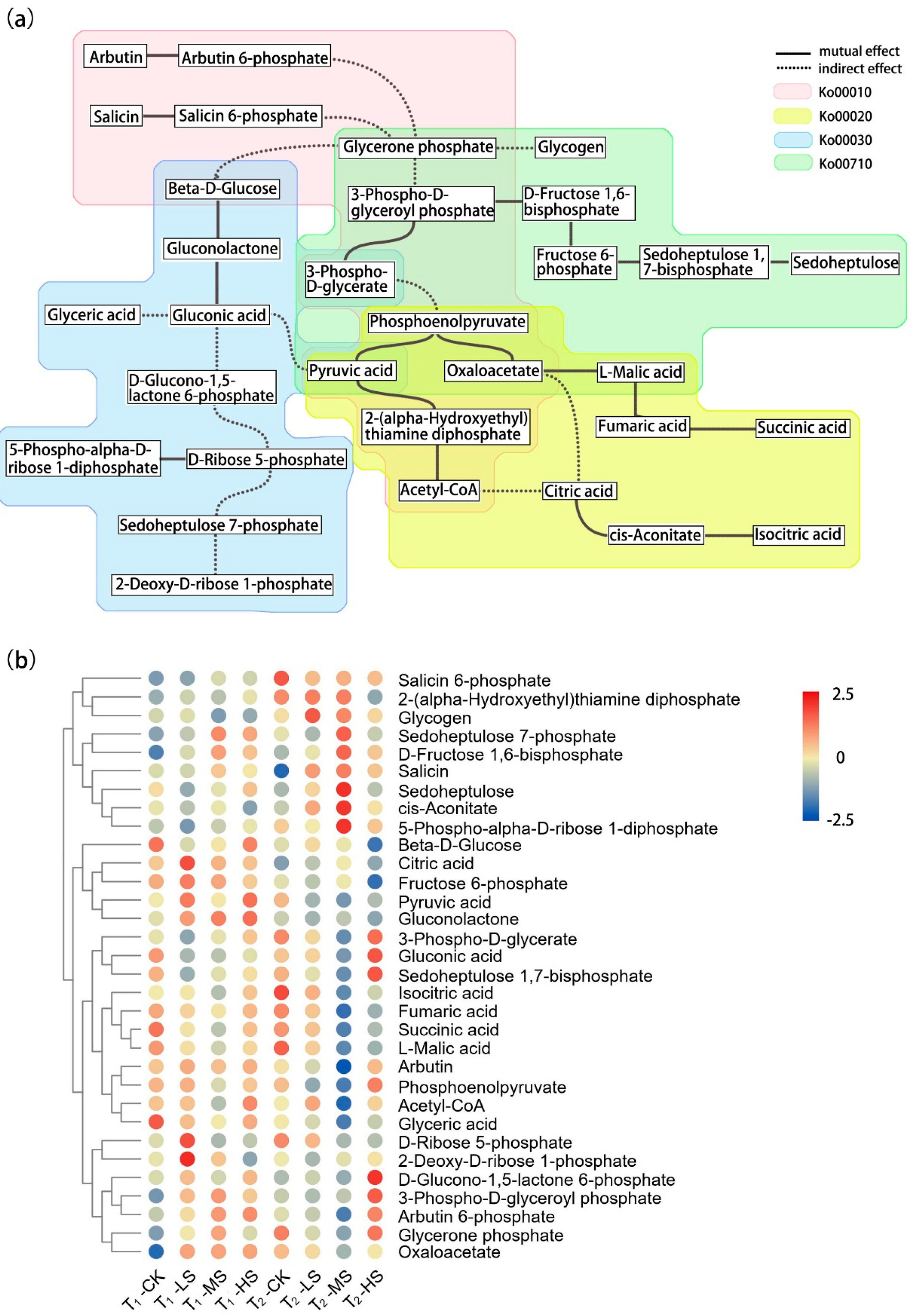
2.2. Metabolomic Response to Salt Stress
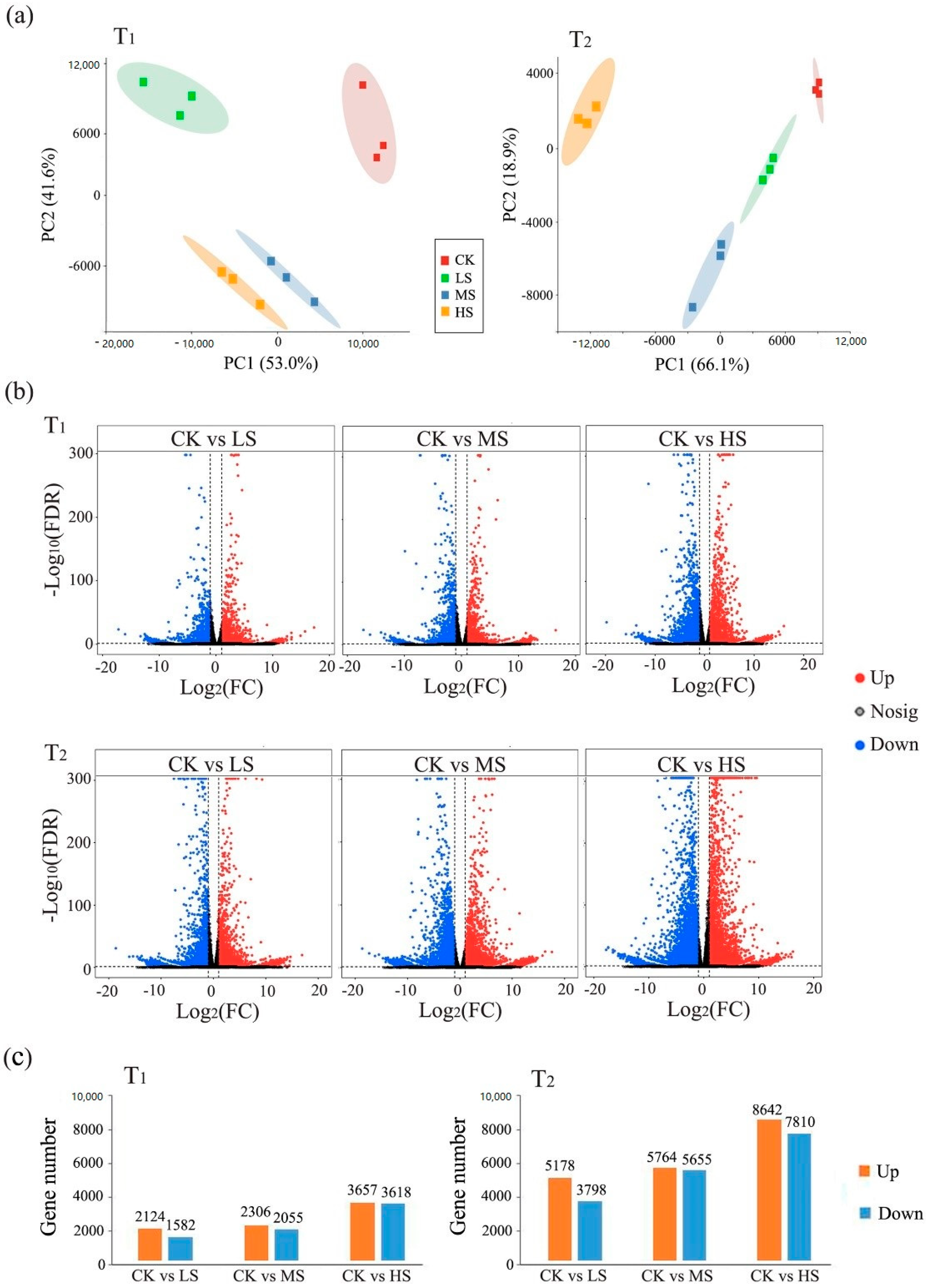
2.3. Transcriptomic Response to Salt Stress
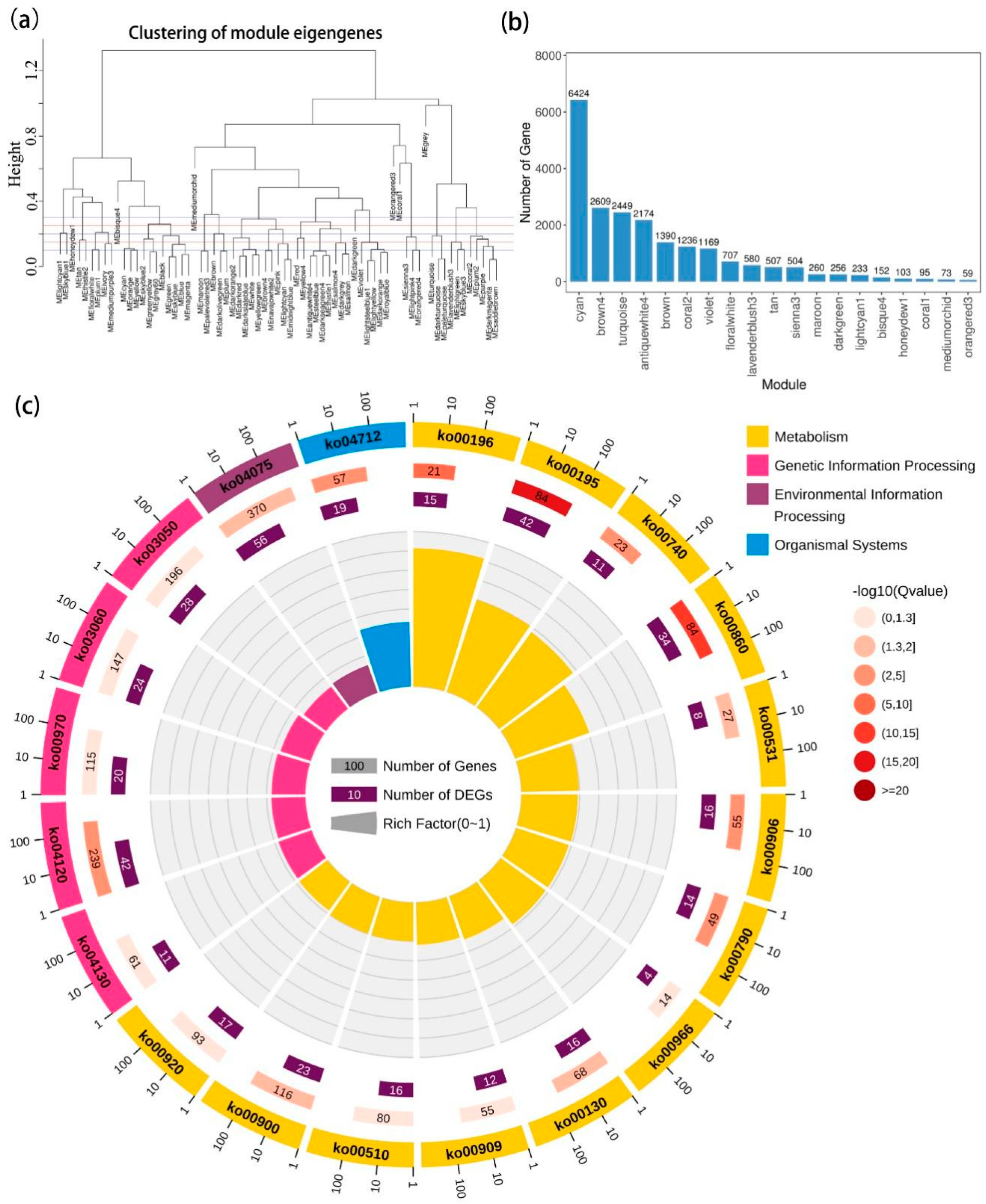
2.4. Gene Co-Expression Network Analysis
3. Discussion
3.1. Effects of Salt Stress on Photosynthetic Performance
3.2. Effects of Salt Stress on Carbon Metabolism
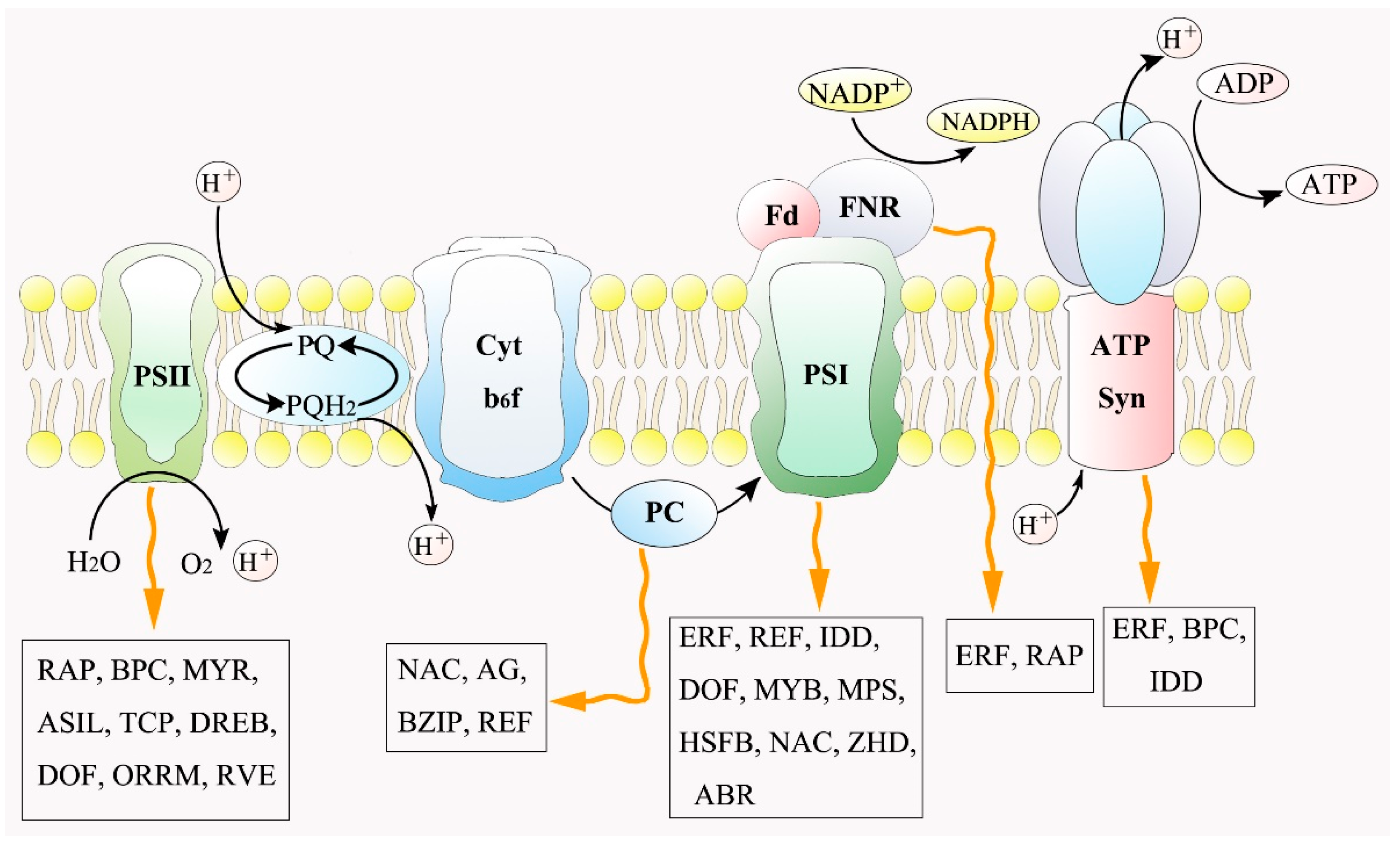
3.3. Transcriptional Regulation in Photosynthetic System to Salt Stress
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Photosynthetic and Fluorescence Parameters Determination
4.3. Carbon and Nitrogen Measurement
4.4. Metabolite Extraction and LC-MS/MS Analysis
4.5. Transcriptome Data Acquisition and Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Högberg, P.; Nordgren, A.; Buchmann, N.; Taylor, A.; Ekblad, A.; Högberg, M.N.; Nyberg, G.; Ottosson-Löfvenius, M.; Read, D.J. Large-Scale forest girdling shows that current photosynthesis drives soil respiration. Nature 2001, 411, 789–792. [Google Scholar] [CrossRef]
- Friso, G.; Giacomelli, L.; Ytterberg, A.J.; Peltier, J.B.; Rudella, A.; Sun, Q.; van Wijk, K.J. In-Depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 2004, 16, 478–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodd, A.N.; Belbin, F.E.; Frank, A.; Webb, A.A. Interactions between circadian clocks and photosynthesis for the temporal and spatial coordination of metabolism. Front. Plant Sci. 2015, 6, 245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.M.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawkar, G.; Kang, C.; Maibam, P.; Park, J.; Jung, Y.; Chae, H.; Chi, Y.H.; Jung, I.; Kim, W.Y.; Yun, D.J.; et al. HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 201609844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Gelderen, K.; Kang, C.; Paalman, R.; Keuskamp, D.; Hayes, S.; Pierik, R. Far-red light detection in the shoot regulates lateral root development through the HY5 transcription factor. Plant Cell 2018, 30, 101–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coruzzi, G.; Bush, D.R. Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol. 2001, 125, 61–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, W.; Sun, X.; Zhang, L. Intracellular signaling from plastids to the nucleus. Annu. Rev. Plant Biol. 2013, 64, 559–582. [Google Scholar] [CrossRef] [Green Version]
- Estavillo, G.; Crisp, P.; Pornsiriwong, W.; Wirtz, M.; Collinge, D.; Carrie, C.; Giraud, E.; Whelan, J.; David, P.; Javot, H.; et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 2011, 23, 3992–4012. [Google Scholar] [CrossRef] [Green Version]
- Moore, B.; Cheng, S.H.; Sims, D.; Seemann, J. The biochemical and molecular basis for acclimation to elevated CO2. Plant Cell Environ. 2002, 22, 567–582. [Google Scholar] [CrossRef]
- Cho, Y.H.; Yoo, S.D.; Sheen, J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 2006, 127, 579–589. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.G.; Long, S.; Ort, D. Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 2010, 61, 235–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flowers, T.J.; Gaur, P.M.; Gowda, C.; Krishnamurthy, L.; Samineni, S.; Siddique, K.; Turner, N.C.; Vadez, V.; Varshney, R.K.; Colmer, T.D. Salt sensitivity in chickpea. Plant Cell Environ. 2010, 33, 490–509. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Roychoudhury, A. Gene regulation at transcriptional and post transcriptional levels to combat salt stress in plants. Physiol. Plant. 2021, 173, 1556–1572. [Google Scholar] [CrossRef]
- Giovagnetti, V.; Ruban, A.V. Discerning the effects of photoinhibition and photoprotection on the rate of oxygen evolution in Arabidopsis leaves. J. Photochem. Photobiol. B Biol. 2015, 152, 272–278. [Google Scholar] [CrossRef]
- Goussi, R.; Manaa, A.; Derbali, W.; Cantamessa, S.; Abdelly, C.; Barbato, R. Comparative analysis of salt stress, duration and intensity, on the chloroplast ultrastructure and photosynthetic apparatus in Thellungiella salsuginea. J. Photochem. Photobiol. B Biol. 2018, 183, 275–287. [Google Scholar] [CrossRef]
- Chan, K.X.; Phua, S.Y.; Crisp, P.; McQuinn, R.; Pogson, B. Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu. Rev. Plant Biol. 2016, 67, 25–53. [Google Scholar] [CrossRef]
- Peng, W.; Hendron, R.W.; Kelly, S. Transcriptional control of photosynthetic capacity: Conservation and divergence from Arabidopsis to rice. New Phytol. 2017, 216, 32–45. [Google Scholar] [CrossRef] [Green Version]
- Fang, S.Z.; Yang, W.X.; Chu, X.L.; Shang, X.L.; She, C.Q.; Fu, X.X. Provenance and temporal variations in selected flavonoids in leaves of Cyclocarya paliurus. Food Chem. 2011, 124, 1382–1386. [Google Scholar] [CrossRef]
- Xie, J.H.; Dong, C.J.; Nie, S.P.; Li, F.; Wang, Z.J.; Shen, M.Y.; Xie, M.Y. Extraction, chemical composition and antioxidant activity of flavonoids from Cyclocarya paliurus (Batal)Iljinskaja leaves. Food Chem. 2015, 186, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.H.; Xie, M.Y.; Nie, S.P.; Shen, M.Y.; Wang, Y.X.; Li, C. Isolation, chemical composition and antioxidant activities of a water-soluble polysaccharide from Cyclocarya paliurus (Batal)Iljinskaja. Food Chem. 2010, 119, 1626–1632. [Google Scholar] [CrossRef]
- Qin, J.; Yue, X.; Fang, S.; Qian, M.; Zhou, S.; Shang, X.; Yang, W. Responses of nitrogen metabolism, photosyntheticparameter and growth to nitrogen fertilization in Cyclocarya paliurus. For. Ecol. Manag. 2021, 502, 119715. [Google Scholar] [CrossRef]
- Yao, R.L.; Wang, Y.; Fang, S.Z. Calcium alleviation of sodium toxicity in salt-treated Cyclocarya paliurus seedlings. Afr. J. Agric. Res. 2011, 6, 5967–5971. [Google Scholar] [CrossRef]
- Yao, R.L.; Fang, S.Z.; Nichols, D.J. A strategy of Ca2+ alleviating Na+ toxicity in salt-treated Cyclocarya paliurus seedlings: Photosynthetic and nutritional responses. Plant Growth Regul. 2012, 68, 351–359. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, V.; Shahzad, B.; Ramakrishnan, M.; Singh Sidhu, G.P.; Bali, A.S.; Handa, N.; Kapoor, D.; Yadav, P.; Khanna, K.; et al. Photosynthetic response of plants under different abiotic stresses: A review. J. Plant Growth Regul. 2020, 39, 509–531. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Z.J.; Fang, S.Z.; Liu, Y.; Shang, X.L. Integrative analysis of metabolome and transcriptome reveals molecular regulatory mechanism of flavonoid biosynthesis in Cyclocarya paliurus under salt stress. Ind. Crops Prod. 2021, 170, 113823. [Google Scholar] [CrossRef]
- Hussain, S.; Luro, F.; Costantino, G.; Ollitrault, P.; Morillon, R. Physiological analysis of salt stress behaviour of citrus species and genera: Low chloride accumulation as an indicator of salt tolerance. S. Afr. J. Bot. 2012, 81, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Bethke, P.; Drew, M. Stomatal and nonstomatal components to inhibition of photosynthesis in leaves of Capsicum annuum during progressive exposure to NaCl salinity. Plant Physiol. 1992, 99, 219–226. [Google Scholar] [CrossRef]
- Jiao, L.; Wang, L.; Zhou, Q.; Huang, X. Stomatal and non-stomatal factors regulated the photosynthesis of soybean seedlings in the present of exogenous bisphenol A. Ecotoxicol. Environ. Saf. 2017, 145, 150–160. [Google Scholar] [CrossRef]
- Flexas, J.; Diaz-Espejo, A.; Galmés, J.; Kaldenhoff, R.; Medrano, H.; Ribas-Carbo, M. Rapid variations of mesophyll conductance in response to changes in CO2 concentration around leaves. Plant Cell Environ. 2007, 30, 1284–1298. [Google Scholar] [CrossRef]
- Wellburn, A.R. The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Kowalewska, G.; Szymczak-Żyła, M. Influence of selected abiotic factors on the decomposition of chlorophylls. Oceanologia 2001, 43, 315–328. [Google Scholar] [CrossRef] [Green Version]
- Agong, S.G.; Kingetsu, M.; Yoshida, Y.; Yazawa, S.; Masuda, M. Response of tomato genotypes to induced salt stress. Afr. Crop Sci. J. 2003, 11, 133–142. [Google Scholar] [CrossRef]
- Tiwari, J.; Munshi, A.; Kumar, R.; Pandey, R.; Arora, A.; Bhat, J.; Sureja, A. Effect of salt stress on cucumber: Na+–K+ ratio, osmolyte concentration, phenols and chlorophyll content. Acta Physiol. Plant 2009, 32, 103–114. [Google Scholar] [CrossRef]
- Maxwell, K.; Johnson, G. Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef]
- Scarascia-Mugnozza, G.; Angelis, P.; Matteucci, G.; Valentini, R. Long-term exposure to elevated [CO2] in a natural Quercus ilex L. community: Net photosynthesis and photochemical efficiency of PSII at different levels of water stress. Plant Cell Environ. 1996, 19, 643–654. [Google Scholar] [CrossRef]
- Liu, J.; Shi, D. Photosynthesis, chlorophyll fluorescence, inorganic ion and organic acid accumulations of sunflower in responses to salt and salt-alkaline mixed stress. Photosynthetica 2010, 48, 127–134. [Google Scholar] [CrossRef]
- Gilmore, A. Mechanistic aspects of xanthophyll cycle-dependent photoprotection in higher plant chloroplasts and leaves. Physiol. Plant. 2006, 99, 197–209. [Google Scholar] [CrossRef]
- Nianwei, Q.; Lu, Q.; Lu, C. Photosynthesis, photosystem II efficiency and the xanthophyll cycle in the salt-adapted halophyte Atriplex centralasiatica. New Phytol. 2003, 159, 479–486. [Google Scholar] [CrossRef]
- Li, G.; Wan, S.; Zhou, J.; Yang, Z.; Qin, P. Leaf chlorophyll fluorescence, hyperspectral reflectance, pigments content, malondialdehyde and proline accumulation responses of castor bean (Ricinus communis L.) seedlings to salt stress levels. Ind. Crops Prod. 2010, 31, 13–19. [Google Scholar] [CrossRef]
- Sagun, J.; Badger, M.; Chow, W.; Ghannoum, O. Mehler reaction plays a role in C3 and C4 photosynthesis under shade and low CO2. Photosynth. Res. 2021, 149, 171–185. [Google Scholar] [CrossRef]
- Kozuleva, M.; Petrova, A.; Milrad, Y.; Semenov, A.; Ivanov, B.; Redding, K.; Yacoby, I. Phylloquinone is the principal Mehler reaction site within photosystem I in high light. Plant Physiol. 2021, 186, 1848–1858. [Google Scholar] [CrossRef]
- Ensminger, I.; Busch, F.; Huner, N. Photostasis and cold acclimation: Sensing low temperature through photosynthesis. Physiol. Plant. 2006, 126, 28–44. [Google Scholar] [CrossRef]
- Sharkey, T. Estimating the rate of photorespiration in leaves. Physiol. Plant. 1988, 73, 147–152. [Google Scholar] [CrossRef]
- Flügel, F.; Timm, S.; Arrivault, S.; Florian, A.; Stitt, M.; Fernie, A.R.; Bauwe, H. The photorespiratory metabolite 2-phosphoglycolate regulates photosynthesis and starch accumulation in Arabidopsis. Plant Cell 2017, 29, 2537–2551. [Google Scholar] [CrossRef] [Green Version]
- Schwarte, S.; Bauwe, H. Identification of the photorespiratory 2-phosphoglycolate phosphatase, PGLP1, in Arabidopsis. Plant Physiol. 2007, 144, 1580–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowes, G.; Ogren, W.; Hageman, R.H. Phosphoglycolate production catalyzed by ribulose diphosphate carboxylase. Biochem. Biophys. Res. Commun. 1971, 45, 716–722. [Google Scholar] [CrossRef]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.; Campbell, K.; Cheung, E.; Olin, M.; Grüning, N.-M.; Krüger, A.; Alam, M.T.; et al. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. 2015, 90, 927–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medeiros, D.B.; Martins, S.C.V.; Cavalcanti, J.H.F.; Daloso, D.M.; Martinoia, E.; Nunes-Nesi, A.; DaMatta, F.M.; Fernie, A.R.; Araújo, W.L. Enhanced photosynthesis and growth in atquac1 knockout mutants are due to altered organic acid accumulation and an increase in both stomatal and mesophyll conductance. Plant Physiol. 2016, 170, 86–101. [Google Scholar] [CrossRef] [Green Version]
- Meyer, S.; Mumm, P.; Imes, D.; Endler, A.; Weder, B.; AL-Rasheid, K.; Geiger, D.; Marten, I.; Martinoia, E.; Hedrich, R. AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 2010, 63, 1054–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santelia, D.; Lawson, T. Rethinking guard cell metabolism. Plant Physiol. 2016, 172, 1371–1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigelt, K.; Küster, H.; Radchuk, R.; Müller, M.; Weichert, H.; Fait, A.; Fernie, A.R.; Saalbach, I.; Weber, H. Increasing amino acid supply in pea embryos reveals specific interactions of N and C metabolism, and highlights the importance of mitochondrial metabolism. Plant J. 2008, 55, 909–926. [Google Scholar] [CrossRef]
- Hu, W.; Lu, Z.; Meng, F.; Li, X.; Cong, R.; Ren, T.; Lu, J. Potassium modulates central carbon metabolism to participate in regulating CO2 transport and assimilation in Brassica napus leaves. Plant Sci. 2021, 307, 110891. [Google Scholar] [CrossRef] [PubMed]
- Bose, J.; Munns, R.; Shabala, S.; Gilliham, M.; Pogson, B.; Tyerman, S.D. Chloroplast function and ion regulation in plants growing on saline soils: Lessons from halophytes. J. Exp. Bot. 2017, 68, 3129–3143. [Google Scholar] [CrossRef]
- Kunz, H.-H.; Gierth, M.; Herdean, A.; Satoh-Cruz, M.; Kramer, D.M.; Spetea, C.; Schroeder, J.I. Plastidial transporters KEA1, -2, and -3 are essential for chloroplast osmoregulation, integrity, and pH regulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 7480–7485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef] [Green Version]
- Munns, R.; James, R.A.; Läuchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef] [Green Version]
- Shams, M.; Yildirim, E. Variations in response of CaPAO and CaATG8c genes, hormone, photosynthesis and antioxidative system in pepper genotypes under salinity stress. Sci. Hortic. 2021, 282, 110041. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar] [CrossRef] [Green Version]
- Dossa, K.; Wei, X.; Li, D.; Fonceka, D.; Zhang, Y.; Wang, L.; Yu, J.; Boshou, L.; Diouf, D.; Cissé, N.; et al. Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress. BMC Plant Biol. 2016, 16, 171. [Google Scholar] [CrossRef] [Green Version]
- Shu, Y.; Liu, Y.; Zhang, J.; Song, L.; Guo, C. Genome-wide analysis of the AP2/ERF superfamily genes and their responses to abiotic stress in Medicago truncatula. Front. Plant Sci. 2015, 6, 1247. [Google Scholar] [CrossRef] [Green Version]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Et Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Wang, M.; Zhuang, J.; Zou, Z.; Li, Q.; Xin, H.; Li, X. Overexpression of a Camellia sinensis DREB transcription factor gene (CsDREB) increases salt and drought tolerance in transgenic Arabidopsis thaliana. J. Plant Biol. 2017, 60, 452–461. [Google Scholar] [CrossRef]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [Green Version]
- Qi, S.-L.; Lin, Q.-F.; Feng, X.-J.; Han, H.-L.; Liu, J.; Zhang, L.; Wu, S.; Le, J.; Blumwald, E.; Hua, X.-J. IDD16 negatively regulates stomatal initiation via trans-repression of SPCH in Arabidopsis. Plant Biotechnol. J. 2019, 17, 1446–1457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.; Yan, M.; Li, L.; He, J.; Zhou, S.; Li, Z.; Niu, C.; Bao, C.; Zhi, F.; Ma, F.; et al. The apple DNA-binding one zinc-finger protein MdDof54 promotes drought resistance. Hortic. Res. 2020, 7, 195. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.Z.; Wang, J.Y.; Wei, Z.Y.; Zhu, Z.X. Methods to break seed dormancy in Cyclocarya paliurus (Batal)Iljinskaja. Sci. Hortic. 2006, 110, 305–309. [Google Scholar] [CrossRef]
- Asghar, T.; Jamil, Y.; Iqbal, M.; Zia-ul-Haq; Abbas, M. Laser light and magnetic field stimulation effect on biochemical, enzymes activities and chlorophyll contents in soybean seeds and seedlings during early growth stages. J. Photochem. Photobiol. B Biol. 2016, 165, 283–290. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Bailey, T.; Bodén, M.; Buske, F.; Frith, M.; Grant, C.; Clementi, L.; Ren, J.; Li, W.; Noble, W. Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.; Wang, J.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]

| Sampling Time | Treatment | Photosynthetic Parameters | |||
|---|---|---|---|---|---|
| Ci (μmol·mol−1) | Tr (mmol·m−2·s−1) | Gs (mol·m−2·s−1) | Pn (μmol·m−2·s−1) | ||
| T1 | CK | 312.67 ± 24.01 c | 2.03 ± 0.15 a | 80.67 ± 1.53 a | 12.13 ± 1.10 Aa |
| LS | 342.00 ± 6.93 b | 1.70 ± 0.10 b | 72.00 ± 4.58 b | 8.77 ± 129 Ab | |
| MS | 351.33 ± 11.37 b | 1.50 ± 0.09 bc | 69.67 ± 2.08 b | 5.27 ± 1.27 Ac | |
| HS | 388.33 ± 07.02 a | 1.40 ± 0.12 c | 48.67 ± 5.03 c | 3.77 ± 0.89 Ac | |
| T2 | CK | 273.33 ± 08.15 c | 2.07 ± 0.26 a | 83.00 ± 4.58 a | 9.83 ± 0.06 Ba |
| LS | 342.67 ± 23.59 b | 1.43 ± 0.15 b | 47.33 ± 4.04 b | 4.40 ± 0.20 Bb | |
| MS | 378.33 ± 05.69 a | 1.23 ± 0.12 b | 37.67 ± 5.03 c | 2.93 ± 0.06 Bc | |
| HS | 387.33 ± 03.06 a | 1.10 ± 0.20 b | 35.33 ± 3.05 c | 2.63 ± 0.06 Bd | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Zhang, Z.; Fang, S.; Liu, Y.; Shang, X. Metabolome and Transcriptome Analyses Unravel the Molecular Regulatory Mechanisms Involved in Photosynthesis of Cyclocarya paliurus under Salt Stress. Int. J. Mol. Sci. 2022, 23, 1161. https://doi.org/10.3390/ijms23031161
Zhang L, Zhang Z, Fang S, Liu Y, Shang X. Metabolome and Transcriptome Analyses Unravel the Molecular Regulatory Mechanisms Involved in Photosynthesis of Cyclocarya paliurus under Salt Stress. International Journal of Molecular Sciences. 2022; 23(3):1161. https://doi.org/10.3390/ijms23031161
Chicago/Turabian StyleZhang, Lei, Zijie Zhang, Shengzuo Fang, Yang Liu, and Xulan Shang. 2022. "Metabolome and Transcriptome Analyses Unravel the Molecular Regulatory Mechanisms Involved in Photosynthesis of Cyclocarya paliurus under Salt Stress" International Journal of Molecular Sciences 23, no. 3: 1161. https://doi.org/10.3390/ijms23031161
APA StyleZhang, L., Zhang, Z., Fang, S., Liu, Y., & Shang, X. (2022). Metabolome and Transcriptome Analyses Unravel the Molecular Regulatory Mechanisms Involved in Photosynthesis of Cyclocarya paliurus under Salt Stress. International Journal of Molecular Sciences, 23(3), 1161. https://doi.org/10.3390/ijms23031161







