Mesothelin Expression Is Not Associated with the Presence of Cancer Stem Cell Markers SOX2 and ALDH1 in Ovarian Cancer
Abstract
1. Introduction
2. Results
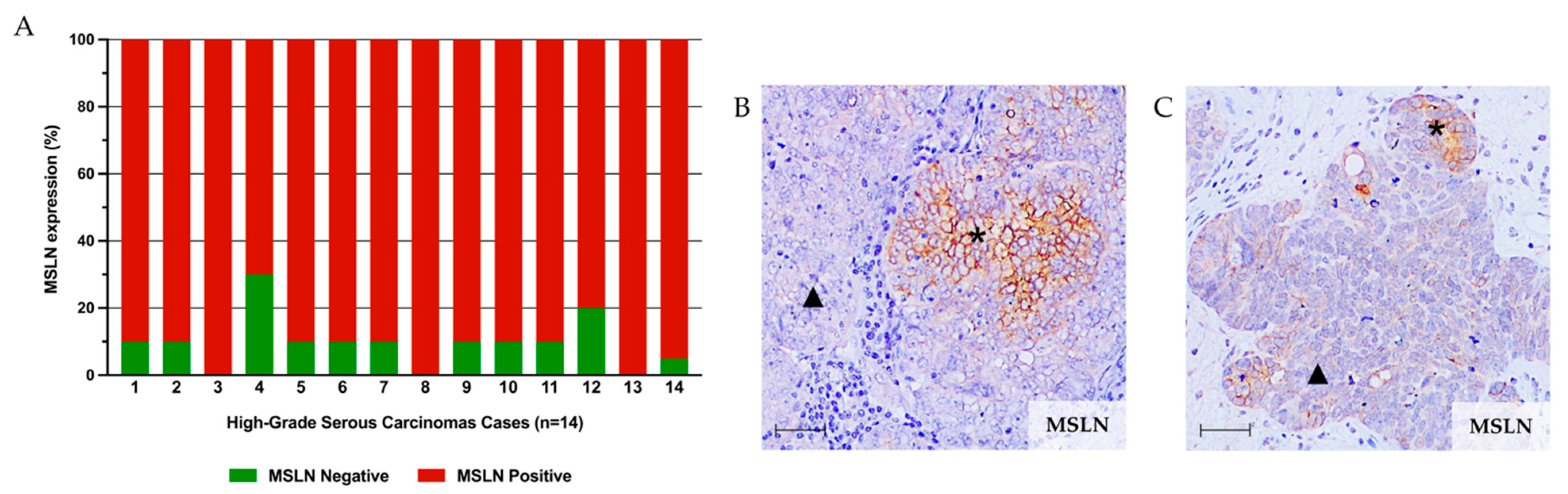
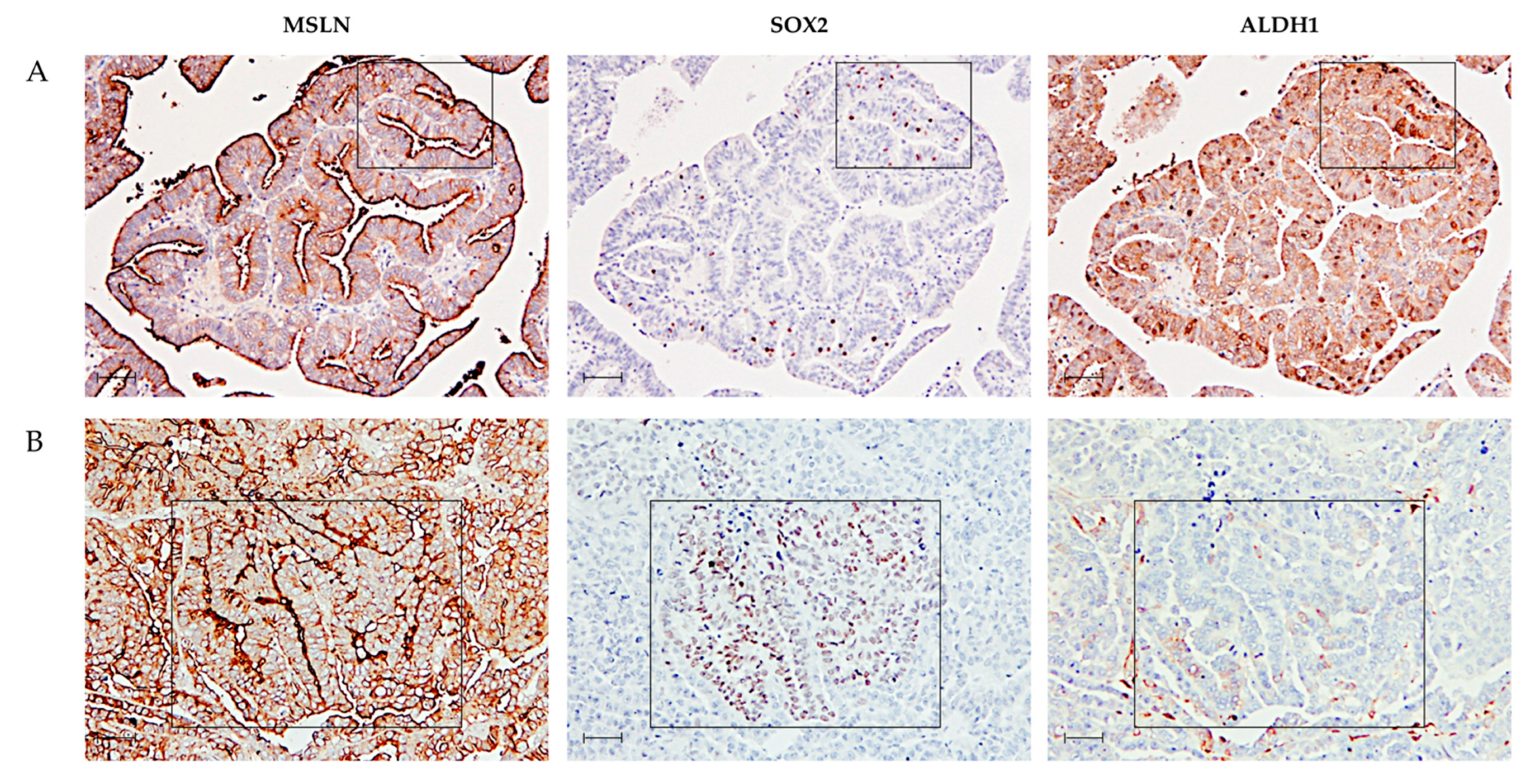
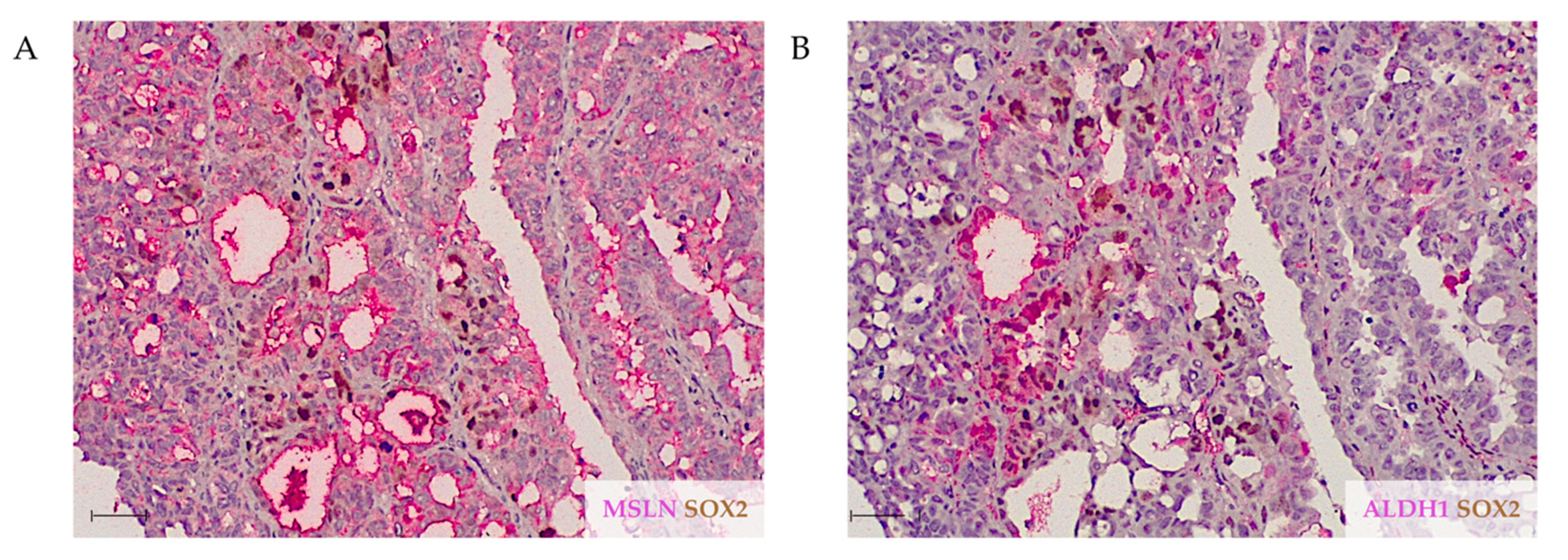
2.1. MSLN Is Not Associated with Expression of CSC Markers in HGSC
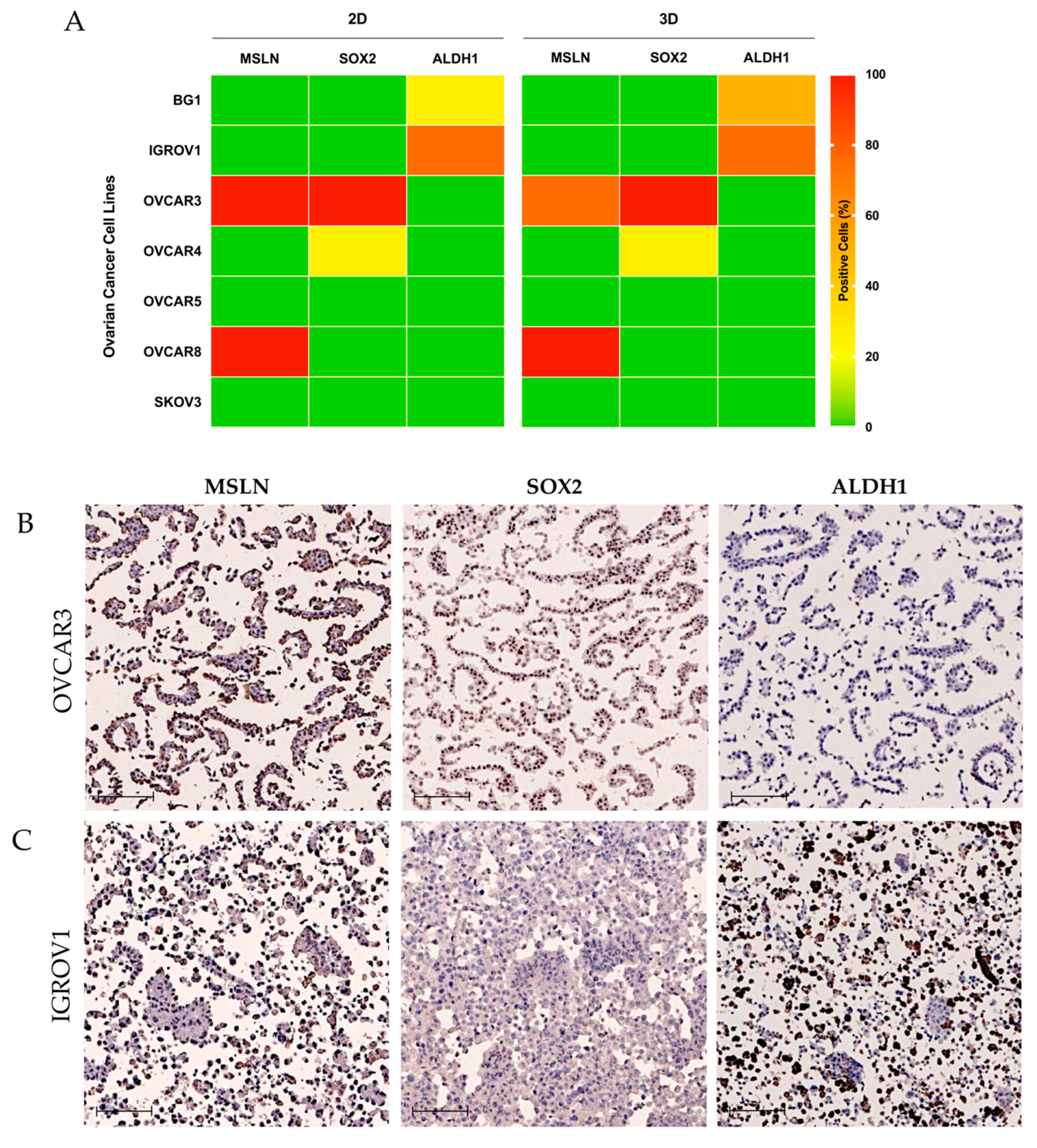
2.2. MSLN Is Not Associated with Expression of CSC Markers in OC Cell Lines
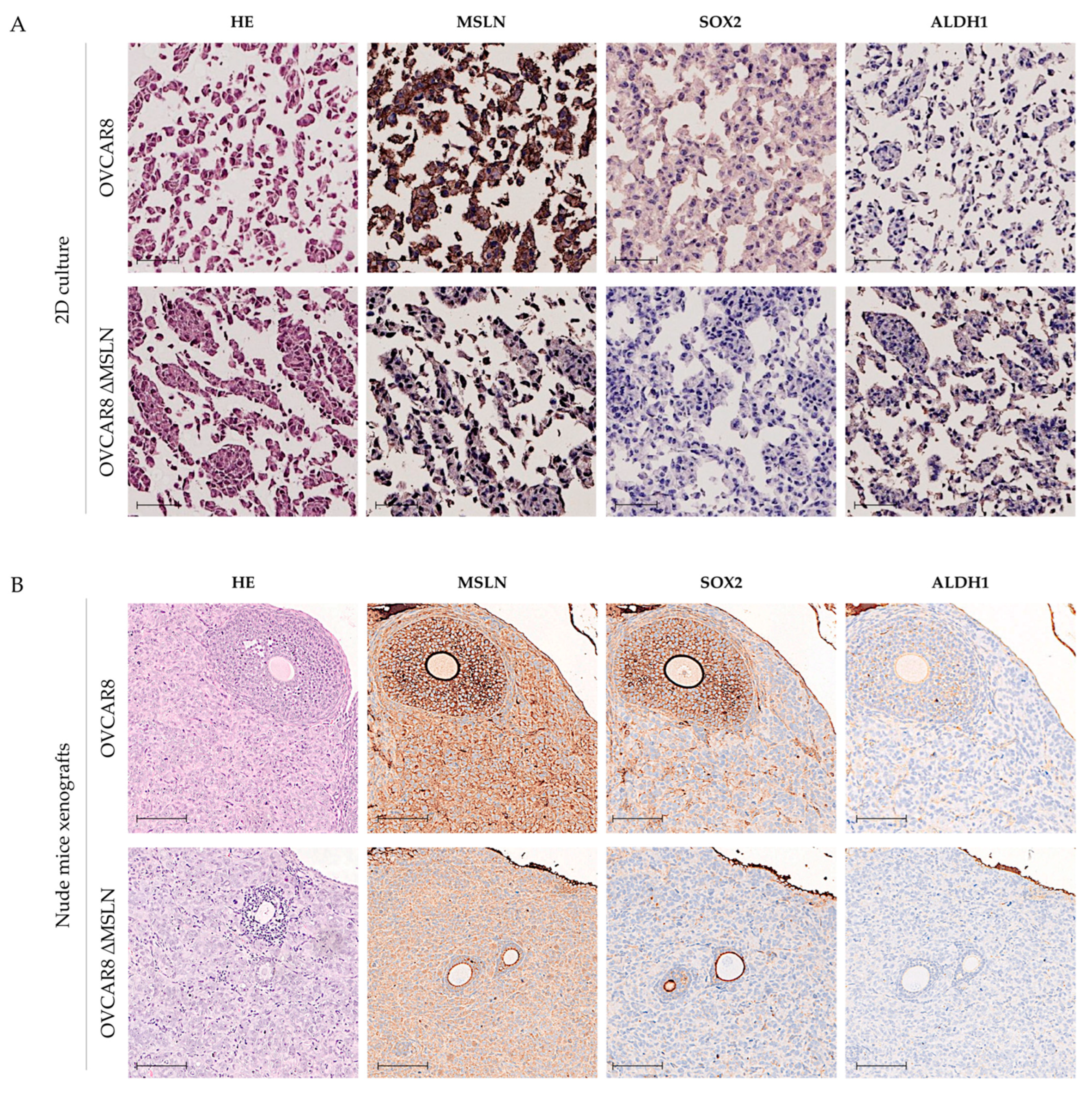
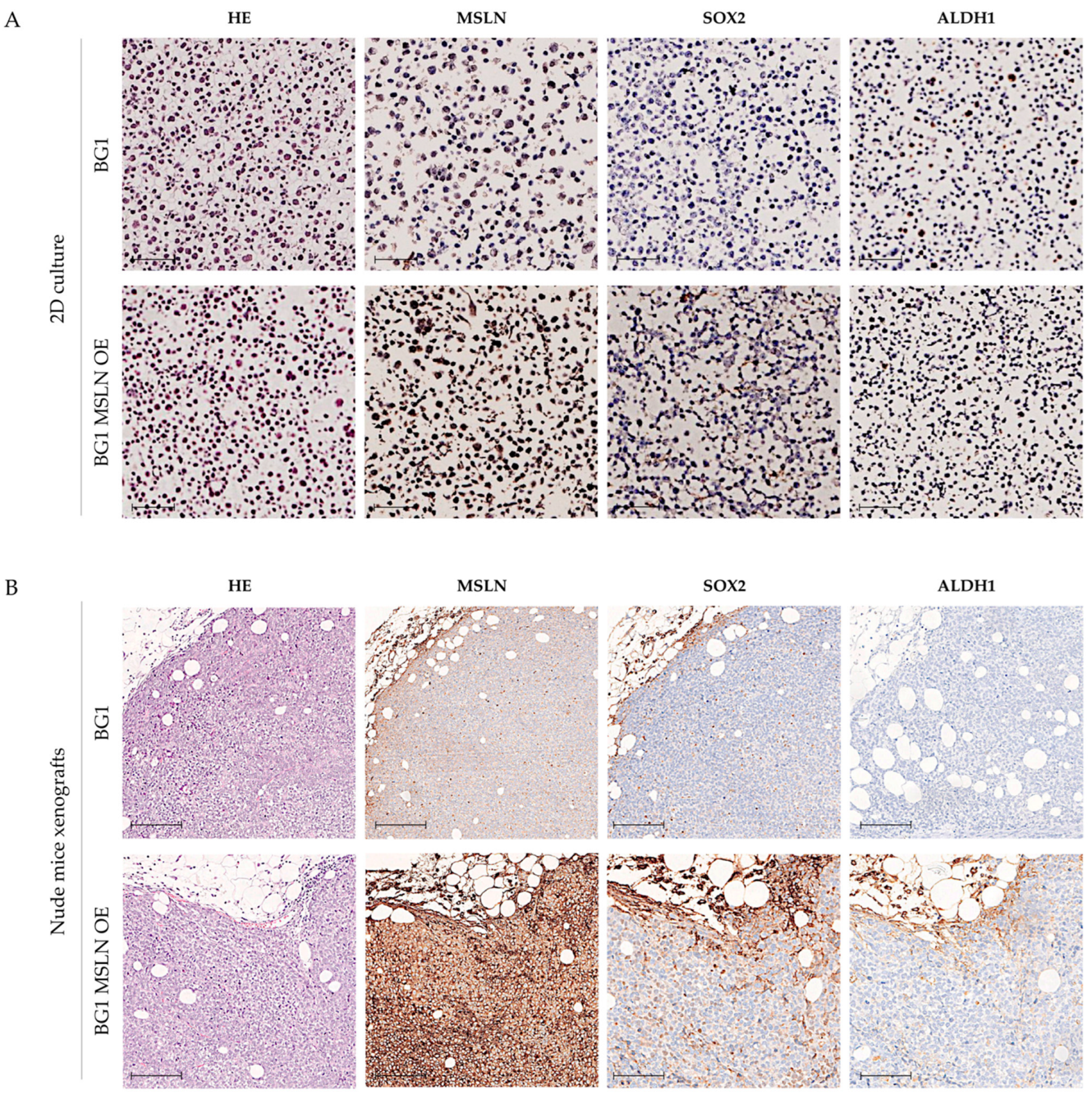
2.3. Genetic Engineering of MSLN Does Not Affect CSC Marker Expression
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. Cell Lines
4.3. Nude Mice Xenografts
4.4. Cell/Tissue Microarray Construction
4.5. Immunohistochemistry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Chang, K.; Pastan, I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc. Natl. Acad. Sci. USA 1996, 93, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Weidemann, S.; Gagelmann, P.; Gorbokon, N.; Lennartz, M.; Menz, A.; Luebke, A.; Kluth, M.; Hube-Magg, C.; Blessin, N.; Fraune, C.; et al. Mesothelin Expression in Human Tumors: A Tissue Microarray Study on 12,679 Tumors. Biomedicines 2021, 9, 397. [Google Scholar] [CrossRef]
- Coelho, R.; Ricardo, S.; Amaral, A.L.; Huang, Y.-L.; Nunes, M.; Neves, J.P.; Mendes, N.; López, M.N.; Bartosch, C.; Ferreira, V.; et al. Regulation of invasion and peritoneal dissemination of ovarian cancer by mesothelin manipulation. Oncogenesis 2020, 9, 61. [Google Scholar] [CrossRef]
- Clevers, H. The cancer stem cell: Premises, promises and challenges. Nat. Med. 2011, 17, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Kusoglu, A.; Avci, C.B. Cancer stem cells: A brief review of the current status. Gene 2019, 681, 80–85. [Google Scholar] [CrossRef]
- Nowicki, A.; Kulus, M.; Wieczorkiewicz, M.; Pieńkowski, W.; Stefańska, K.; Skupin-Mrugalska, P.; Bryl, R.; Mozdziak, P.; Kempisty, B.; Piotrowska-Kempisty, H. Ovarian Cancer and Cancer Stem Cells—Cellular and Molecular Characteristics, Signaling Pathways, and Usefulness as a Diagnostic Tool in Medicine and Oncology. Cancers 2021, 13, 4178. [Google Scholar] [CrossRef] [PubMed]
- Abelson, S.; Shamai, Y.; Berger, L.; Shouval, R.; Skorecki, K.; Tzukerman, M. Intratumoral Heterogeneity in the Self-Renewal and Tumorigenic Differentiation of Ovarian Cancer. Stem Cells 2012, 30, 415–424. [Google Scholar] [CrossRef]
- Garson, K.; Vanderhyden, B.C. Epithelial ovarian cancer stem cells: Underlying complexity of a simple paradigm. Reproduction 2015, 149, R59–R70. [Google Scholar] [CrossRef]
- Roy, L.; Dahl, K.C. Can Stemness and Chemoresistance Be Therapeutically Targeted via Signaling Pathways in Ovarian Cancer? Cancers 2018, 10, 241. [Google Scholar] [CrossRef]
- Hu, Y.; Fu, L. Targeting cancer stem cells: A new therapy to cure cancer patients. Am. J. Cancer Res. 2012, 2, 340–356. [Google Scholar]
- Fischer, A.K.; Pham, D.L.; Bösmüller, H.; Lengerke, C.; Wagner, P.; Bachmann, C.; Beschorner, C.; Perner, S.; Kommoss, S.; Fend, F.; et al. Comprehensive in situ analysis of ALDH1 and SOX2 reveals increased expression of stem cell markers in high-grade serous carcinomas compared to low-grade serous carcinomas and atypical proliferative serous tumors. Virchows Arch. 2019, 475, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Bareiss, P.M.; Paczulla, A.; Wang, H.; Schairer, R.; Wiehr, S.; Kohlhofer, U.; Rothfuss, O.C.; Fischer, A.; Perner, S.; Staebler, A.; et al. SOX2 expression associates with stem cell state in human ovarian carcinoma. Cancer Res. 2013, 73, 5544–5555. [Google Scholar] [CrossRef] [PubMed]
- Weina, K.; Utikal, J. SOX2 and cancer: Current research and its implications in the clinic. Clin. Transl. Med. 2014, 3, 19. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.; Diaz, E.; Reya, T. Stem cells in cancer initiation and progression. J. Cell Biol. 2020, 219, e201911053. [Google Scholar] [CrossRef] [PubMed]
- Pádua, D.; Barros, R.; Amaral, A.L.; Mesquita, P.; Freire, A.F.; Sousa, M.; Maia, A.F.; Caiado, I.; Fernandes, H.; Pombinho, A.; et al. A SOX2 Reporter System Identifies Gastric Cancer Stem-Like Cells Sensitive to Monensin. Cancers 2020, 12, 495. [Google Scholar] [CrossRef]
- Ye, F.; Li, Y.; Hu, Y.; Zhou, C.; Hu, Y.; Chen, H. Expression of Sox2 in human ovarian epithelial carcinoma. J. Cancer Res. Clin. Oncol. 2011, 137, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chang, D.Y.; Mercado-Uribe, I.; Liu, J. Sex-determining region Y-box 2 expression predicts poor prognosis in human ovarian carcinoma. Hum. Pathol. 2012, 43, 1405–1412. [Google Scholar] [CrossRef]
- Singh, S.; Brocker, C.; Koppaka, V.; Chen, Y.; Jackson, B.C.; Matsumoto, A.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic. Biol. Med. 2013, 56, 89–101. [Google Scholar] [CrossRef]
- Kuroda, T.; Hirohashi, Y.; Torigoe, T.; Yasuda, K.; Takahashi, A.; Asanuma, H.; Morita, R.; Mariya, T.; Asano, T.; Mizuuchi, M.; et al. ALDH1-High Ovarian Cancer Stem-Like Cells Can Be Isolated from Serous and Clear Cell Adenocarcinoma Cells, and ALDH1 High Expression Is Associated with Poor Prognosis. PLoS ONE 2013, 8, e65158. [Google Scholar] [CrossRef]
- Novak, D.; Hüser, L.; Elton, J.J.; Umansky, V.; Altevogt, P.; Utikal, J. SOX2 in development and cancer biology. Semin. Cancer Biol. 2020, 67, 74–82. [Google Scholar] [CrossRef]
- Moreb, J.S.; Baker, H.V.; Chang, L.-J.; Amaya, M.; Lopez, M.C.; Ostmark, B.; Chou, W. ALDH isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells. Mol. Cancer 2008, 7, 87. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Hou, Y.; Huang, Z.; Cai, J.; Wang, Z. SOX2 is required to maintain cancer stem cells in ovarian cancer. Cancer Sci. 2017, 108, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Kaipio, K.; Chen, P.; Roering, P.; Huhtinen, K.; Mikkonen, P.; Östling, P.; Lehtinen, L.; Mansuri, N.; Korpela, T.; Potdar, S.; et al. ALDH1A1-related stemness in high-grade serous ovarian cancer is a negative prognostic indicator but potentially targetable by EGFR/mTOR-PI3K/aurora kinase inhibitors. J. Pathol. 2020, 250, 159–169. [Google Scholar] [CrossRef]
- Bååth, M.; Westbom-Fremer, S.; De La Fuente, L.M.; Ebbesson, A.; Davis, J.; Malander, S.; Måsbäck, A.; Kannisto, P.; Hedenfalk, I. SOX2 is a promising predictor of relapse and death in advanced stage high-grade serous ovarian cancer patients with residual disease after debulking surgery. Mol. Cell. Oncol. 2020, 7, 1805094. [Google Scholar] [CrossRef] [PubMed]
- Coelho, R.; Marcos-Silva, L.; Mendes, N.; Pereira, D.; Brito, C.; Jacob, F.; Steentoft, C.; Mandel, U.; Clausen, H.; David, L.; et al. Mucins and Truncated O-Glycans Unveil Phenotypic Discrepancies between Serous Ovarian Cancer Cell Lines and Primary Tumours. Int. J. Mol. Sci. 2018, 19, 2045. [Google Scholar] [CrossRef]
- Chang, M.-C.; Chen, C.-A.; Chen, P.-J.; Chen, Y.-L.; Mao, T.-L.; Lin, H.-W.; Chiang, W.-H.L.; Cheng, W.-F. Mesothelin enhances invasion of ovarian cancer by inducing MMP-7 through MAPK/ERK and JNK pathways. Biochem. J. 2012, 442, 293–302. [Google Scholar] [CrossRef]
- Shen, J.; Sun, X.; Zhou, J. Insights Into the Role of Mesothelin as a Diagnostic and Therapeutic Target in Ovarian Carcinoma. Front. Oncol. 2020, 10, 1263. [Google Scholar] [CrossRef] [PubMed]
- Hanaoka, T.; Hasegawa, K.; Kato, T.; Sato, S.; Kurosaki, A.; Miyara, A.; Nagao, S.; Seki, H.; Yasuda, M.; Fujiwara, K. Correlation Between Tumor Mesothelin Expression and Serum Mesothelin in Patients with Epithelial Ovarian Carcinoma: A Potential Noninvasive Biomarker for Mesothelin-targeted Therapy. Mol. Diagn. Ther. 2017, 21, 187–198. [Google Scholar] [CrossRef]
- Cheng, W.-F.; Huang, C.-Y.; Chang, M.-C.; Hu, Y.-H.; Chiang, Y.-C.; Chen, Y.-L.; Hsieh, C.-Y.; Chen, C.-A. High mesothelin correlates with chemoresistance and poor survival in epithelial ovarian carcinoma. Br. J. Cancer 2009, 100, 1144–1153. [Google Scholar] [CrossRef]
- Yildiz, Y.; Kabadayi, G.; Yigit, S.; Kucukzeybek, Y.; Alacacioglu, A.; Varol, U.; Taskaynatan, H.; Salman, T.; Oflazoglu, U.; Akyol, M.; et al. High expression of mesothelin in advanced serous ovarian cancer is associated with poor prognosis. J. BUON 2019, 24, 1549–1554. [Google Scholar]
- He, X.; Wang, L.; Riedel, H.; Wang, K.; Yang, Y.; Dinu, C.Z.; Rojanasakul, Y. Mesothelin promotes epithelial-to-mesenchymal transition and tumorigenicity of human lung cancer and mesothelioma cells. Mol. Cancer 2017, 16, 1–13. [Google Scholar] [CrossRef]
- Matsuzawa, F.; Kamachi, H.; Mizukami, T.; Einama, T.; Kawamata, F.; Fujii, Y.; Fukai, M.; Kobayashi, N.; Hatanaka, Y.; Taketomi, A. Mesothelin blockage by Amatuximab suppresses cell invasiveness, enhances gemcitabine sensitivity and regulates cancer cell stemness in mesothelin-positive pancreatic cancer cells. BMC Cancer 2021, 21, 1–16. [Google Scholar] [CrossRef]
- Ottevanger, P.B. Ovarian cancer stem cells more questions than answers. Semin. Cancer Biol. 2017, 44, 67–71. [Google Scholar] [CrossRef]
- Pieterse, Z.; Padilla, M.A.; Singomat, T.; Binju, M.; Madjid, B.D.; Yu, Y.; Kaur, P. Ovarian cancer stem cells and their role in drug resistance. Int. J. Biochem. Cell Biol. 2019, 106, 117–126. [Google Scholar] [CrossRef]
- Sabini, C.; Sorbi, F.; Cunnea, P.; Fotopoulou, C. Ovarian cancer stem cells: Ready for prime time? Arch Gynecol. Obstet. 2020, 301, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Tjhay, F.; Motohara, T.; Tayama, S.; Narantuya, D.; Fujimoto, K.; Guo, J.; Sakaguchi, I.; Honda, R.; Tashiro, H.; Katabuchi, H. CD 44 variant 6 is correlated with peritoneal dissemination and poor prognosis in patients with advanced epithelial ovarian cancer. Cancer Sci. 2015, 106, 1421–1428. [Google Scholar] [CrossRef] [PubMed]
- Samardzija, C.; Luwor, R.B.; Quinn, M.A.; Kannourakis, G.; Findlay, G.K.; Ahmed, N. Coalition of Oct4A and beta1 integrins in facilitating metastasis in ovarian cancer. BMC Cancer 2016, 16, 432. [Google Scholar] [CrossRef][Green Version]
- Parte, S.C.; Batra, S.K.; Kakar, S.S. Characterization of stem cell and cancer stem cell populations in ovary and ovarian tumors. J. Ovarian Res. 2018, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Li, B.; Fang, Y.; Liu, Y.; Wang, Y.; Li, J.; Zhou, W.; Wang, X. Overexpression of Class III β-tubulin, Sox2, and nuclear Survivin is predictive of taxane resistance in patients with stage III ovarian epithelial cancer. BMC Cancer 2015, 15, 1–11. [Google Scholar] [CrossRef]
- Li, Y.; Chen, K.; Li, L.; Li, R.; Zhang, J.; Ren, W. Overexpression of SOX2 is involved in paclitaxel resistance of ovarian cancer via the PI3K/Akt pathway. Tumor Biol. 2015, 36, 9823–9828. [Google Scholar] [CrossRef]
- Wang, X.; Ji, X.; Chen, J.; Yan, D.; Zhang, Z.; Wang, Q.; Xi, X.; Feng, Y. SOX2 Enhances the Migration and Invasion of Ovarian Cancer Cells via Src Kinase. PLoS ONE 2014, 9, e99594. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Han, X.; Jin, C.; Tian, W.; Yu, W.; Ding, D.; Cheng, L.; Huang, B.; Jiang, H.; Lin, B. SOX2 Targets Fibronectin 1 to Promote Cell Migration and Invasion in Ovarian Cancer: New Molecular Leads for Therapeutic Intervention. OMICS A J. Integr. Biol. 2013, 17, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Chefetz, I.; Grimley, E.; Yang, K.; Hong, L.; Vinogradova, E.V.; Suciu, R.; Kovalenko, I.; Karnak, D.; Morgan, C.A.; Chtcherbinine, M.; et al. A Pan-ALDH1A Inhibitor Induces Necroptosis in Ovarian Cancer Stem-like Cells. Cell Rep. 2019, 26, 3061–3075. [Google Scholar] [CrossRef] [PubMed]
- Landen, C.N.; Goodman, B.; Katre, A.A.; Steg, A.D.; Nick, A.M.; Stone, R.L.; Miller, L.; Mejia, P.V.; Jennings, N.B.; Gershenson, D.M.; et al. Targeting Aldehyde Dehydrogenase Cancer Stem Cells in Ovarian Cancer. Mol. Cancer Ther. 2010, 9, 3186–3199. [Google Scholar] [CrossRef] [PubMed]
- Steg, A.D.; Bevis, K.S.; Katre, A.A.; Ziebarth, A.; Dobbin, Z.C.; Alvarez, R.D.; Zhang, K.; Conner, M.; Landen, C.N. Stem Cell Pathways Contribute to Clinical Chemoresistance in Ovarian Cancer. Clin. Cancer Res. 2012, 18, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Ruscito, I.; Darb-Esfahani, S.; Kulbe, H.; Bellati, F.; Zizzari, I.G.; Koshkaki, H.R.; Napoletano, C.; Caserta, D.; Rughetti, A.; Kessler, M.; et al. The prognostic impact of cancer stem-like cell biomarker aldehyde dehydrogenase-1 (ALDH1) in ovarian cancer: A meta-analysis. Gynecol. Oncol. 2018, 150, 151–157. [Google Scholar] [CrossRef]
- Domcke, S.; Sinha, R.; Levine, D.A.; Sander, C.; Schultz, N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013, 4, 2126. [Google Scholar] [CrossRef] [PubMed]
- Avninder, S.; Ylaya, K.; Hewitt, S. Tissue microarray: A simple technology that has revolutionized research in pathology. J. Postgrad. Med. 2008, 54, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Mirlacher, M.; Sauter, G. Tissue microarrays. Methods Mol. Med. 2004, 97, 377–389. [Google Scholar]
- Nunes, M.; Silva, P.M.A.; Coelho, R.; Pinto, C.; Resende, A.; Bousbaa, H.; Almeida, G.M.; Ricardo, S. Generation of Two Paclitaxel-Resistant High-Grade Serous Carcinoma Cell Lines With Increased Expression of P-Glycoprotein. Front. Oncol. 2021, 11, 752127. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nunes, M.; Pacheco, F.; Coelho, R.; Leitão, D.; Ricardo, S.; David, L. Mesothelin Expression Is Not Associated with the Presence of Cancer Stem Cell Markers SOX2 and ALDH1 in Ovarian Cancer. Int. J. Mol. Sci. 2022, 23, 1016. https://doi.org/10.3390/ijms23031016
Nunes M, Pacheco F, Coelho R, Leitão D, Ricardo S, David L. Mesothelin Expression Is Not Associated with the Presence of Cancer Stem Cell Markers SOX2 and ALDH1 in Ovarian Cancer. International Journal of Molecular Sciences. 2022; 23(3):1016. https://doi.org/10.3390/ijms23031016
Chicago/Turabian StyleNunes, Mariana, Francisca Pacheco, Ricardo Coelho, Dina Leitão, Sara Ricardo, and Leonor David. 2022. "Mesothelin Expression Is Not Associated with the Presence of Cancer Stem Cell Markers SOX2 and ALDH1 in Ovarian Cancer" International Journal of Molecular Sciences 23, no. 3: 1016. https://doi.org/10.3390/ijms23031016
APA StyleNunes, M., Pacheco, F., Coelho, R., Leitão, D., Ricardo, S., & David, L. (2022). Mesothelin Expression Is Not Associated with the Presence of Cancer Stem Cell Markers SOX2 and ALDH1 in Ovarian Cancer. International Journal of Molecular Sciences, 23(3), 1016. https://doi.org/10.3390/ijms23031016





