The Modular Architecture of Metallothioneins Facilitates Domain Rearrangements and Contributes to Their Evolvability in Metal-Accumulating Mollusks
Abstract
1. Introduction
2. Results
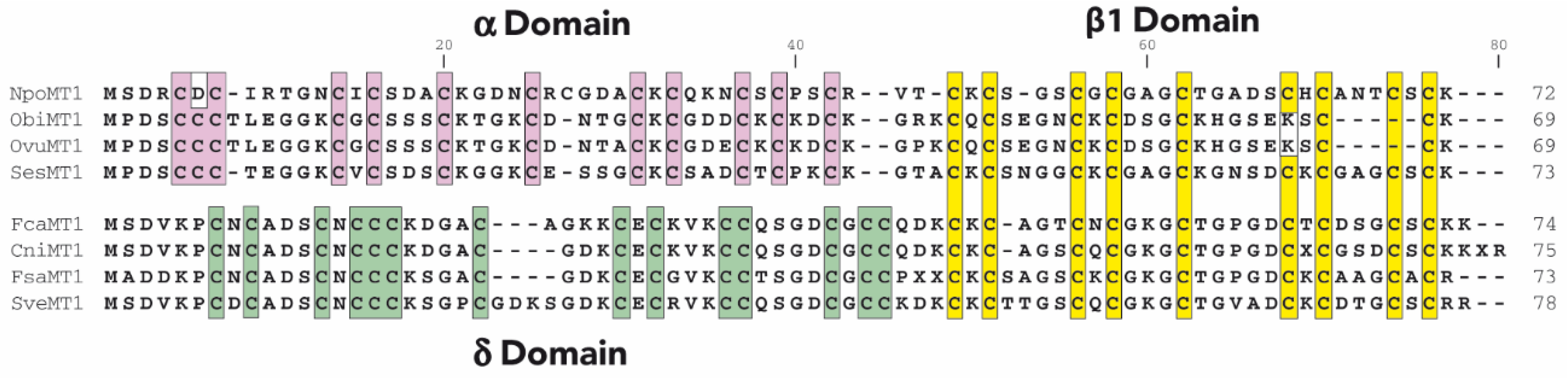
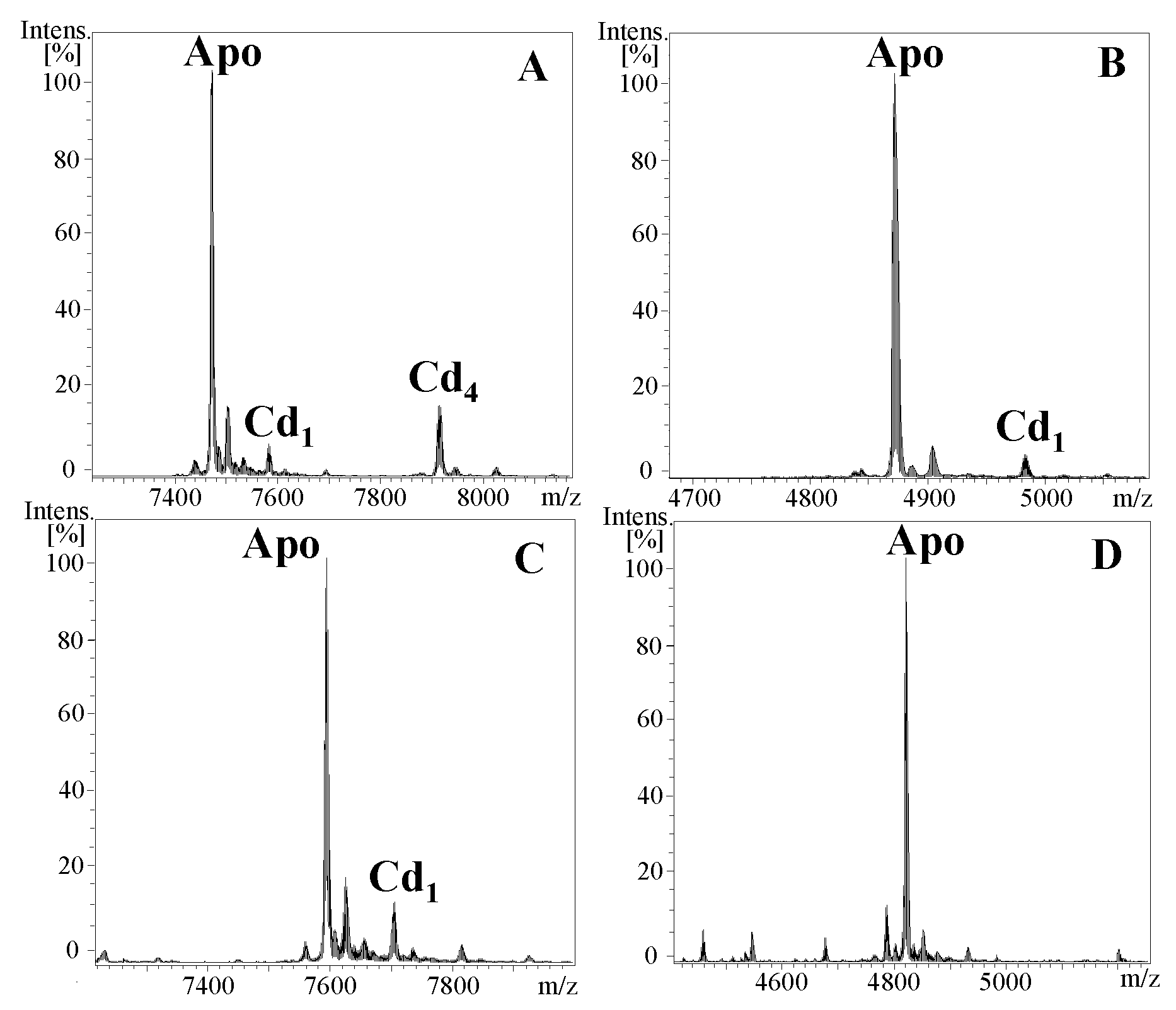
2.1. Characterization of Cephalopoda and Caudofoveata MTs: NpoMT1 and FcaMT1
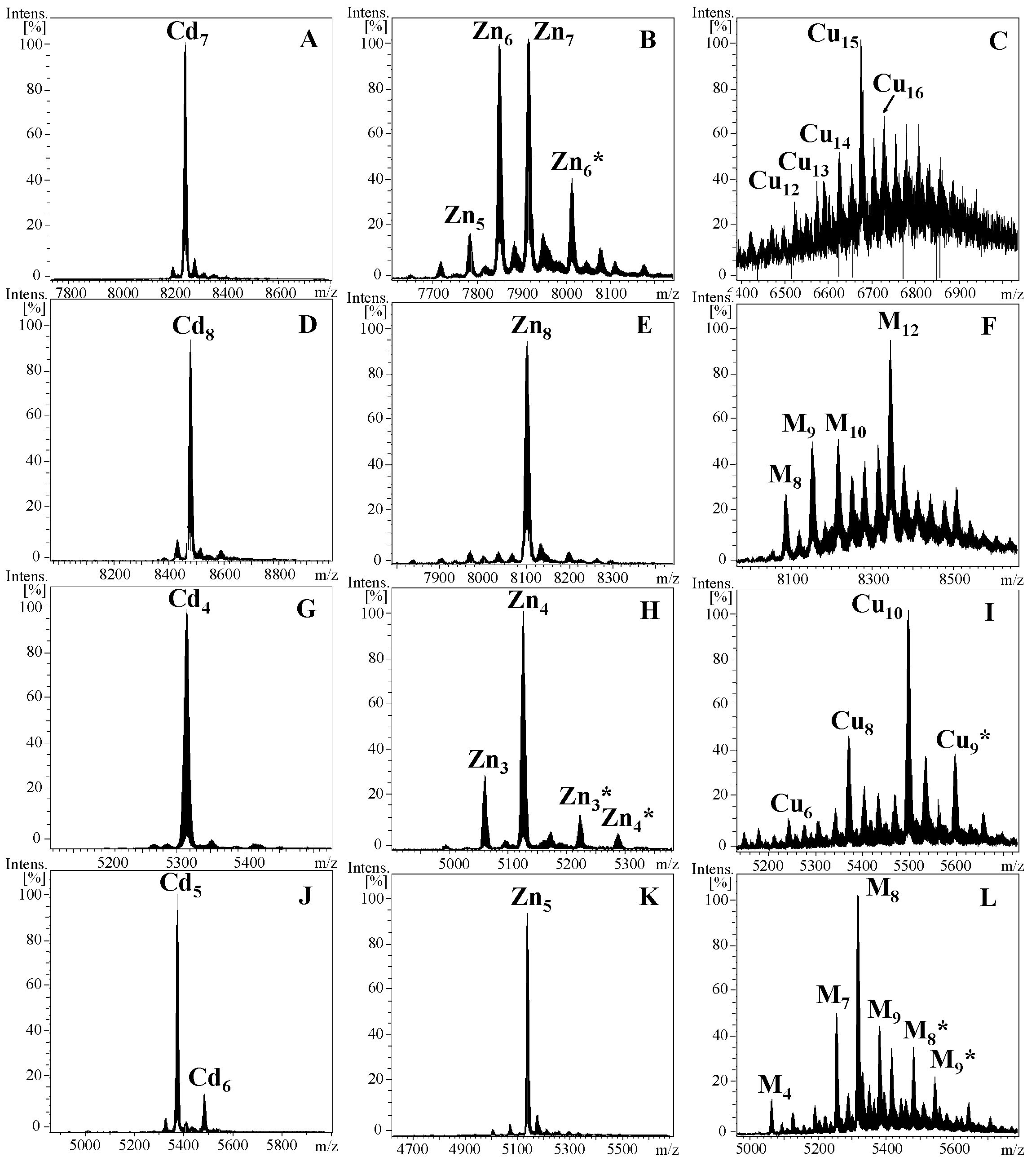
2.2. Characterization of the α and δ Domains of Cephalopoda and Caudofoveata MTs
3. Discussion
3.1. Cephalopoda NpoMT1 and Caudofoveata FcaMT1, and the α and δ Domains
3.2. Biological Implications of the Cephalopoda and Caudofoveata MTs
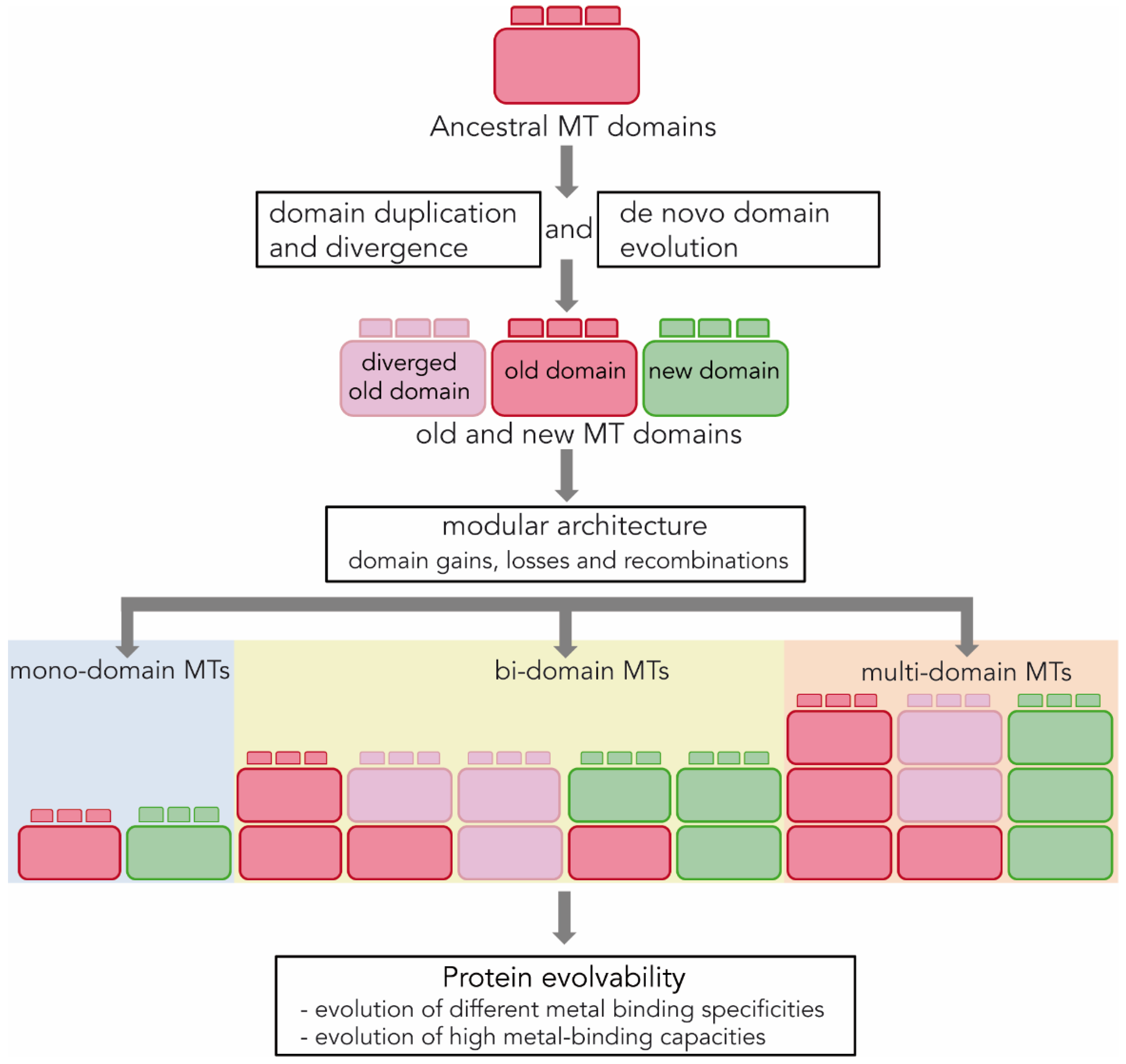
3.3. Origin and Evolution of MT Domains
3.4. MT Modularity and Evolvability
4. Materials and Methods
4.1. Production and Purification of Recombinant Metal-MT Complexes
4.2. Analysis of Metal-MT Complexes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wetlaufer, D.B. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc. Natl. Acad. Sci. USA 1973, 70, 697–701. [Google Scholar] [CrossRef] [PubMed]
- Janin, J.; Chothia, C. Domains in proteins: Definitions, location, and structural principles. Methods Enzym. 1985, 115, 420–430. [Google Scholar] [CrossRef]
- Heringa, J. Protein domains. In Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2005. [Google Scholar]
- Apic, G.; Gough, J.; Teichmann, S.A. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 2001, 310, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Levitt, M. Nature of the protein universe. Proc. Natl. Acad. Sci. USA 2009, 106, 11079–11084. [Google Scholar] [CrossRef]
- Wang, M.; Caetano-Anolles, G. The evolutionary mechanics of domain organization in proteomes and the rise of modularity in the protein world. Structure 2009, 17, 66–78. [Google Scholar] [CrossRef]
- Bjorklund, A.K.; Ekman, D.; Light, S.; Frey-Skott, J.; Elofsson, A. Domain rearrangements in protein evolution. J. Mol. Biol. 2005, 353, 911–923. [Google Scholar] [CrossRef]
- Ekman, D.; Björklund, A.K.; Frey-Skött, J.; Elofsson, A. Multi-domain proteins in the three kingdoms of life: Orphan domains and other unassigned regions. J. Mol. Biol. 2005, 348, 231–243. [Google Scholar] [CrossRef]
- Kummerfeld, S.K.; Teichmann, S.A. Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet. 2005, 21, 25–30. [Google Scholar] [CrossRef]
- Fong, J.H.; Geer, L.Y.; Panchenko, A.R.; Bryant, S.H. Modeling the evolution of protein domain architectures using maximum parsimony. J. Mol. Biol. 2007, 366, 307–315. [Google Scholar] [CrossRef]
- Dohmen, E.; Klasberg, S.; Bornberg-Bauer, E.; Perrey, S.; Kemena, C. The modular nature of protein evolution: Domain rearrangement rates across eukaryotic life. BMC Evol. Biol. 2020, 20, 30. [Google Scholar] [CrossRef]
- Moore, A.D.; Björklund, A.K.; Ekman, D.; Bornberg-Bauer, E.; Elofsson, A. Arrangements in the modular evolution of proteins. Trends Biochem. Sci. 2008, 33, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Forslund, S.K.; Kaduk, M.; Sonnhammer, E.L.L. Evolution of Protein Domain Architectures. Methods Mol. Biol. 2019, 1910, 469–504. [Google Scholar] [CrossRef] [PubMed]
- Kersting, A.R.; Bornberg-Bauer, E.; Moore, A.D.; Grath, S. Dynamics and adaptive benefits of protein domain emergence and arrangements during plant genome evolution. Genome Biol. Evol. 2012, 4, 316–329. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.D.; Bornberg-Bauer, E. The dynamics and evolutionary potential of domain loss and emergence. Mol. Biol. Evol. 2012, 29, 787–796. [Google Scholar] [CrossRef]
- Toll-Riera, M.; Alba, M.M. Emergence of novel domains in proteins. BMC Evol. Biol. 2013, 13, 47. [Google Scholar] [CrossRef]
- Margolin, J.F.; Friedman, J.R.; Meyer, W.K.; Vissing, H.; Thiesen, H.J.; Rauscher, F.J., 3rd. Krüppel-associated boxes are potent transcriptional repression domains. Proc. Natl. Acad. Sci. USA 1994, 91, 4509–4513. [Google Scholar] [CrossRef]
- Vasak, M.; Meloni, G. Chemistry and biology of mammalian metallothioneins. J. Biol. Inorg. Chem. 2011, 16, 1067–1078. [Google Scholar] [CrossRef]
- Ziller, A.; Fraissinet-Tachet, L. Metallothionein diversity and distribution in the tree of life: A multifunctional protein. Metallomics 2018, 10, 1549–1559. [Google Scholar] [CrossRef]
- Capdevila, M.; Atrian, S. Metallothionein protein evolution: A miniassay. J. Biol. Inorg. Chem. 2011, 16, 977–989. [Google Scholar] [CrossRef]
- Calatayud, S.; Garcia-Risco, M.; Pedrini-Martha, V.; Eernisse, D.J.; Dallinger, R.; Palacios, O.; Capdevila, M.; Albalat, R. Modularity in Protein Evolution: Modular Organization and De Novo Domain Evolution in Mollusk Metallothioneins. Mol. Biol. Evol. 2021, 38, 424–436. [Google Scholar] [CrossRef]
- Jenny, M.J.; Payton, S.L.; Baltzegar, D.A.; Lozier, J.D. Phylogenetic Analysis of Molluscan Metallothioneins: Evolutionary Insight from Crassostrea virginica. J. Mol. Evol. 2016, 83, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.K.; Kim, E.J. Diversification and domain evolution of molluskan metallothioneins: A mini review. Fish. Aquat. Sci. 2017, 20, 1–18. [Google Scholar] [CrossRef]
- Garcia-Risco, M.; Calatayud, S.; Pedrini-Martha, V.; Albalat, R.; Dallinger, R.; Palacios, O.; Capdevila, M. Metal-Specificity Divergence between Metallothioneins of Nerita peloronta (Neritimorpha, Gastropoda) Sets the Starting Point for a Novel Chemical MT Classification Proposal. Int. J. Mol. Sci. 2021, 22, 13114. [Google Scholar] [CrossRef]
- Cols, N.; Romero-Isart, N.; Capdevila, M.; Oliva, B.; Gonzalez-Duarte, P.; Gonzalez-Duarte, R.; Atrian, S. Binding of excess cadmium(II) to Cd7-metallothionein from recombinant mouse Zn7-metallothionein 1. UV-VIS absorption and circular dichroism studies and theoretical location approach by surface accessibility analysis. J. Inorg. Biochem. 1997, 68, 157–166. [Google Scholar] [CrossRef]
- Espart, A.; Marin, M.; Gil-Moreno, S.; Palacios, O.; Amaro, F.; Martin-Gonzalez, A.; Gutierrez, J.C.; Capdevila, M.; Atrian, S. Hints for metal-preference protein sequence determinants: Different metal binding features of the five tetrahymena thermophila metallothioneins. Int. J. Biol. Sci. 2015, 11, 456–471. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Risco, M.; Gonzalez, A.; Calatayud, S.; Lopez-Jaramillo, F.; Pedrini-Martha, V.; Albalat, R.; Dallinger, R.; Dominguez-Vera, J.M.; Palacios, O.; Capdevila, M. Glycosilation in Recombinant Metallothioneins. Chem. Commun. 2022, 58, 13755–13758. [Google Scholar] [CrossRef] [PubMed]
- Palacios, O.; Atrian, S.; Capdevila, M. Zn- and Cu-thioneins: A functional classification for metallothioneins? J. Biol. Inorg. Chem. 2011, 16, 991–1009. [Google Scholar] [CrossRef]
- Raimundo, J.; Costa, P.M.; Vale, C.; Costa, M.H.; Moura, I. Metallothioneins and trace elements in digestive gland, gills, kidney and gonads of Octopus vulgaris. Comp. Biochem. Physiol. C Toxicol. Pharm. 2010, 152, 139–146. [Google Scholar] [CrossRef]
- Dallinger, R.; Zerbe, O.; Baumann, C.; Egger, B.; Capdevila, M.; Palacios, O.; Albalat, R.; Calatayud, S.; Ladurner, P.; Schlick-Steiner, B.C.; et al. Metallomics reveals a persisting impact of cadmium on the evolution of metal-selective snail metallothioneins. Metallomics 2020, 12, 702–720. [Google Scholar] [CrossRef]
- Palacios, O.; Pagani, A.; Perez-Rafael, S.; Egg, M.; Hockner, M.; Brandstatter, A.; Capdevila, M.; Atrian, S.; Dallinger, R. Shaping mechanisms of metal specificity in a family of metazoan metallothioneins: Evolutionary differentiation of mollusc metallothioneins. BMC Biol. 2011, 9, 4. [Google Scholar] [CrossRef]
- Bustamante, P.; Cosson, R.P.; Gallien, I.; Caurant, F.; Miramand, P. Cadmium detoxification processes in the digestive gland of cephalopods in relation to accumulated cadmium concentrations. Mar. Environ. Res. 2002, 53, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Penicaud, V.; Lacoue-Labarthe, T.; Bustamante, P. Metal bioaccumulation and detoxification processes in cephalopods: A review. Environ. Res 2017, 155, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Bilbao, E.; Lozano, G.; Gutierrez, A.J.; Hardisson, A.; Rubio, C.; Paz, S.; Weller, D.G. The influence of the degassing phase of the Tagoro submarine volcano (Canary Islands) on the metal content of three species of cephalopods. Mar. Pollut. Bull. 2022, 182, 113964. [Google Scholar] [CrossRef] [PubMed]
- Passos, F.D.; Fassina, P.V.; Corrêa, P.V.F.; Miranda, M.S. Separated from the cradle: A new species of Falcidens (Mollusca, Aplacophora, Caudofoveata) reveals weird patterns of distribution in the deep-sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 2022, 186, 103825. [Google Scholar] [CrossRef]
- Calatayud, S.; Garcia-Risco, M.; Capdevila, M.; Canestro, C.; Palacios, O.; Albalat, R. Modular Evolution and Population Variability of Oikopleura dioica Metallothioneins. Front. Cell Dev. Biol. 2021, 9, 702688. [Google Scholar] [CrossRef] [PubMed]
- Calatayud, S.; Garcia-Risco, M.; Palacios, O.; Capdevila, M.; Canestro, C.; Albalat, R. Tunicates Illuminate the Enigmatic Evolution of Chordate Metallothioneins by Gene Gains and Losses, Independent Modular Expansions, and Functional Convergences. Mol. Biol. Evol. 2021, 38, 4435–4448. [Google Scholar] [CrossRef] [PubMed]
- Blevins, W.R.; Ruiz-Orera, J.; Messeguer, X.; Blasco-Moreno, B.; Villanueva-Canas, J.L.; Espinar, L.; Diez, J.; Carey, L.B.; Alba, M.M. Uncovering de novo gene birth in yeast using deep transcriptomics. Nat. Commun. 2021, 12, 604. [Google Scholar] [CrossRef]
- Blindauer, C.A. Metallothioneins. In Binding, Transport and Storage of Metal Ions in Biological Cells; Royal Society of Chemistry: Cambridge, UK, 2014; Volume 2, pp. 606–665. [Google Scholar]
- Isani, G.; Carpene, E. Metallothioneins, unconventional proteins from unconventional animals: A long journey from nematodes to mammals. Biomolecules 2014, 4, 435–457. [Google Scholar] [CrossRef]
- Ruiz-Orera, J.; Messeguer, X.; Subirana, J.A.; Alba, M.M. Long non-coding RNAs as a source of new peptides. eLife 2014, 3, e03523. [Google Scholar] [CrossRef]
- Ruiz-Orera, J.; Villanueva-Canas, J.L.; Alba, M.M. Evolution of new proteins from translated sORFs in long non-coding RNAs. Exp. Cell Res. 2020, 391, 111940. [Google Scholar] [CrossRef]
- Reinhardt, J.A.; Wanjiru, B.M.; Brant, A.T.; Saelao, P.; Begun, D.J.; Jones, C.D. De Novo ORFs in Drosophila Are Important to Organismal Fitness and Evolved Rapidly from Previously Non-coding Sequences. PLoS Genet. 2013, 9, e1003860. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Song, R.; Regev, A.; Struhl, K. Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife 2015, 4, e08890. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Najarian, R.; Haslinger, A.; Valenzuela, P.; Welch, J.; Fogel, S. Primary structure and transcription of an amplified genetic locus: The CUP1 locus of yeast. Proc. Natl. Acad. Sci. USA 1984, 81, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Wong, W.H.; Bornberg-Bauer, E.; Chan, H.S. Recombinatoric exploration of novel folded structures: A heteropolymer-based model of protein evolutionary landscapes. Proc. Natl. Acad. Sci. USA 2002, 99, 809–814. [Google Scholar] [CrossRef] [PubMed]
- Rorick, M.M.; Wagner, G.P. Protein structural modularity and robustness are associated with evolvability. Genome Biol. Evol. 2011, 3, 456–475. [Google Scholar] [CrossRef]
- Niederwanger, M.; Calatayud, S.; Zerbe, O.; Atrian, S.; Albalat, R.; Capdevila, M.; Palacios, O.; Dallinger, R. Biomphalaria glabrata Metallothionein: Lacking Metal Specificity of the Protein and Missing Gene Upregulation Suggest Metal Sequestration by Exchange Instead of through Selective Binding. Int. J. Mol. Sci. 2017, 18, 1457. [Google Scholar] [CrossRef]
- Pedrini-Martha, V.; Köll, S.; Dvorak, M.; Dallinger, R. Cadmium Uptake, MT Gene Activation and Structure of Large-Sized Multi-Domain Metallothioneins in the Terrestrial Door Snail Alinda biplicata (Gastropoda, Clausiliidae). Int. J. Mol. Sci. 2020, 21, 1631. [Google Scholar] [CrossRef]
- Calatayud, S.; Garcia-Risco, M.; Rojas, N.S.; Espinosa-Sanchez, L.; Artime, S.; Palacios, O.; Canestro, C.; Albalat, R. Metallothioneins of the urochordate Oikopleura dioica have Cys-rich tandem repeats, large size and cadmium-binding preference. Metallomics 2018, 10, 1585–1594. [Google Scholar] [CrossRef]
- Bongers, J.; Walton, C.D.; Richardson, D.E.; Bell, J.U. Micromolar protein concentrations and metalloprotein stoichiometries obtained by inductively coupled plasma atomic emission spectrometric determination of sulfur. Anal. Chem. 1988, 60, 2683–2686. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calatayud, S.; Garcia-Risco, M.; Pedrini-Martha, V.; Niederwanger, M.; Dallinger, R.; Palacios, Ò.; Capdevila, M.; Albalat, R. The Modular Architecture of Metallothioneins Facilitates Domain Rearrangements and Contributes to Their Evolvability in Metal-Accumulating Mollusks. Int. J. Mol. Sci. 2022, 23, 15824. https://doi.org/10.3390/ijms232415824
Calatayud S, Garcia-Risco M, Pedrini-Martha V, Niederwanger M, Dallinger R, Palacios Ò, Capdevila M, Albalat R. The Modular Architecture of Metallothioneins Facilitates Domain Rearrangements and Contributes to Their Evolvability in Metal-Accumulating Mollusks. International Journal of Molecular Sciences. 2022; 23(24):15824. https://doi.org/10.3390/ijms232415824
Chicago/Turabian StyleCalatayud, Sara, Mario Garcia-Risco, Veronika Pedrini-Martha, Michael Niederwanger, Reinhard Dallinger, Òscar Palacios, Mercè Capdevila, and Ricard Albalat. 2022. "The Modular Architecture of Metallothioneins Facilitates Domain Rearrangements and Contributes to Their Evolvability in Metal-Accumulating Mollusks" International Journal of Molecular Sciences 23, no. 24: 15824. https://doi.org/10.3390/ijms232415824
APA StyleCalatayud, S., Garcia-Risco, M., Pedrini-Martha, V., Niederwanger, M., Dallinger, R., Palacios, Ò., Capdevila, M., & Albalat, R. (2022). The Modular Architecture of Metallothioneins Facilitates Domain Rearrangements and Contributes to Their Evolvability in Metal-Accumulating Mollusks. International Journal of Molecular Sciences, 23(24), 15824. https://doi.org/10.3390/ijms232415824








