Substrate Recognition Properties from an Intermediate Structural State of the UreA Transporter
Abstract
1. Introduction

2. Results
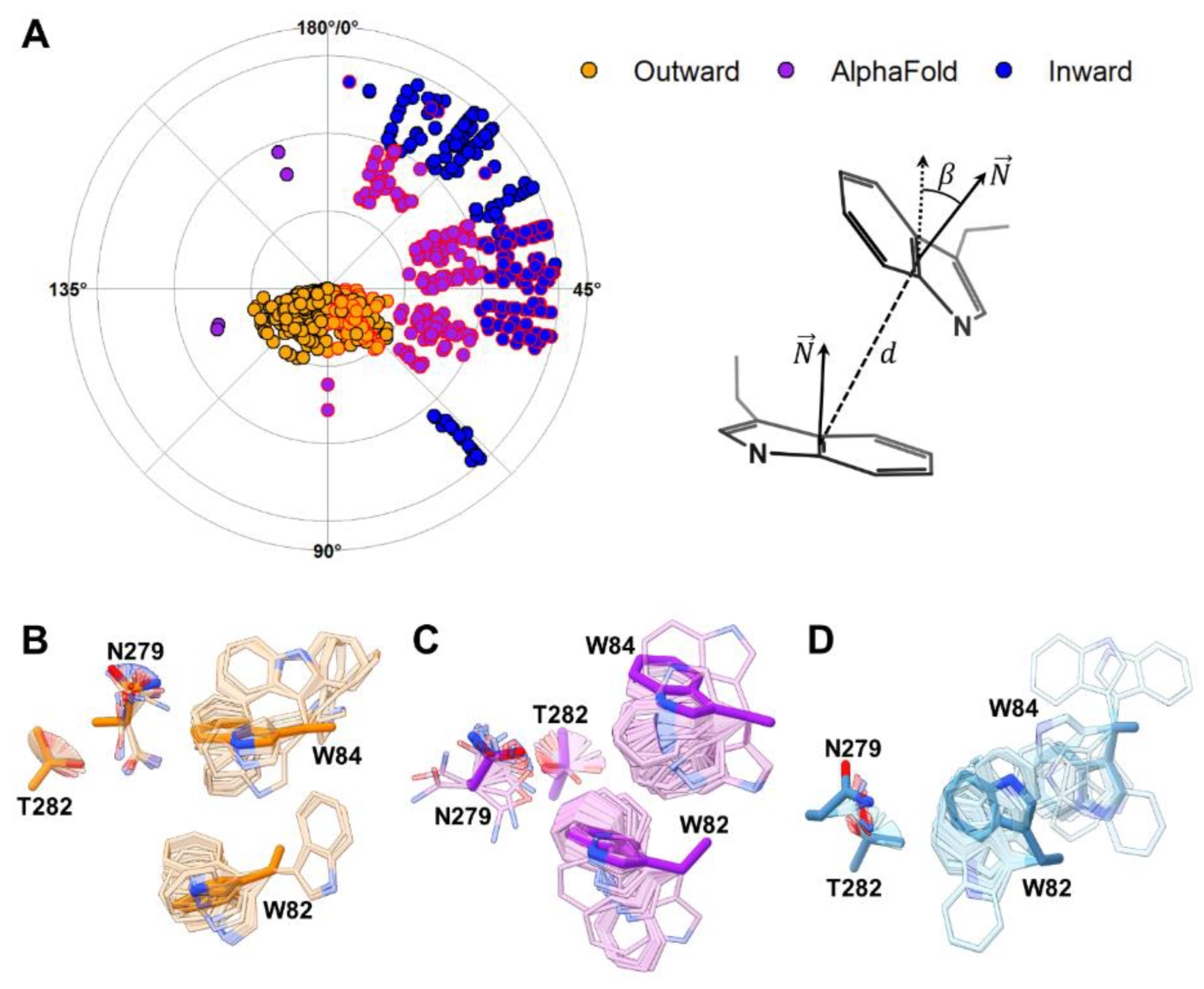
2.1. Characterization of a Theoretical Intermediate State of UreA and Its Binding Site
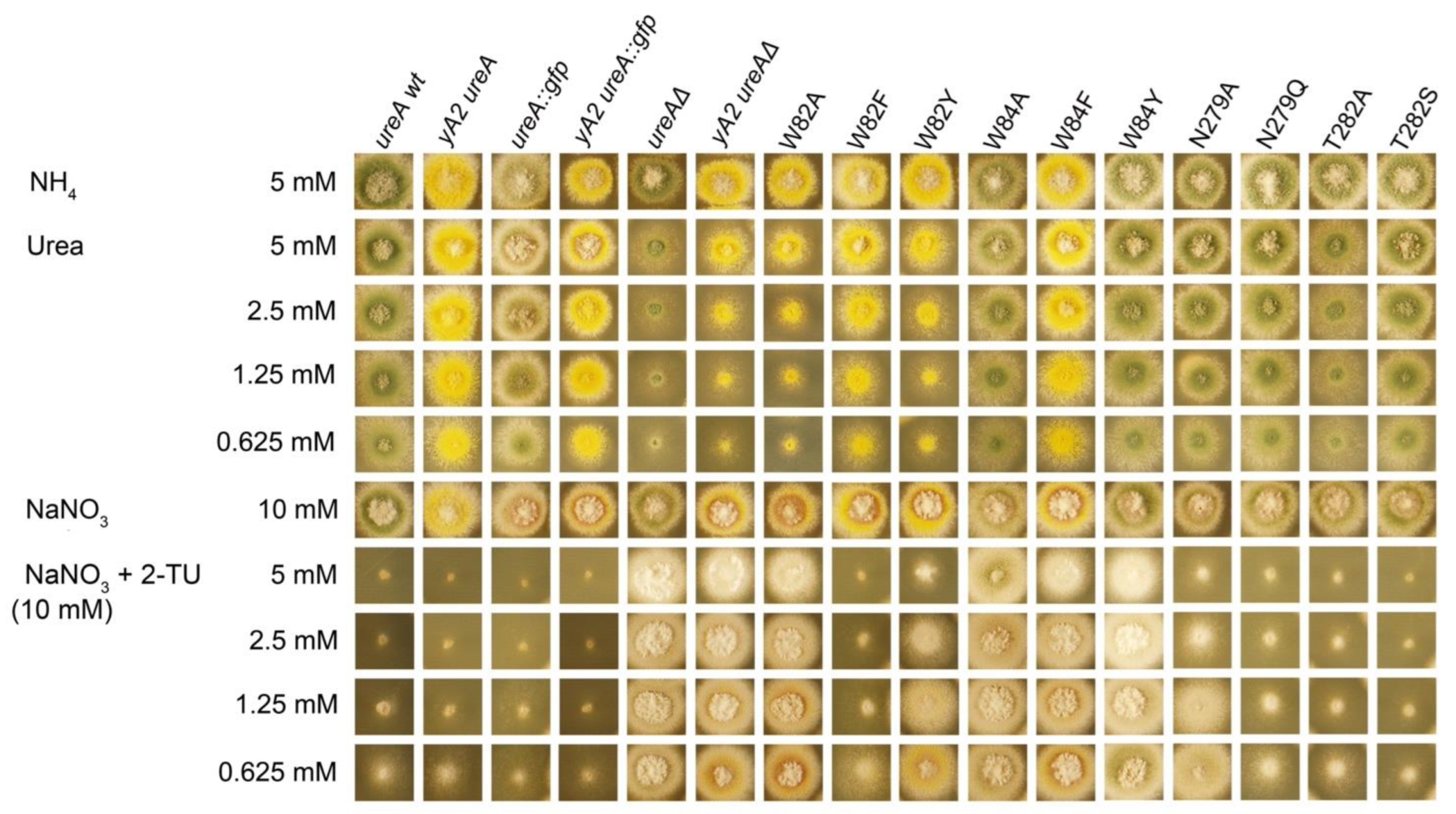
2.2. Site-Directed Mutagenesis of Identified Residues in the Putative Binding Site
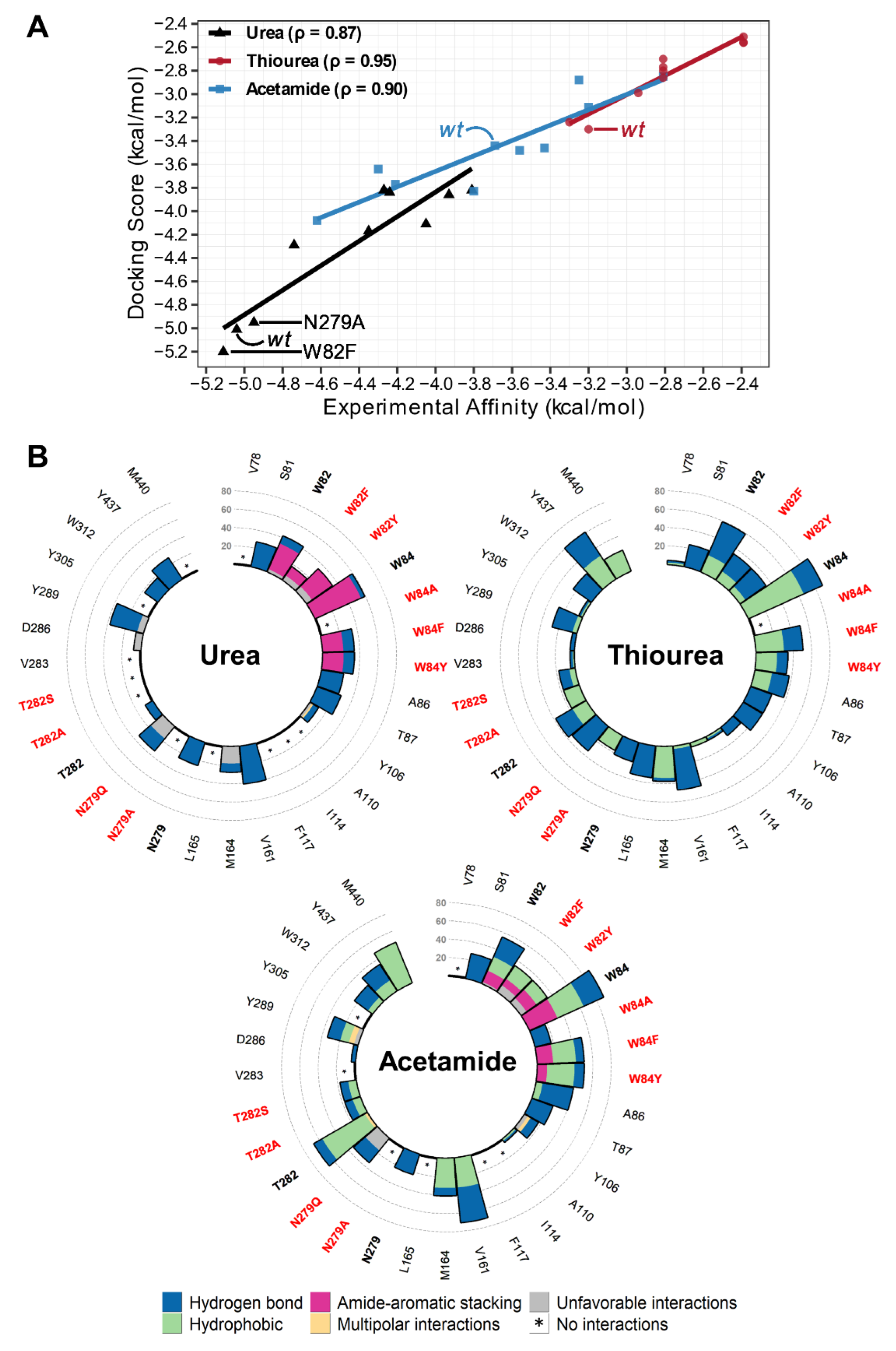
2.3. Insights into Ligand Binding by the Intermediate State of UreA
3. Discussion
4. Materials and Methods
4.1. Structure Prediction
4.2. Molecular Docking
4.3. Structural Analysis
4.4. A. nidulans Strains, Media and Transformation Procedures
4.5. Construction of ureA Mutants by Site-Directed Mutagenesis
4.6. [14C]-Urea Uptake Measurements
4.7. Protein Extraction and Western Blot Assays
4.8. Epifluorescence Microscopy
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Omara, P.; Aula, L.; Oyebiyi, F.; Raun, W.R. World Cereal Nitrogen Use Efficiency Trends: Review and Current Knowledge. Agrosystems Geosci. Environ. 2019, 2, 1–8. [Google Scholar] [CrossRef]
- International Fertilizer Association. Available online: https://www.ifastat.org/ (accessed on 9 November 2022).
- Beier, M.P.; Kojima, S. The Function of High-Affinity Urea Transporters in Nitrogen-Deficient Conditions. Physiol. Plant. 2021, 171, 802–808. [Google Scholar] [CrossRef] [PubMed]
- Abreu, C.; Sanguinetti, M.; Amillis, S.; Ramon, A. UreA, the Major Urea/H+ Symporter in Aspergillus nidulans. Fungal Genet. Biol. 2010, 47, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Levin, E.J.; Quick, M.; Zhou, M. Crystal Structure of a Bacterial Homologue of the Kidney Urea Transporter. Nature 2009, 462, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Levin, E.J.; Cao, Y.; Enkavi, G.; Quick, M.; Pan, Y.; Tajkhorshid, E.; Zhou, M. Structure and Permeation Mechanism of a Mammalian Urea Transporter. Proc. Natl. Acad. Sci. USA 2012, 109, 11194–11199. [Google Scholar] [CrossRef] [PubMed]
- Sands, J.M. Mammalian Urea Transporters. Annu. Rev. Physiol. 2003, 65, 543–566. [Google Scholar] [CrossRef] [PubMed]
- Sachs, G.; Kraut, J.A.; Wen, Y.; Feng, J.; Scott, D.R. Urea Transport in Bacteria: Acid Acclimation by Gastric Helicobacter Spp. J. Membr. Biol. 2006, 212, 71–82. [Google Scholar] [CrossRef]
- ElBerry, H.M.; Majumdar, M.L.; Cunningham, T.S.; Sumrada, R.A.; Cooper, T.G. Regulation of the Urea Active Transporter Gene (DUR3) in Saccharomyces Cerevisiae. J. Bacteriol. 1993, 175, 4688–4698. [Google Scholar] [CrossRef]
- Morel, M.; Jacob, C.; Fitz, M.; Wipf, D.; Chalot, M.; Brun, A. Characterization and Regulation of PiDur3, a Permease Involved in the Acquisition of Urea by the Ectomycorrhizal Fungus Paxillus Involutus. Fungal Genet. Biol. 2008, 45, 912–921. [Google Scholar] [CrossRef]
- Navarathna, D.H.M.L.P.; Das, A.; Morschhäuser, J.; Nickerson, K.W.; Roberts, D.D. Dur3 Is the Major Urea Transporter in Candida Albicans and Is Co-Regulated with the Urea Amidolyase Dur1,2. Microbiology 2011, 157, 270–279. [Google Scholar] [CrossRef]
- Mérigout, P.; Lelandais, M.; Bitton, F.; Renou, J.P.; Briand, X.; Meyer, C.; Daniel-Vedele, F. Physiological and Transcriptomic Aspects of Urea Uptake and Assimilation in Arabidopsis Plants. Plant Physiol. 2008, 147, 1225–1238. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.W.; Sun, A.L.; Li, D.Q.; Athman, A.; Gilliham, M.; Liu, L.H. Molecular Identification and Functional Analysis of a Maize (Zea Mays) DUR3 Homolog That Transports Urea with High Affinity. Planta 2015, 241, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.H.; Köhler, B.; Cao, F.Q.; Liu, G.W.; Gong, Y.Y.; Sheng, S.; Song, Q.C.; Cheng, X.Y.; Garnett, T.; Okamoto, M.; et al. Rice DUR3 Mediates High-Affinity Urea Transport and Plays an Effective Role in Improvement of Urea Acquisition and Utilization When Expressed in Arabidopsis. New Phytol. 2012, 193, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Diallinas, G.; Martzoukou, O. Transporter Membrane Traffic and Function: Lessons from a Mould. FEBS J. 2019, 286, 4861–4875. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.; Amillis, S.; Pantano, S.; Scazzocchio, C.; Ramón, A. Modelling and Mutational Analysis of Aspergillus nidulans UreA, a Member of the Subfamily of Urea/H+ Transporters in Fungi and Plants. Open Biol. 2014, 4, 140070. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sanguinetti, M.; Iriarte, A.; Amillis, S.; Marín, M.; Musto, H.; Ramón, A. A Pair of Non-Optimal Codons Are Necessary for the Correct Biosynthesis of the Aspergillus nidulans Urea Transporter, UreA. R. Soc. Open Sci. 2019, 6, 190773. [Google Scholar] [CrossRef]
- Pateman, J.A.; Dunn, E.; Mackay, E.M. Urea and Thiourea Transport in Aspergillus nidulans. Biochem. Genet. 1982, 20, 777–790. [Google Scholar] [CrossRef]
- Tange, Y.; Niwa, O. Identification of the Ure1+ Gene Encoding Urease in Fission Yeast. Curr. Genet. 1997, 32, 244–246. [Google Scholar] [CrossRef]
- Witte, C.P. Urea Metabolism in Plants. Plant Sci. 2011, 180, 431–438. [Google Scholar] [CrossRef]
- Mackay, E.M.; Pateman, J.A. The Regulation of Urease Activity in Aspergillus nidulans. Biochem. Genet. 1982, 20, 763–776. [Google Scholar] [CrossRef]
- Faham, S.; Watanabe, A.; Besserer, G.M.; Cascio, D.; Specht, A.; Hirayama, B.A.; Wright, E.M.; Abramson, J. The Crystal Structure of a Sodium Galactose Transporter Reveals Mechanistic Insights into Na+/Sugar Symport. Science 2008, 321, 810–814. [Google Scholar] [CrossRef] [PubMed]
- Weyand, S.; Shimamura, T.; Yajima, S.; Suzuki, S.N.I.; Mirza, O.; Krusong, K.; Carpenter, E.P.; Rutherford, N.G.; Hadden, J.M.; O’Reilly, J.; et al. Structure and Molecular Mechanism of a Nucleobase-Cation-Symport-1 Family Transporter. Science 2008, 322, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. Common Folds and Transport Mechanisms of Secondary Active Transporters. Annu. Rev. Biophys. 2013, 42, 51–72. [Google Scholar] [CrossRef] [PubMed]
- Diallinas, G. Biochemistry: An Almost-Complete Movie. Science 2008, 322, 1644–1645. [Google Scholar] [CrossRef]
- Forrest, L.R.; Zhang, Y.W.; Jacobs, M.T.; Gesmonde, J.; Xie, L.; Honig, B.H.; Rudnick, G. Mechanism for Alternating Access in Neurotransmitter Transporters. Proc. Natl. Acad. Sci. USA 2008, 105, 10338–10343. [Google Scholar] [CrossRef]
- Shimamura, T.; Weyand, S.; Beckstein, O.; Rutherford, N.G.; Hadden, J.M.; Sharpies, D.; Sansom, M.S.P.; Iwata, S.; Henderson, P.J.F.; Cameron, A.D. Molecular Basis of Alternating Access Membrane Transport by the Sodium-Hydantoin Transporter Mhp1. Science 2010, 328, 470–473. [Google Scholar] [CrossRef]
- Drew, D.; Boudker, O. Shared Molecular Mechanisms of Membrane Transporters. Annu. Rev. Biochem. 2016, 85, 543–572. [Google Scholar] [CrossRef]
- Niu, Y.; Cui, W.; Liu, R.; Wang, S.; Ke, H.; Lei, X.; Chen, L. Structural Mechanism of SGLT1 Inhibitors. Nat. Commun. 2022, 13, 6440. [Google Scholar] [CrossRef]
- Paz, A.; Claxton, D.P.; Kumar, J.P.; Kazmier, K.; Bisignano, P.; Sharma, S.; Nolte, S.A.; Liwag, T.M.; Nayak, V.; Wright, E.M. Conformational Transitions of the Sodium-Dependent Sugar Transporter, VSGLT. Proc. Natl. Acad. Sci. USA 2018, 115, E2742–E2751. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Jumper, J.; Hassabis, D. Protein Structure Predictions to Atomic Accuracy with AlphaFold. Nat. Methods 2022, 19, 11–12. [Google Scholar] [CrossRef] [PubMed]
- del Alamo, D.; Govaerts, C.; Mchaourab, H.S. AlphaFold2 Predicts the Inward-Facing Conformation of the Multidrug Transporter LmrP. Proteins Struct. Funct. Bioinform. 2021, 89, 1226–1228. [Google Scholar] [CrossRef] [PubMed]
- Del Alamo, D.; Sala, D.; McHaourab, H.S.; Meiler, J. Sampling Alternative Conformational States of Transporters and Receptors with AlphaFold2. Elife 2022, 11, e75751. [Google Scholar] [CrossRef] [PubMed]
- Heo, L.; Feig, M. Multi-State Modeling of G-Protein Coupled Receptors at Experimental Accuracy. Proteins Struct. Funct. Bioinform. 2022, 90, 1873–1885. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Porter, L.L. AlphaFold2 Fails to Predict Protein Fold Switching. Protein Sci. 2022, 31, e4353. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Fassio, A.V.; Shub, L.; Ponzoni, L.; McKinley, J.; O’Meara, M.J.; Ferreira, R.S.; Keiser, M.J.; de Melo Minardi, R.C. Prioritizing Virtual Screening with Interpretable Interaction Fingerprints. J. Chem. Inf. Model. 2022, 62, 4300–4318. [Google Scholar] [CrossRef]
- Mitrovic, D.; McComas, S.E.; Alleva, C.; Bonaccorsi, M.; Drew, D.; Delemotte, L. Reconstructing the Transport Cycle in the Sugar Porter Superfamily Using Coevolution-Powered Machine Learning. bioRxiv 2022. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Zhu, Z.; Song, C. Exploring the Alternative Conformation of a Known Protein Structure Based on Contact Map Prediction. bioRxiv 2022. [Google Scholar] [CrossRef]
- Tong, A.B.; Burch, J.D.; McKay, D.; Bustamante, C.; Crackower, M.A.; Wu, H. Could AlphaFold Revolutionize Chemical Therapeutics? Nat. Struct. Mol. Biol. 2021, 28, 771–772. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.M.; Laskowski, R.A.; Borkakoti, N. AlphaFold Heralds a Data-Driven Revolution in Biology and Medicine. Nat. Med. 2021, 27, 1666–1669. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Thornton, J.M. The Impact of AlphaFold2 One Year On. Nat. Methods 2022, 19, 15–20. [Google Scholar] [CrossRef]
- Huang, N.; Shoichet, B.K. Exploiting Ordered Waters in Molecular Docking. J. Med. Chem. 2008, 51, 4862–4865. [Google Scholar] [CrossRef]
- Barillari, C.; Taylor, J.; Viner, R.; Essex, J.W. Classification of Water Molecules in Protein Binding Sites. J. Am. Chem. Soc. 2007, 129, 2577–2587. [Google Scholar] [CrossRef]
- Levin, E.J.; Zhou, M. Structure of Urea Transporters. In Sub-Cellular Biochemistry; Yang, B., Sands, J.M., Eds.; Springer: Dordrecht, The Netherlands, 2014; Volume 73, pp. 65–78. ISBN 978-94-017-9343-8. [Google Scholar]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Quiroga, R.; Villarreal, M.A. Vinardo: A Scoring Function Based on Autodock Vina Improves Scoring, Docking, and Virtual Screening. PLoS ONE 2016, 11, e0155183. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Core R Team. A Language and Environment for Statistical Computing. R Found. Stat. Comput. 2019, 2. Available online: https://www.R--project.org (accessed on 9 November 2022).
- Scazzocchio, C.; Arst, H.N. The Nature of an Initiator Constitutive Mutation in Aspergillus nidulans. Nature 1978, 274, 177–179. [Google Scholar] [CrossRef] [PubMed]
- Cove, D.J. The Induction and Repression of Nitrate Reductase in the Fungus Aspergillus nidulans. Biochim. Biophys. Acta 1966, 113, 51–56. [Google Scholar] [CrossRef]
- Oakley, B.R.; Szewczyk, E.; Nayak, T.; Oakley, C.E.; Edgerton, H.; Xiong, Y.; Taheri-Talesh, N.; Osmani, S.A. Fusion Pcr and Gene Targeting in Aspergillus nidulans. Nat. Protoc. 2006, 1, 3111–3120. [Google Scholar] [CrossRef]
- Krypotou, E.; Diallinas, G. Transport Assays in Filamentous Fungi: Kinetic Characterization of the UapC Purine Transporter of Aspergillus nidulans. Fungal Genet. Biol. 2014, 63, 1–8. [Google Scholar] [CrossRef]
- Yung-Chi, C.; Prusoff, W.H. Relationship between the Inhibition Constant (KI) and the Concentration of Inhibitor Which Causes 50 per Cent Inhibition (I50) of an Enzymatic Reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar] [CrossRef]
- Apostolaki, A.; Harispe, L.; Calcagno-Pizarelli, A.M.; Vangelatos, I.; Sophianopoulou, V.; Arst, H.N.; Peñalva, M.A.; Amillis, S.; Scazzocchio, C. Aspergillus nidulans CkiA Is an Essential Casein Kinase I Required for Delivery of Amino Acid Transporters to the Plasma Membrane. Mol. Microbiol. 2012, 84, 530–549. [Google Scholar] [CrossRef]
- Edelstein, A.D.; Tsuchida, M.A.; Amodaj, N.; Pinkard, H.; Vale, R.D.; Stuurman, N. Advanced Methods of Microscope Control Using ΜManager Software. J. Biol. Methods 2014, 1, e10. [Google Scholar] [CrossRef]

| Allele | Location in Protein | Vo (%) | Urea Km (µM) | Thiourea Ki (µM) | Acetamide Ki (µM) |
|---|---|---|---|---|---|
| wt | 100 | 26 ± 2 | 528 ± 42 | 237 ± 14 | |
| W82F * | TMS2 | 50 ± 8 | 23 ± 5 | 447 ± 24 | 198 ± 12 |
| W82Y | TMS 2 | 25 ± 2 | 42 ± 5 | >1000 | 101 ± 16 |
| W84A | TMS 2 | 20 ± 2 | 193 ± 12 | >2000 | 489 ± 39 |
| W84F | TMS 2 | 15 ± 2 | 96 ±13 | >2000 | 524 ± 42 |
| W84Y | TMS 2 | 46 ± 4 | 159 ± 11 | >2000 | >1000 |
| N279A | TMS 7 | 100 ± 5 | 30 ± 3 | >1000 | 51 ± 5 |
| N279Q | TMS 7 | 70 ± 3 | 91 ± 6 | 813 ± 59 | 87 ± 9 |
| T282A | TMS 7 | 14 ± 2 | 80 ± 27 | >1000 | 361 ± 85 |
| T282S | TMS 7 | 65 ± 3 | 130 ± 22 | >1000 | 290 ± 12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanguinetti, M.; Silva Santos, L.H.; Dourron, J.; Alamón, C.; Idiarte, J.; Amillis, S.; Pantano, S.; Ramón, A. Substrate Recognition Properties from an Intermediate Structural State of the UreA Transporter. Int. J. Mol. Sci. 2022, 23, 16039. https://doi.org/10.3390/ijms232416039
Sanguinetti M, Silva Santos LH, Dourron J, Alamón C, Idiarte J, Amillis S, Pantano S, Ramón A. Substrate Recognition Properties from an Intermediate Structural State of the UreA Transporter. International Journal of Molecular Sciences. 2022; 23(24):16039. https://doi.org/10.3390/ijms232416039
Chicago/Turabian StyleSanguinetti, Manuel, Lucianna Helene Silva Santos, Juliette Dourron, Catalina Alamón, Juan Idiarte, Sotiris Amillis, Sergio Pantano, and Ana Ramón. 2022. "Substrate Recognition Properties from an Intermediate Structural State of the UreA Transporter" International Journal of Molecular Sciences 23, no. 24: 16039. https://doi.org/10.3390/ijms232416039
APA StyleSanguinetti, M., Silva Santos, L. H., Dourron, J., Alamón, C., Idiarte, J., Amillis, S., Pantano, S., & Ramón, A. (2022). Substrate Recognition Properties from an Intermediate Structural State of the UreA Transporter. International Journal of Molecular Sciences, 23(24), 16039. https://doi.org/10.3390/ijms232416039







