Rumen Epithelial Development- and Metabolism-Related Genes Regulate Their Micromorphology and VFAs Mediating Plateau Adaptability at Different Ages in Tibetan Sheep
Abstract
1. Introduction
2. Results
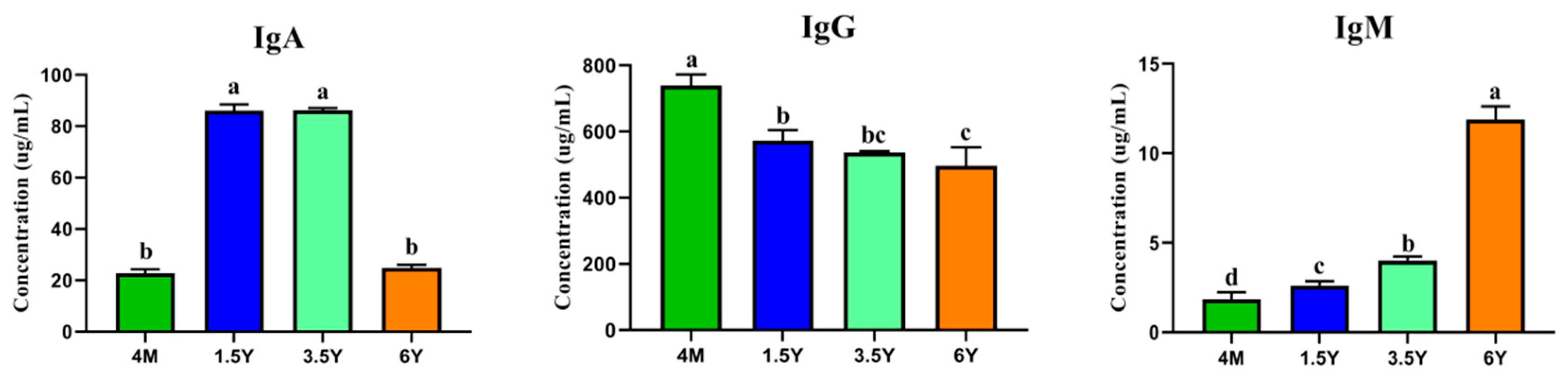
2.1. Serum Immune Levels
2.2. Rumen Fermentation Characteristics
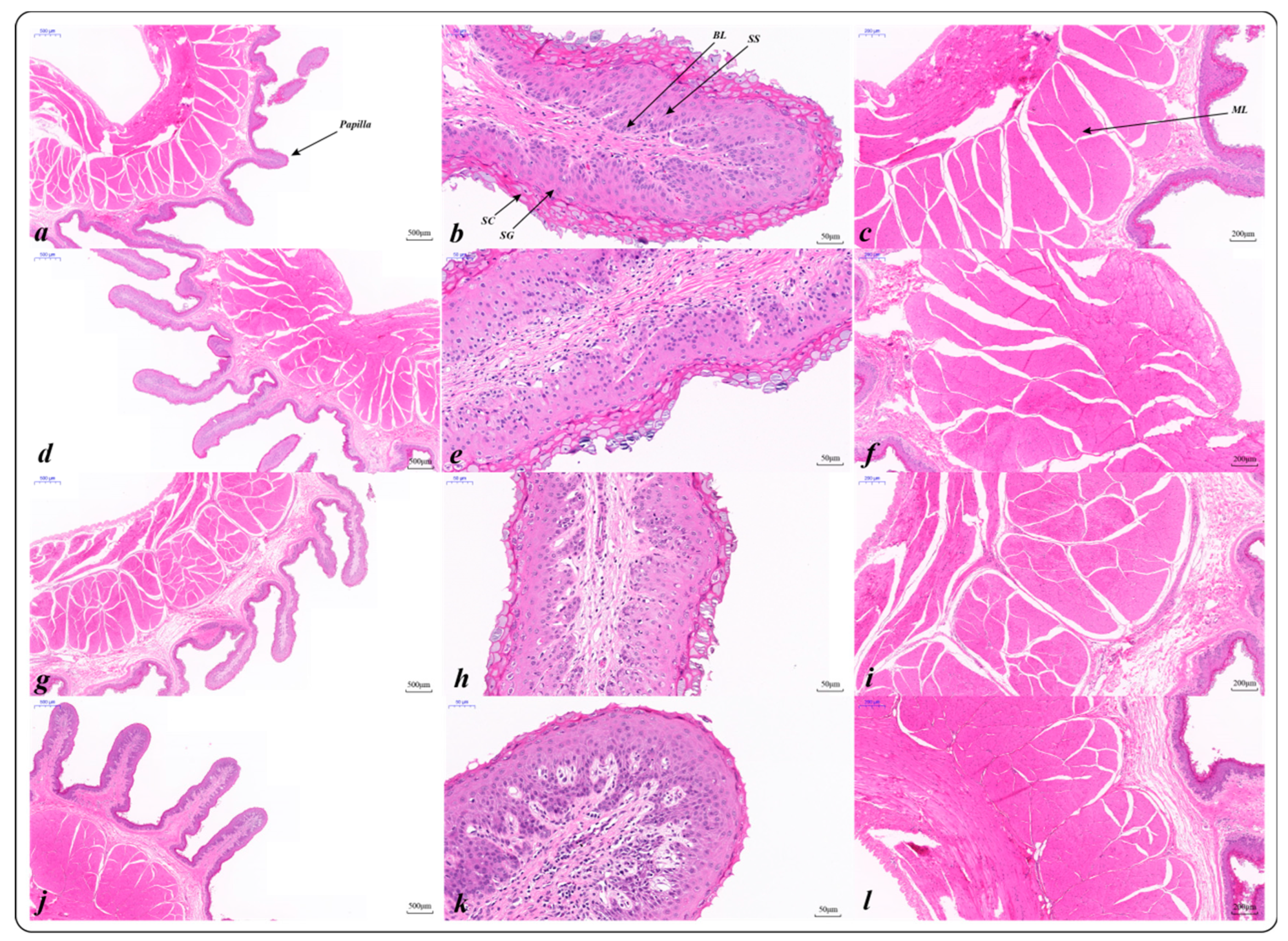
2.3. Rumen Epithelial Development Structure
2.3.1. Histological Characteristics of the Rumen Epithelium (HE)
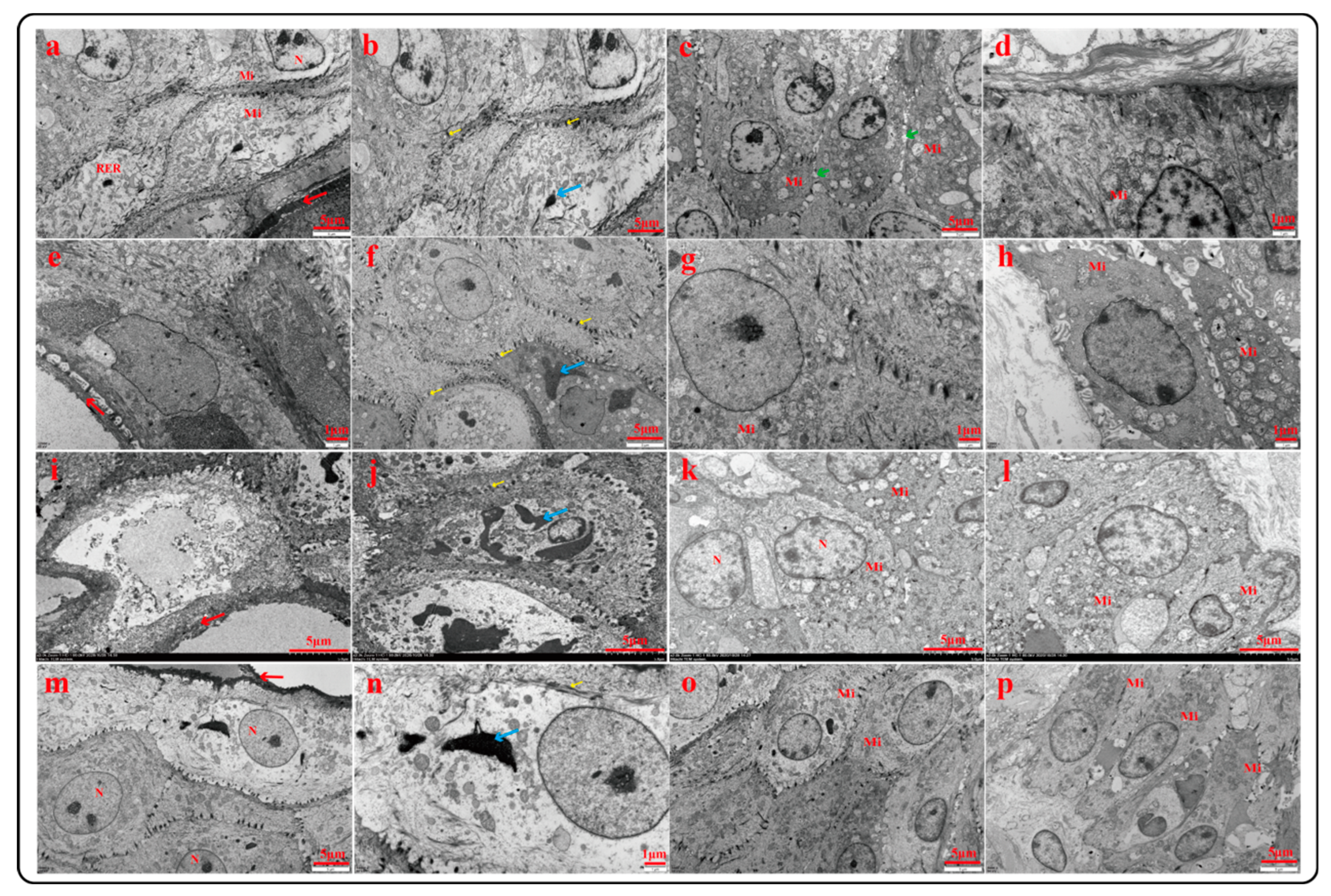
2.3.2. Ultrastructural Characteristics of Rumen Epithelium (TEM)
2.4. mRNA Expression Profile of the Rumen Epithelium
2.4.1. Differential mRNA Analysis
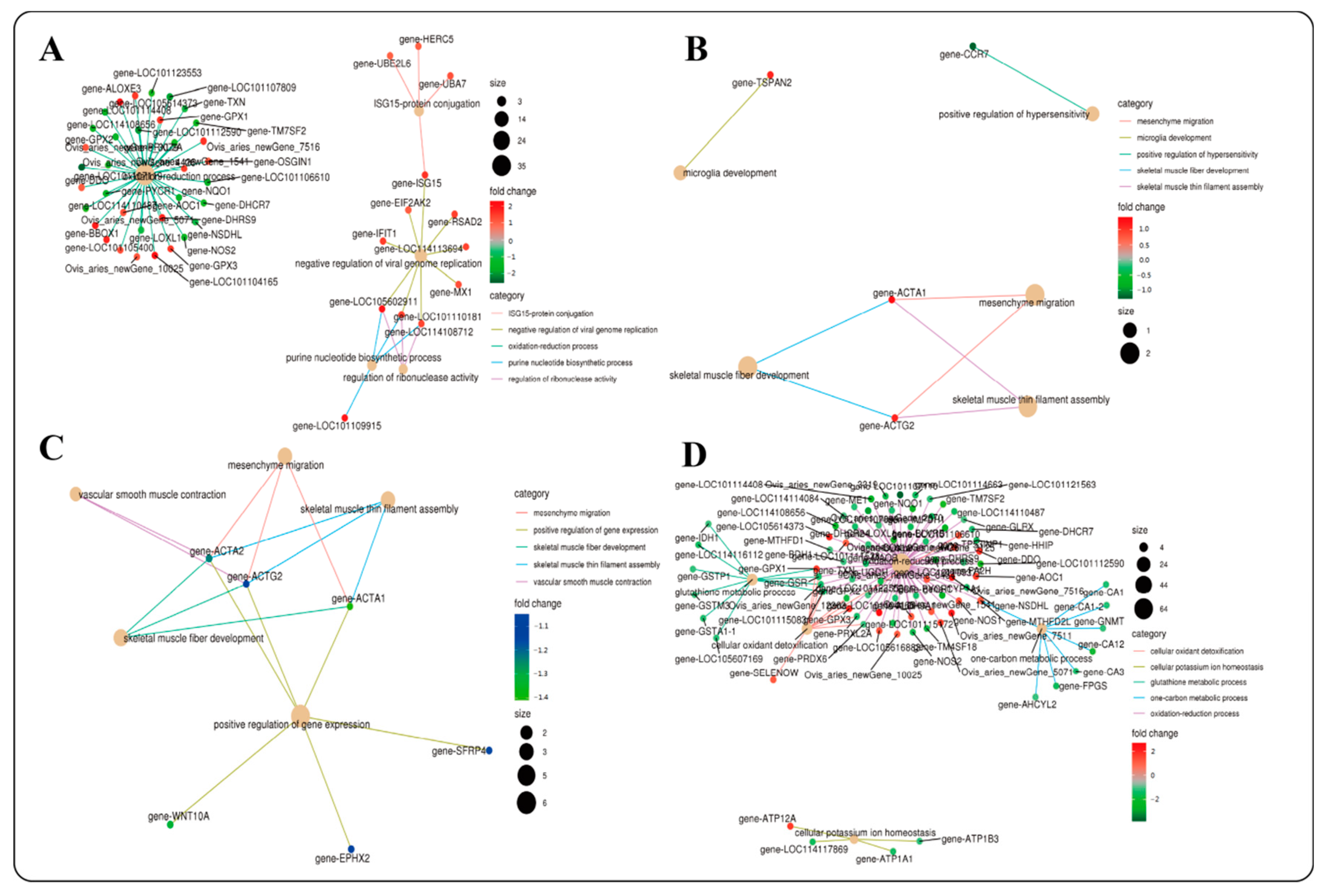
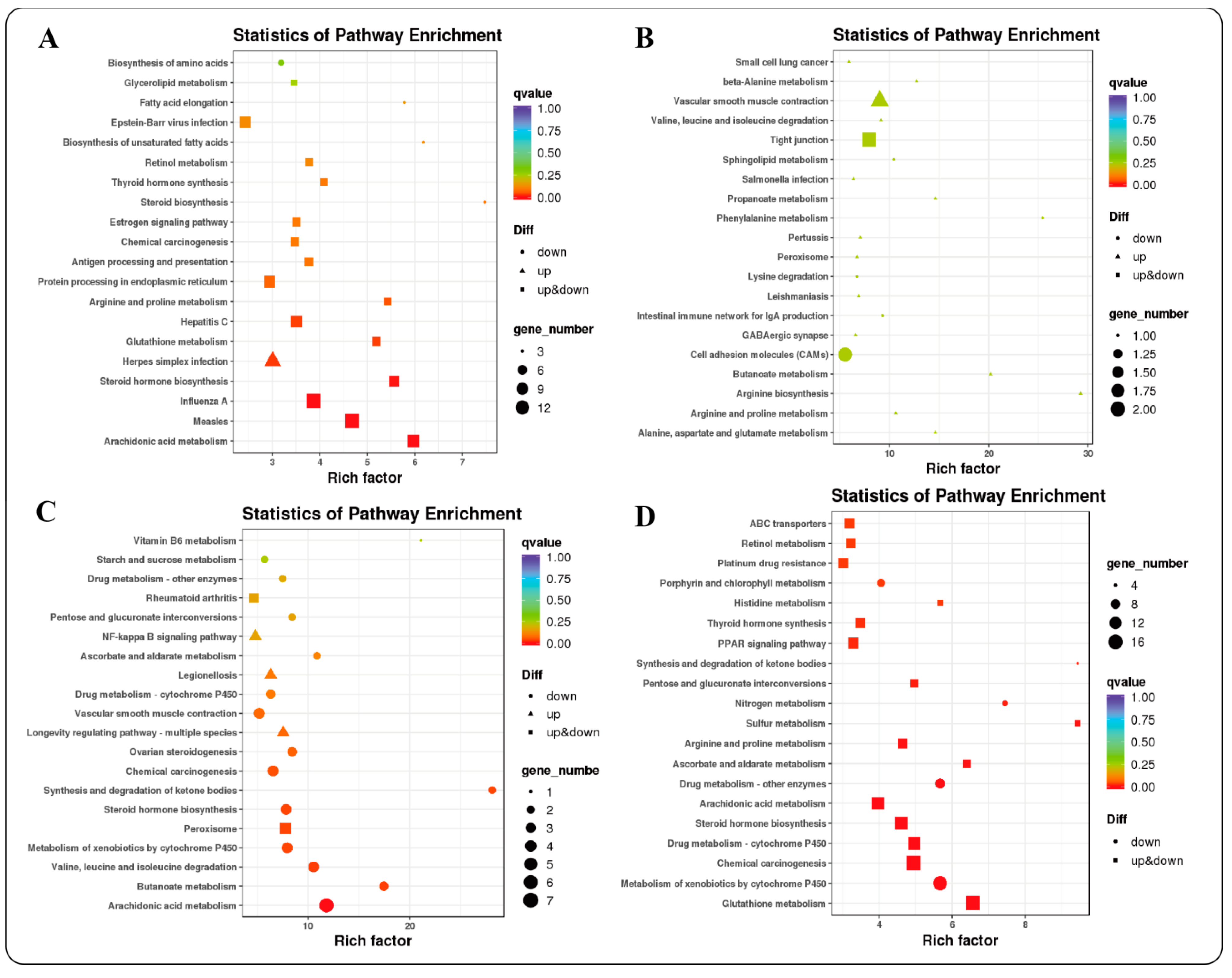
2.4.2. GO and KEGG Functional Enrichment Analysis of DEGs
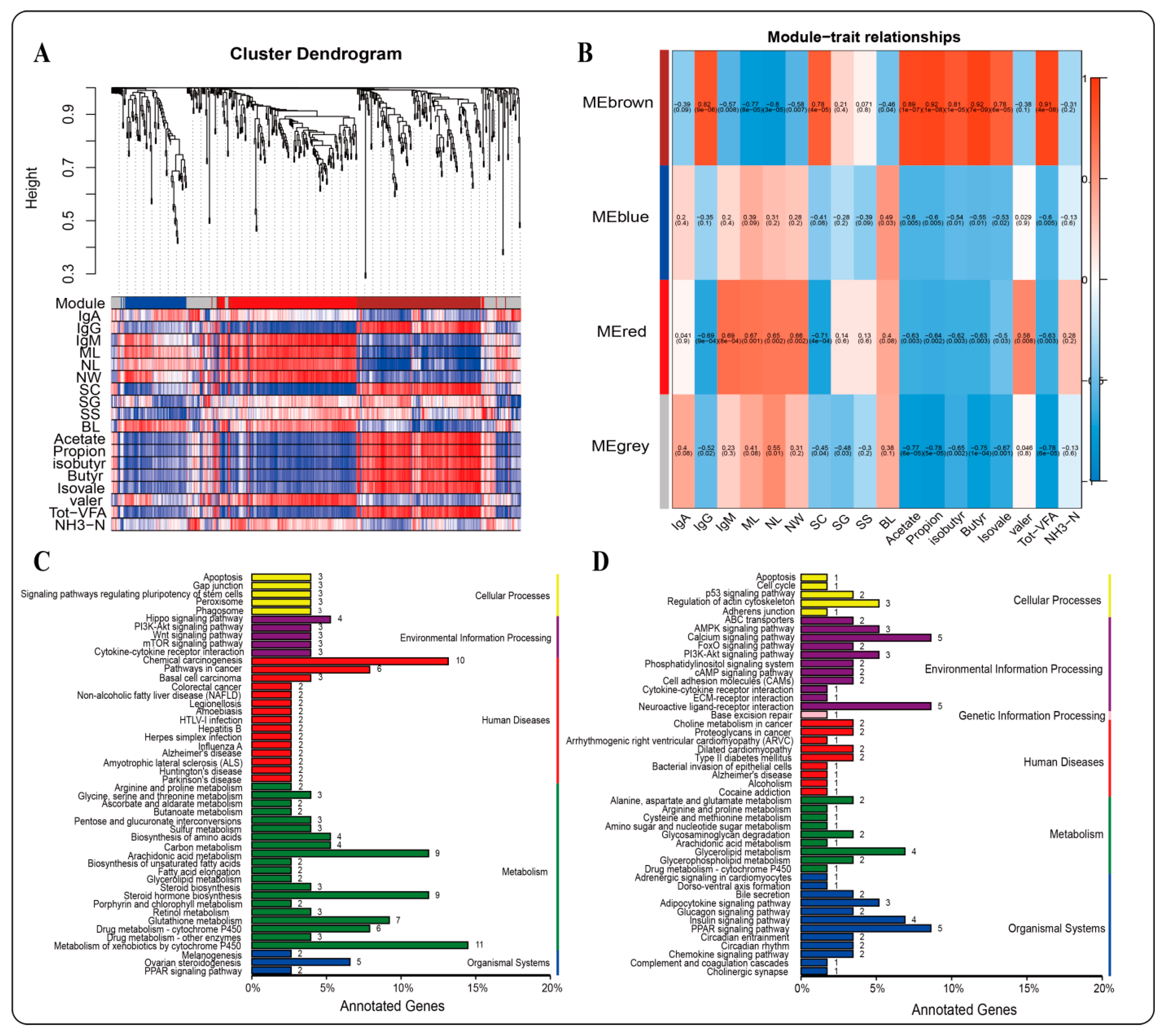
2.5. WGCNA
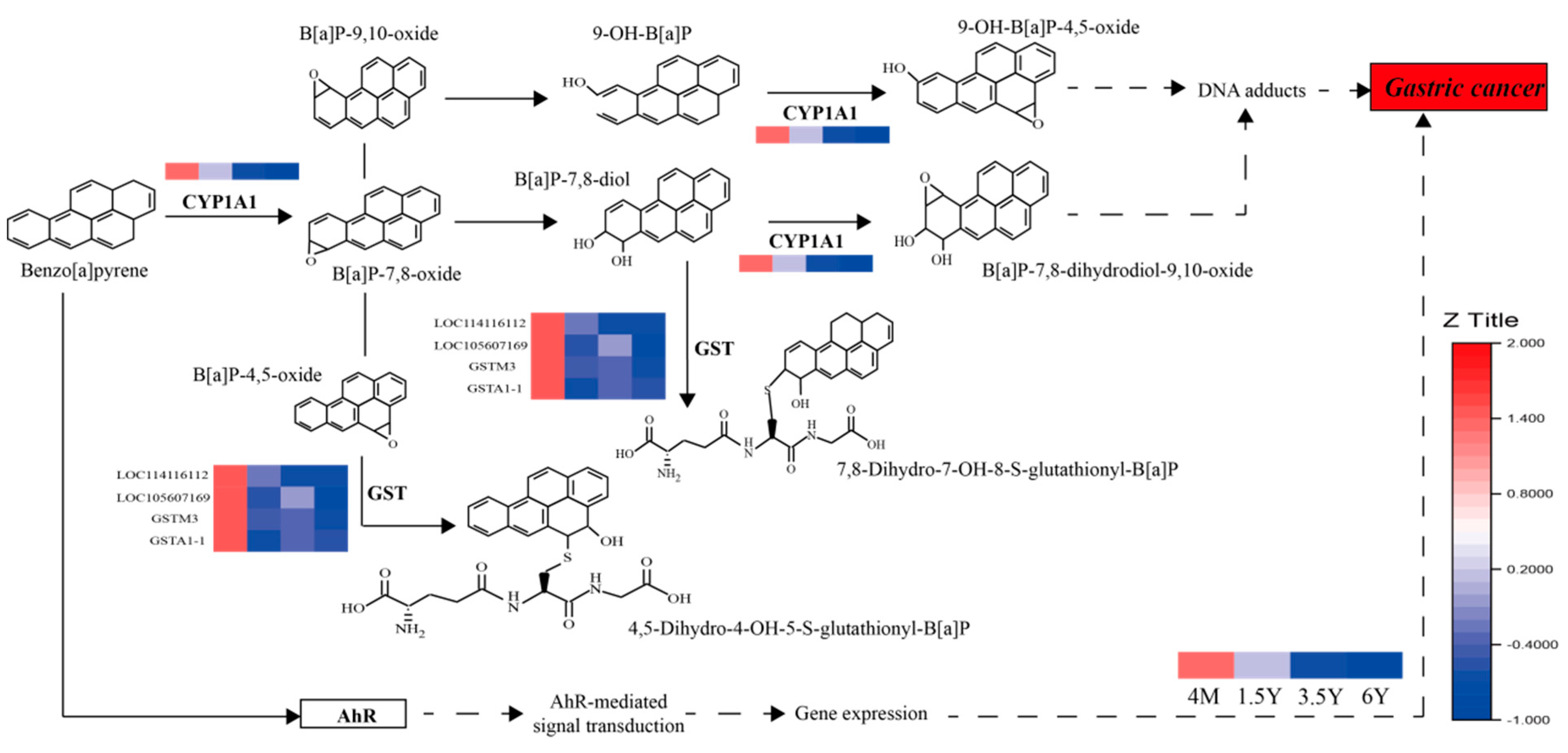
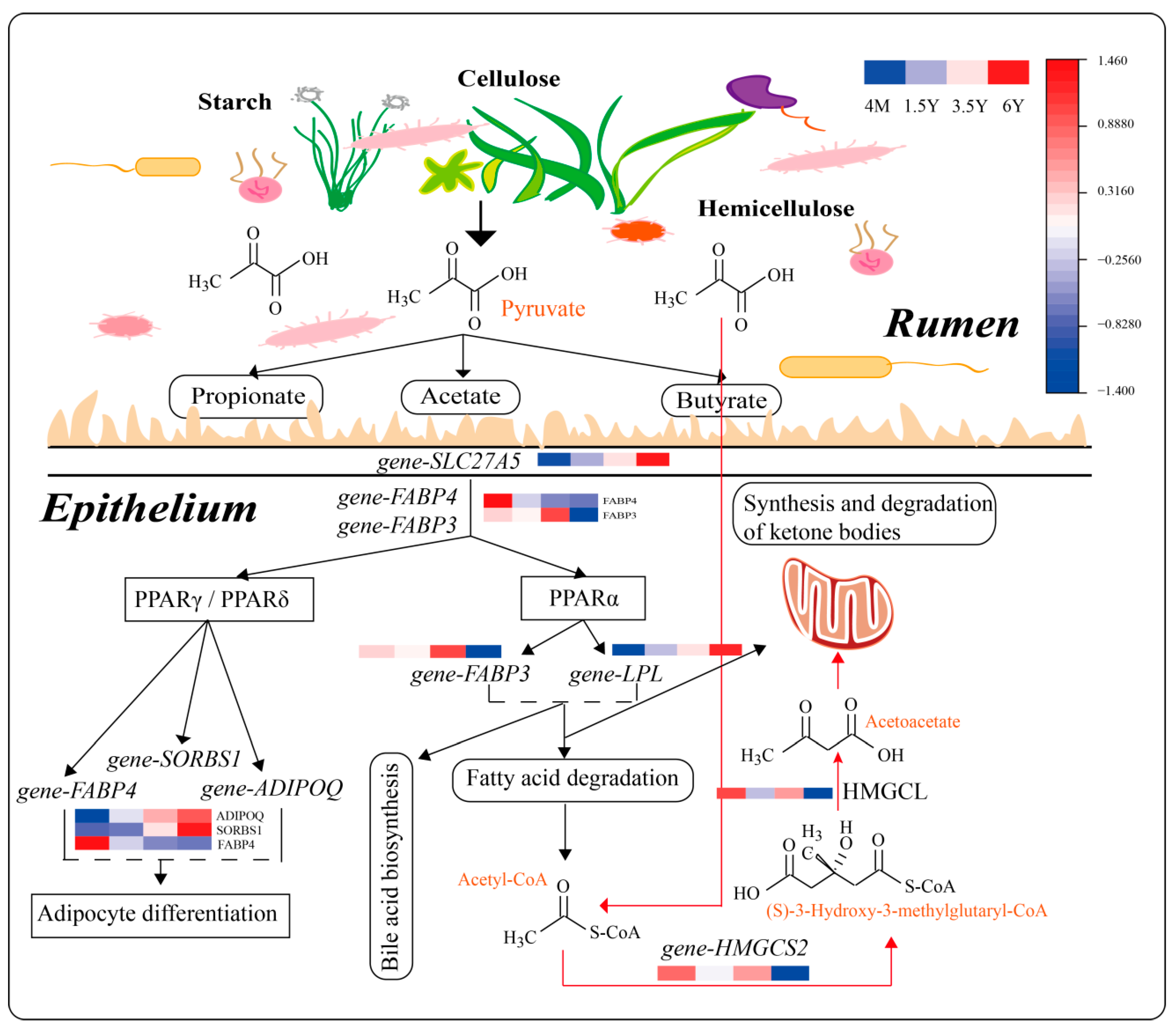
3. Discussion
4. Materials and Methods
4.1. Experimental Design and Sample Collection
4.2. Serum Immune Index Determination
4.3. Determination of the Rumen Fermentation Parameters
4.4. Micromorphological Structure of the Rumen
4.5. RNA Extraction and Transcriptome Sequencing
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, Y.; Munday, J.S.; Perrott, M.; Wang, G.; Liu, X. Association of Age with the Expression of Hypoxia-Inducible Factors HIF-1alpha, HIF-2alpha, HIF-3alpha and VEGF in Lung and Heart of Tibetan Sheep. Animals 2019, 9, 673. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Wang, W.; Degen, A.; Guo, Y.; Kang, J.; Liu, P.; Ding, L.; Shang, Z.; Fievez, V.; Zhou, J.; et al. Tibetan sheep have a high capacity to absorb and to regulate metabolism of SCFA in the rumen epithelium to adapt to low energy intake. Br. J. Nutr. 2020, 123, 721–736. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, D.; Wang, L.; Hao, J.; Wang, J.; Zhou, X.; Wang, W.; Qiu, Q.; Huang, X.; Zhou, J.; et al. Convergent Evolution of Rumen Microbiomes in High-Altitude Mammals. Curr. Biol. 2016, 26, 1873–1879. [Google Scholar] [CrossRef]
- Liu, X.; Sha, Y.; Dingkao, R.; Zhang, W.; Lv, W.; Wei, H.; Shi, H.; Hu, J.; Wang, J.; Li, S.; et al. Interactions Between Rumen Microbes, VFAs, and Host Genes Regulate Nutrient Absorption and Epithelial Barrier Function During Cold Season Nutritional Stress in Tibetan Sheep. Front. Microbiol. 2020, 11, 593062. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Xie, F.; Sun, D.; Liu, J.; Zhu, W.; Mao, S. Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome 2019, 7, 83. [Google Scholar] [CrossRef]
- Graham, C.; Simmons, N.L. Functional organization of the bovine rumen epithelium. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 288, R173–R181. [Google Scholar] [CrossRef]
- Shen, H.; Xu, Z.; Shen, Z.; Lu, Z. The Regulation of Ruminal Short-Chain Fatty Acids on the Functions of Rumen Barriers. Front. Physiol. 2019, 10, 1305. [Google Scholar] [CrossRef]
- Aschenbach, J.R.; Penner, G.B.; Stumpff, F.; Gabel, G. Ruminant Nutrition Symposium: Role of fermentation acid absorption in the regulation of ruminal pH. J. Anim. Sci. 2011, 89, 1092–1107. [Google Scholar] [CrossRef]
- Melo, L.Q.; Costa, S.F.; Lopes, F.; Guerreiro, M.C.; Armentano, L.E.; Pereira, M.N. Rumen morphometrics and the effect of digesta pH and volume on volatile fatty acid absorption. J. Anim. Sci. 2013, 91, 1775–1783. [Google Scholar] [CrossRef]
- Yang, W.; Shen, Z.; Martens, H. An energy-rich diet enhances expression of Na(+)/H(+) exchanger isoform 1 and 3 messenger RNA in rumen epithelium of goat. J. Anim. Sci. 2012, 90, 307–317. [Google Scholar] [CrossRef]
- Li, C.J.; Li, R.W.; Baldwin, R.T.; Blomberg, L.A.; Wu, S.; Li, W. Transcriptomic Sequencing Reveals a Set of Unique Genes Activated by Butyrate-Induced Histone Modification. Gene Regul. Syst. Biol. 2016, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Steele, M.A.; Schiestel, C.; AlZahal, O.; Dionissopoulos, L.; Laarman, A.H.; Matthews, J.C.; McBride, B.W. The periparturient period is associated with structural and transcriptomic adaptations of rumen papillae in dairy cattle. J. Dairy Sci. 2015, 98, 2583–2595. [Google Scholar] [CrossRef] [PubMed]
- Guilloteau, P.; Zabielski, R.; Hammon, H.M.; Metges, C.C. Nutritional programming of gastrointestinal tract development. Is the pig a good model for man? Nutr. Res. Rev. 2010, 23, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Malhi, M.; Gui, H.; Yao, L.; Aschenbach, J.R.; Gabel, G.; Shen, Z. Increased papillae growth and enhanced short-chain fatty acid absorption in the rumen of goats are associated with transient increases in cyclin D1 expression after ruminal butyrate infusion. J. Dairy Sci. 2013, 96, 7603–7616. [Google Scholar] [CrossRef] [PubMed]
- Penner, G.B.; Steele, M.A.; Aschenbach, J.R.; McBride, B.W. Ruminant Nutrition Symposium: Molecular adaptation of ruminal epithelia to highly fermentable diets. J. Anim. Sci. 2011, 89, 1108–1119. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Kuhla, S.; Zitnan, R.; Seyfert, H.M.; Schneider, F.; Hagemeister, H.; Chudy, A.; Lohrke, B.; Blum, J.W.; Hammon, H.M.; et al. Intraruminal infusion of n-butyric acid induces an increase of ruminal papillae size independent of IGF-1 system in castrated bulls. Arch. Anim. Nutr. 2005, 59, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Xu, T.T.; Liu, Y.J.; Zhu, W.Y.; Mao, S.Y. A high-grain diet causes massive disruption of ruminal epithelial tight junctions in goats. Am J. Physiol. Regul. Integr. Comp. Physiol. 2013, 305, R232–R241. [Google Scholar] [CrossRef]
- Xiang, R.; Oddy, V.H.; Archibald, A.L.; Vercoe, P.E.; Dalrymple, B.P. Epithelial, metabolic and innate immunity transcriptomic signatures differentiating the rumen from other sheep and mammalian gastrointestinal tract tissues. PeerJ 2016, 4, e1762. [Google Scholar] [CrossRef]
- Siciliano-Jones, J.; Murphy, M.R. Production of volatile fatty acids in the rumen and cecum-colon of steers as affected by forage:concentrate and forage physical form. J. Dairy Sci. 1989, 72, 485–492. [Google Scholar] [CrossRef]
- Bergman, E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990, 70, 567–590. [Google Scholar] [CrossRef]
- Xiong, Y.; Miyamoto, N.; Shibata, K.; Valasek, M.A.; Motoike, T.; Kedzierski, R.M.; Yanagisawa, M. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc. Natl. Acad. Sci. USA 2004, 101, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Kang, S.G.; Park, J.H.; Yanagisawa, M.; Kim, C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013, 145, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Zhan, K.; Gong, X.; Chen, Y.; Jiang, M.; Yang, T.; Zhao, G. Short-Chain Fatty Acids Regulate the Immune Responses via G Protein-Coupled Receptor 41 in Bovine Rumen Epithelial Cells. Front. Immunol. 2019, 10, 2042. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Cai, Y.; Li, Z.; Chen, X.; Heller, R.; Wang, N.; Wang, Y.; Zhao, C.; Wang, Y.; Xu, H.; et al. Modes of genetic adaptations underlying functional innovations in the rumen. Sci. China Life Sci. 2021, 64, 1–21. [Google Scholar] [CrossRef]
- Wei, C.; Wang, H.; Liu, G.; Zhao, F.; Kijas, J.W.; Ma, Y.; Lu, J.; Zhang, L.; Cao, J.; Wu, M.; et al. Genome-wide analysis reveals adaptation to high altitudes in Tibetan sheep. Sci. Rep. 2016, 6, 26770. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Cui, X.; Wang, Z.; Chang, S.; Wanapat, M.; Yan, T.; Hou, F. Rumen Microbiota of Tibetan Sheep (Ovis aries) Adaptation to Extremely Cold Season on the Qinghai-Tibetan Plateau. Front. Vet. Sci. 2021, 8, 673822. [Google Scholar] [CrossRef]
- Jia, P.; Cui, K.; Ma, T.; Wan, F.; Wang, W.; Yang, D.; Wang, Y.; Guo, B.; Zhao, L.; Diao, Q. Influence of dietary supplementation with Bacillus licheniformis and Saccharomyces cerevisiae as alternatives to monensin on growth performance, antioxidant, immunity, ruminal fermentation and microbial diversity of fattening lambs. Sci. Rep. 2018, 8, 16712. [Google Scholar] [CrossRef]
- Keyt, B.A.; Baliga, R.; Sinclair, A.M.; Carroll, S.F.; Peterson, M.S. Structure, Function, and Therapeutic Use of IgM Antibodies. Antibodies 2020, 9, 53. [Google Scholar] [CrossRef]
- de Sousa-Pereira, P.; Woof, J.M. IgA: Structure, Function, and Developability. Antibodies 2019, 8, 57. [Google Scholar] [CrossRef]
- Russell, J.B.; Rychlik, J.L. Factors that alter rumen microbial ecology. Science 2001, 292, 1119–1122. [Google Scholar] [CrossRef]
- Gao, Y.; Davis, B.; Zhu, W.; Zheng, N.; Meng, D.; Walker, W.A. Short-chain fatty acid butyrate, a breast milk metabolite, enhances immature intestinal barrier function genes in response to inflammation in vitro and in vivo. Am J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G521–G530. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, T.; Xu, S.; Ma, L.; Han, X.; Wang, X.; Zhang, X.; Hu, L.; Zhao, N.; Chen, Y.; et al. Effect of dietary concentrate to forage ratio on growth performance, rumen fermentation and bacterial diversity of Tibetan sheep under barn feeding on the Qinghai-Tibetan plateau. PeerJ 2019, 7, e7462. [Google Scholar] [CrossRef] [PubMed]
- Gui, H.; Shen, Z. Concentrate diet modulation of ruminal genes involved in cell proliferation and apoptosis is related to combined effects of short-chain fatty acid and pH in rumen of goats. J. Dairy Sci. 2016, 99, 6627–6638. [Google Scholar] [CrossRef] [PubMed]
- Greco, G.; Hagen, F.; Meissner, S.; Shen, Z.; Lu, Z.; Amasheh, S.; Aschenbach, J.R. Effect of individual SCFA on the epithelial barrier of sheep rumen under physiological and acidotic luminal pH conditions. J. Anim. Sci. 2018, 96, 126–142. [Google Scholar] [CrossRef]
- Steele, M.A.; Penner, G.B.; Chaucheyras-Durand, F.; Guan, L.L. Development and physiology of the rumen and the lower gut: Targets for improving gut health. J. Dairy Sci. 2016, 99, 4955–4966. [Google Scholar] [CrossRef]
- Baldwin, R.T.; Connor, E.E. Rumen Function and Development. Vet. Clin. N. Am. Food Anim. Pract. 2017, 33, 427–439. [Google Scholar] [CrossRef]
- Baldwin, R.T.; Jesse, B.W. Technical note: Isolation and characterization of sheep ruminal epithelial cells. J. Anim. Sci. 1991, 69, 3603–3609. [Google Scholar] [CrossRef]
- Horbay, R.; Bilyy, R. Mitochondrial dynamics during cell cycling. Apoptosis 2016, 21, 1327–1335. [Google Scholar] [CrossRef]
- Baldwin, R.T.; Wu, S.; Li, W.; Li, C.; Bequette, B.J.; Li, R.W. Quantification of Transcriptome Responses of the Rumen Epithelium to Butyrate Infusion using RNA-seq Technology. Gene Regul. Syst. Biol. 2012, 6, 67–80. [Google Scholar] [CrossRef]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef]
- Anavi, S.; Tirosh, O. iNOS as a metabolic enzyme under stress conditions. Free Radic. Biol. Med. 2020, 146, 16–35. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Yang, Y.; Wang, Z.; Yang, X.; Gao, Y.; Zhao, Y.; Ge, W.; Liu, J.; Xu, X.; Guan, W.; et al. PER1 interaction with GPX1 regulates metabolic homeostasis under oxidative stress. Redox Biol. 2020, 37, 101694. [Google Scholar] [CrossRef] [PubMed]
- Mirzalieva, O.; Juncker, M.; Schwartzenburg, J.; Desai, S. ISG15 and ISGylation in Human Diseases. Cells 2022, 11, 538. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, N.A.; Paparisto, E.; Barr, S.D.; Spratt, D.E. HERC5 and the ISGylation Pathway: Critical Modulators of the Antiviral Immune Response. Viruses 2021, 13, 1102. [Google Scholar] [CrossRef]
- Thorson, W.; Diaz-Horta, O.; Foster, J.N.; Spiliopoulos, M.; Quintero, R.; Farooq, A.; Blanton, S.; Tekin, M. De novo ACTG2 mutations cause congenital distended bladder, microcolon, and intestinal hypoperistalsis. Hum. Genet. 2014, 133, 737–742. [Google Scholar] [CrossRef]
- Wang, B.; Wu, L.; Chen, J.; Dong, L.; Chen, C.; Wen, Z.; Hu, J.; Fleming, I.; Wang, D.W. Metabolism pathways of arachidonic acids: Mechanisms and potential therapeutic targets. Signal Transduct. Target Ther. 2021, 6, 94. [Google Scholar] [CrossRef]
- Hirabayashi, S.; Tajima, M.; Yao, I.; Nishimura, W.; Mori, H.; Hata, Y. JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol. Cell. Biol. 2003, 23, 4267–4282. [Google Scholar] [CrossRef]
- Lane, M.A.; Baldwin, R.T.; Jesse, B.W. Developmental changes in ketogenic enzyme gene expression during sheep rumen development. J. Anim. Sci. 2002, 80, 1538–1544. [Google Scholar] [CrossRef]
- Connor, E.E.; Li, R.W.; Baldwin, R.L.; Li, C. Gene expression in the digestive tissues of ruminants and their relationships with feeding and digestive processes. Animal 2010, 4, 993–1007. [Google Scholar] [CrossRef]
- Zhao, K.; Chen, Y.H.; Penner, G.B.; Oba, M.; Guan, L.L. Transcriptome analysis of ruminal epithelia revealed potential regulatory mechanisms involved in host adaptation to gradual high fermentable dietary transition in beef cattle. BMC Genom. 2017, 18, 976. [Google Scholar] [CrossRef]
- Schwartz, N.; Verma, A.; Bivens, C.B.; Schwartz, Z.; Boyan, B.D. Rapid steroid hormone actions via membrane receptors. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 2289–2298. [Google Scholar] [CrossRef] [PubMed]
- Dedyukhina, E.G.; Chistyakova, T.I.; Mironov, A.A.; Kamzolova, S.V.; Minkevich, I.G.; Vainshtein, M.B. The effect of pH, aeration, and temperature on arachidonic acid synthesis by Mortierella alpina. Prikl. Biokhim. Mikrobiol. 2015, 51, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Baumann, C.A.; Ribon, V.; Kanzaki, M.; Thurmond, D.C.; Mora, S.; Shigematsu, S.; Bickel, P.E.; Pessin, J.E.; Saltiel, A.R. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature 2000, 407, 202–207. [Google Scholar] [CrossRef] [PubMed]
- CHANEY, A.L.; MARBACH, E.P. Modified reagents for determination of urea and ammonia. Clin. Chem. 1962, 8, 130–132. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, W.; Zeng, P.; Wang, J.; Geng, B.; Yang, J.; Cui, Q. LncTar: A tool for predicting the RNA targets of long noncoding RNAs. Brief. Bioinform. 2015, 16, 806–812. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]

| 4 Months | 1.5 Years | 3.5 Years | 6 Years | p_Value | |
|---|---|---|---|---|---|
| Acetic acid (mmol/L) | 119.66 ± 1.68 a | 30.72 ± 5.54 c | 69.08 ± 4.52 b | 19.30 ± 3.15 d | <0.00 |
| Propionic acid (mmol/L) | 24.63 ± 0.68 a | 3.26 ± 0.87 c | 10.90 ± 1.85 b | 2.70 ± 0.25 c | <0.00 |
| Isobutyric acid (mmol/L) | 1.90 ± 0.22 a | 1.14 ± 0.13 c | 1.53 ± 0.20 b | 0.79 ± 0.07 d | <0.00 |
| Butyric acid (mmol/L) | 18.82 ± 0.52 a | 2.00 ± 0.39 c | 5.91 ± 0.93 b | 1.49 ± 0.32 c | <0.00 |
| Isovaleric acid (mmol/L) | 2.24 ± 0.42 a | 1.27 ± 0.27 c | 1.81 ± 0.26 b | 1.27 ± 0.25 c | <0.00 |
| Pentanoic acid (mmol/L) | 0.09 ± 0.02 bc | 0.05 ± 0.03 c | 0.16 ± 0.09 b | 0.35 ± 0.04 a | <0.00 |
| Total VFAs (mmol/L) | 167.33 ± 1.39 a | 38.43 ± 5.36 c | 89.39 ± 7.18 b | 25.89 ± 3.43 d | <0.00 |
| A/P | 4.86 ± 0.18 b | 10.33 ± 4.86 a | 6.45 ± 0.90 b | 7.14 ± 0.83 ab | 0.023 |
| NH3-N (mg/100 mL) | 6.26 ± 0.04 d | 7.23 ± 0.05 c | 12.26 ± 0.10 a | 7.45 ± 0.04 b | <0.00 |
| 4 Months | 1.5 Years | 3.5 Years | 6 Years | p-Value | |
|---|---|---|---|---|---|
| ML | 1644.00 ± 73.60 d | 1925.24 ± 56.55 c | 2067.68 ± 133.51 b | 2293.48 ± 126.21 a | <0.00 |
| Lp | 1219.98 ± 180.70 b | 1846.56 ± 44.77 a | 2020.96 ± 215.65 a | 1962.40 ± 111.19 a | <0.00 |
| Wp | 368.68 ± 13.85 c | 400.46 ± 22.27 b | 403.36 ± 13.73 b | 536.98 ± 25.06 a | <0.00 |
| SC | 58.72 ± 3.07 c | 44.16 ± 1.41 b | 43.42 ± 2.07 b | 21.52 ± 2.71 a | <0.00 |
| SG | 27.22 ± 3.83 ab | 14.86 ± 0.79 c | 23.44 ± 1.27 b | 29.68 ± 4.80 a | <0.00 |
| SS | 54.82 ± 4.39 | 50.32 ± 4.74 | 50.26 ± 3.60 | 52.72 ± 3.32 | 0.265 |
| BL | 19.44 ± 1.22 b | 21.34 ± 2.48 b | 18.72 ± 1.65 b | 24.62 ± 2.80 a | 0.002 |
| Nutrients (DM Basis) | Dominant Forage Species | |
|---|---|---|
| CP (%) | 10.06 | Poa pratensis L |
| EE (%) | 3.77 | |
| Ash (%) | 4.55 | Elymus nutans Griseb |
| NDF (%) | 70.11 | |
| ADF (%) | 36.17 | Agropyron cristatum (L.) Gaertn |
| HCEL (%) | 33.94 | |
| Ca (%) | 11.50 | Stipa aliena Keng |
| P (%) | 0.65 | |
| Aboveground biomass (g/m2) | 343.52 | Potentilla bifurca Linn. |
| Grass height (cm) | 16.12 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sha, Y.; He, Y.; Liu, X.; Zhao, S.; Hu, J.; Wang, J.; Li, S.; Li, W.; Shi, B.; Hao, Z. Rumen Epithelial Development- and Metabolism-Related Genes Regulate Their Micromorphology and VFAs Mediating Plateau Adaptability at Different Ages in Tibetan Sheep. Int. J. Mol. Sci. 2022, 23, 16078. https://doi.org/10.3390/ijms232416078
Sha Y, He Y, Liu X, Zhao S, Hu J, Wang J, Li S, Li W, Shi B, Hao Z. Rumen Epithelial Development- and Metabolism-Related Genes Regulate Their Micromorphology and VFAs Mediating Plateau Adaptability at Different Ages in Tibetan Sheep. International Journal of Molecular Sciences. 2022; 23(24):16078. https://doi.org/10.3390/ijms232416078
Chicago/Turabian StyleSha, Yuzhu, Yanyu He, Xiu Liu, Shengguo Zhao, Jiang Hu, Jiqing Wang, Shaobin Li, Wenhao Li, Bingang Shi, and Zhiyun Hao. 2022. "Rumen Epithelial Development- and Metabolism-Related Genes Regulate Their Micromorphology and VFAs Mediating Plateau Adaptability at Different Ages in Tibetan Sheep" International Journal of Molecular Sciences 23, no. 24: 16078. https://doi.org/10.3390/ijms232416078
APA StyleSha, Y., He, Y., Liu, X., Zhao, S., Hu, J., Wang, J., Li, S., Li, W., Shi, B., & Hao, Z. (2022). Rumen Epithelial Development- and Metabolism-Related Genes Regulate Their Micromorphology and VFAs Mediating Plateau Adaptability at Different Ages in Tibetan Sheep. International Journal of Molecular Sciences, 23(24), 16078. https://doi.org/10.3390/ijms232416078





