Revisiting the Anti-Cancer Toxicity of Clinically Approved Platinating Derivatives
Abstract
1. Introduction
2. Cisplatin: The First Platinating Agent
2.1. Clinical Usage of Cisplatin
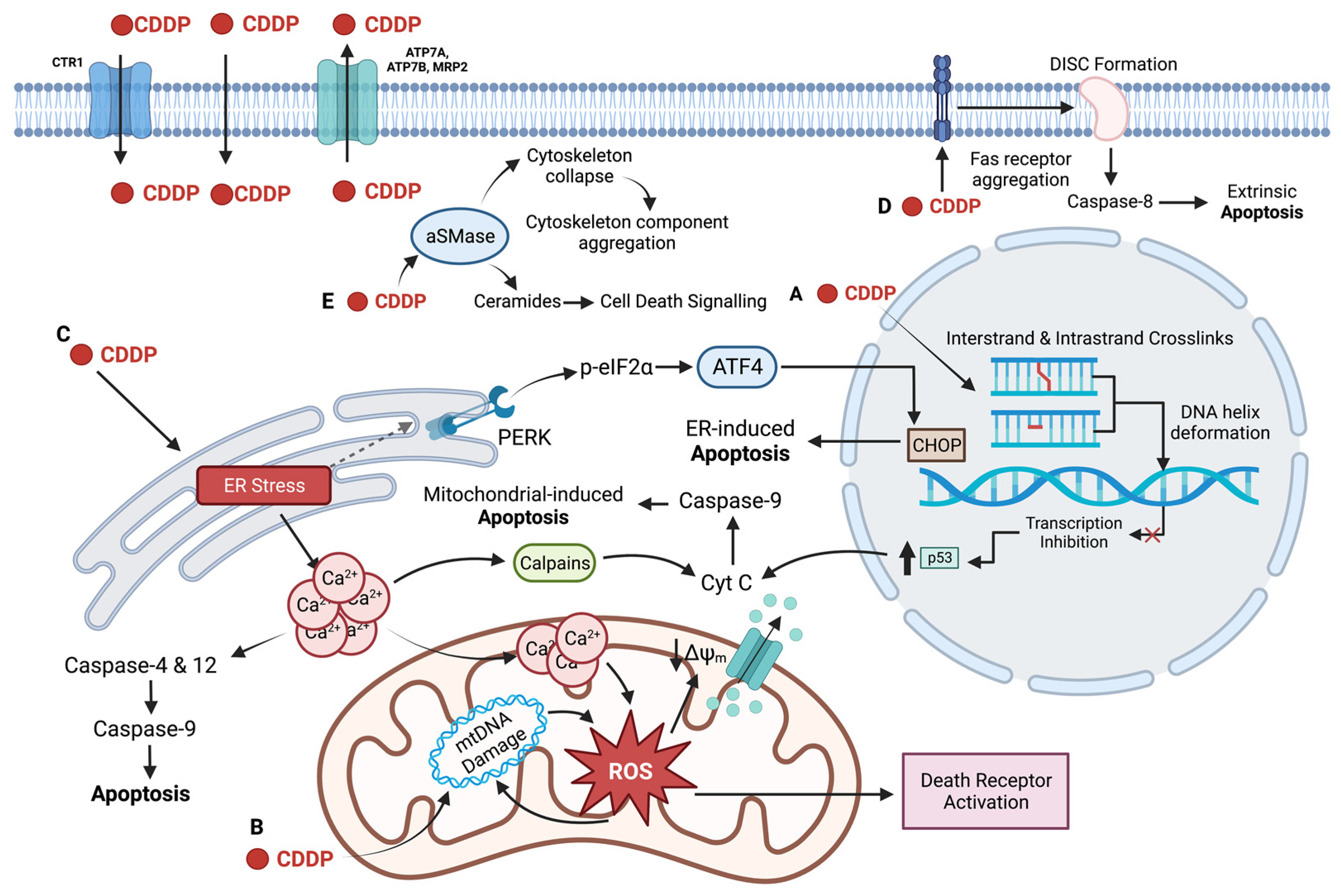
2.2. Cellular Uptake of Cisplatin
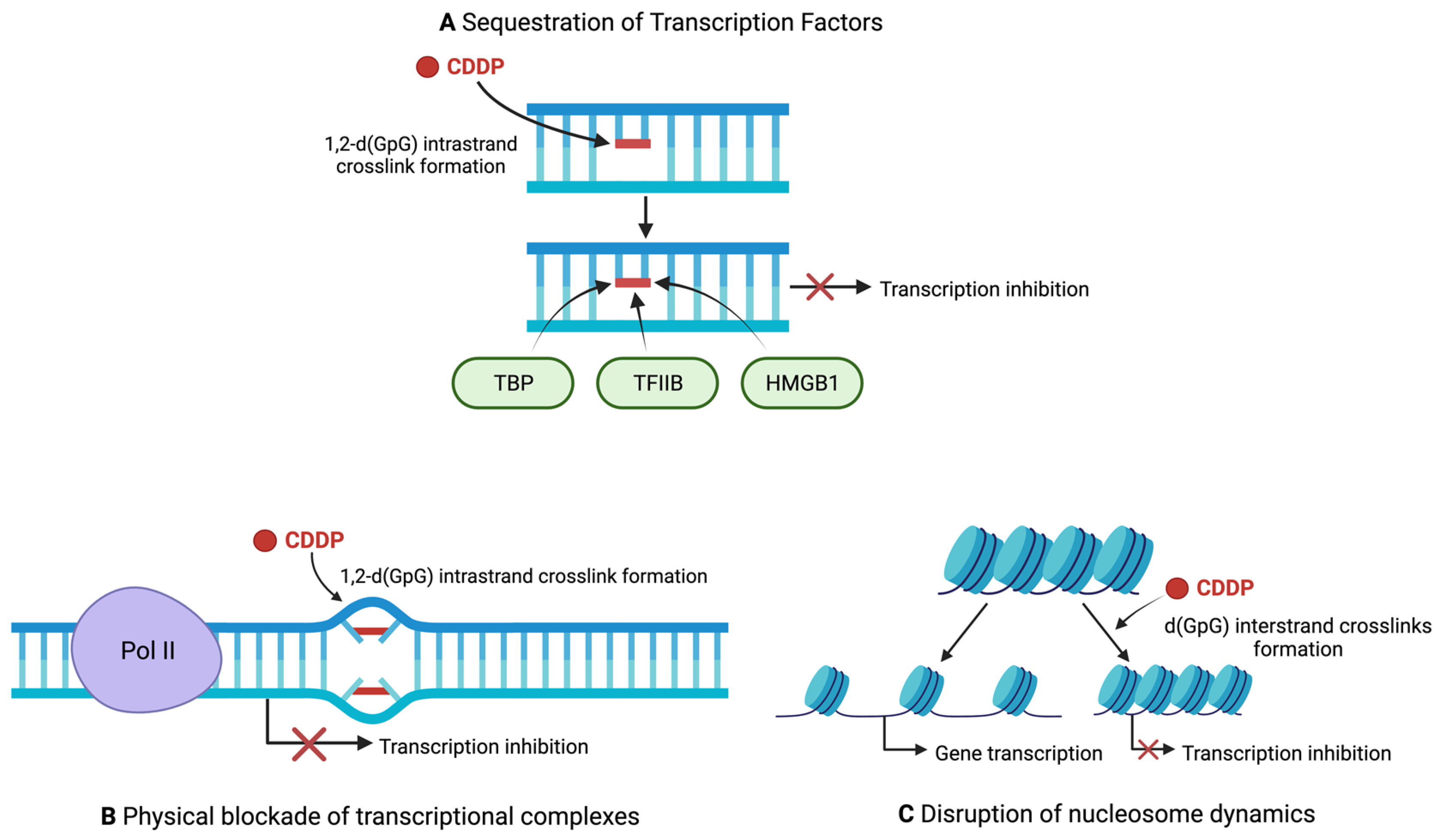
2.3. DNA as the Primary Target of Cisplatin
2.4. Regulation of Transcription by Cisplatin
2.5. Non-Nuclear Targets of Cisplatin
2.6. Limitations of the Use of Cisplatin
3. Carboplatin, the Second-Generation Pt Agent
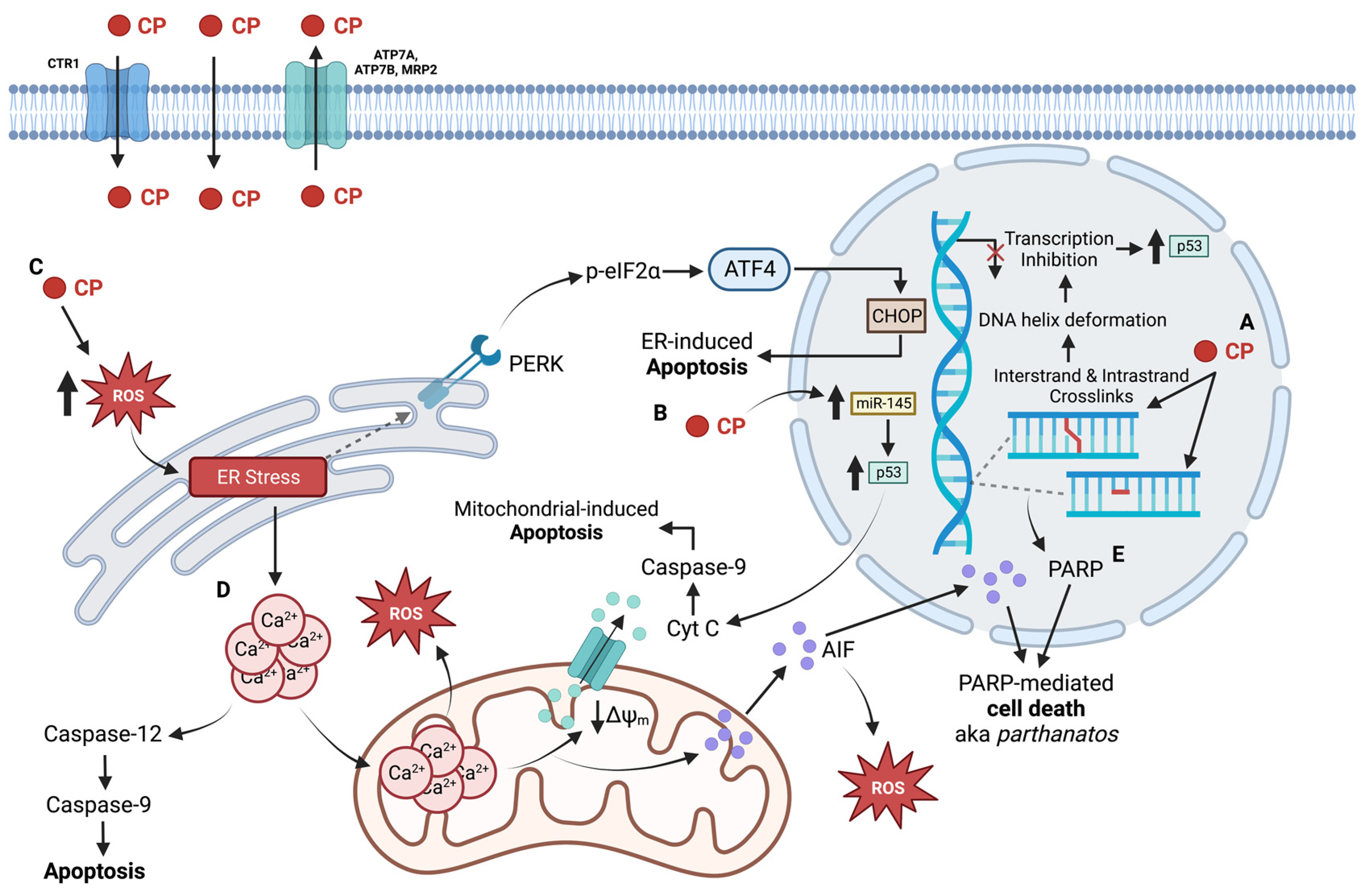
3.1. Clinical Usage of Carboplatin
3.2. The Nuclear Impact of Carboplatin
3.3. Non-Nuclear Targets of Carboplatin
3.4. Limitations for the Use of Carboplatin
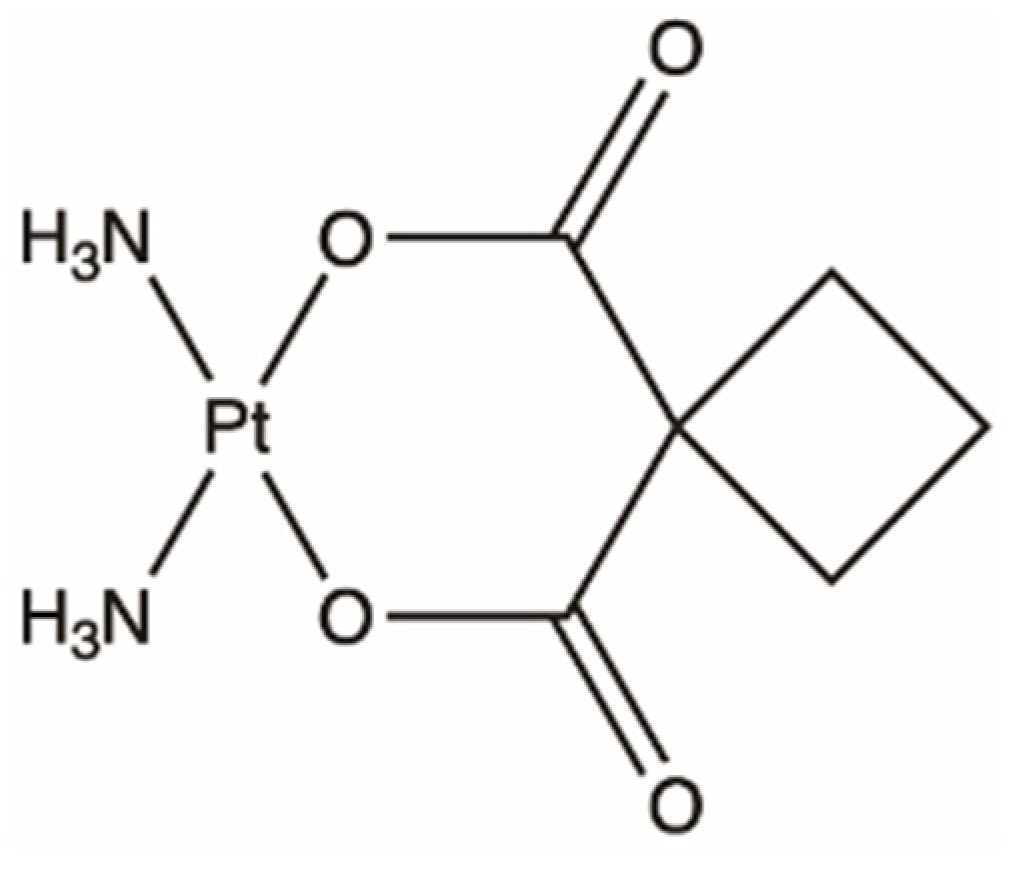
4. Oxaliplatin, a Third Attempt
4.1. Clinical Usage of Oxaliplatin
4.2. Causation of DNA Damage by Oxaliplatin
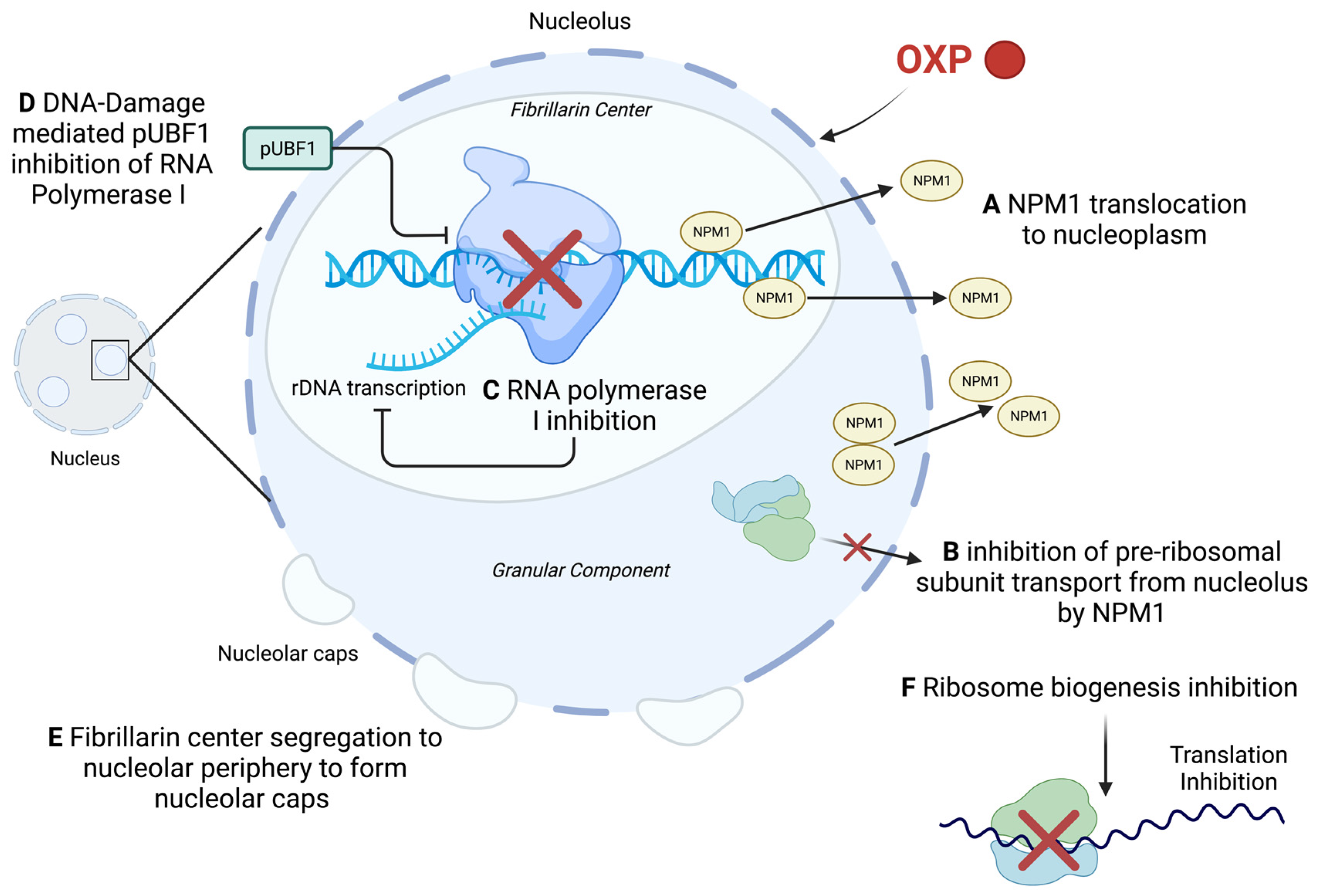
4.3. Effect of Oxaliplatin on Nucleolus Integrity and Ribosome Biogenesis
4.4. Effect of Oxaliplatin on the Mitochondria
4.5. Oxaliplatin and Immunogenic Cell Death
4.6. Limitations of the Use of Oxaliplatin
5. Overall Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AIF | Apoptosis-inducing factor |
| aSMase | Acid sphingomyelinase |
| ATF6 | Activating transcription factor 6 |
| ATP | Adenosine triphosphate |
| BER | Base excision repair |
| CDDP | Cisplatin |
| CHOP | C/EMP homologous protein |
| CP | Carboplatin |
| CRT | Calreticulin |
| CTR1 | High affinity copper uptake protein 1 |
| Cyt C | Cytochrome C |
| DAMP | Damage-associated molecular patterns |
| DC | Dendritic cell |
| DISC | Death-inducing signalling complex |
| DSB | Double strand breaks |
| ER | Endoplasmic reticulum |
| ERCC1 | Cross-complementation group 1 |
| FasR | Fas receptor |
| FDA | US Food and Drug Administration |
| FOLFOX | 5-fluorouracil, leucovorin, oxaliplatin drug combination |
| GEMOX | Gemcitabine, oxaliplatin drug combination |
| GSH | Glutathione |
| HCC | Hepatocellular carcinoma |
| HMGB1 | High-mobility group box 1 |
| HR | Homologous recombination |
| HSOS | Hepatic sinusoidal obstruction syndrome |
| IAP | Inhibitors of apoptosis proteins |
| ICD | Immunogenic cell death |
| ICL | Interstrand crosslink |
| IRE1α | Inositol-requiring protein 1α |
| IROX | Irinotecan, oxaliplatin drug combination |
| miRNA | microRNA |
| MMR | Mismatch repair |
| MRP | Multidrug resistance proteins |
| mtDNA | Mitochondrial DNA |
| nDNA | Nuclear DNA |
| NPM1 | Nucleophosmin |
| Nrf2 | NF-E2-related factor 2 |
| OCT | Organic cation transporter |
| OIPN | Oxaliplatin-induced peripheral neuropathy |
| OXP | Oxaliplatin |
| pEIF2α | Phosphorylated eukaryotic translation factor 2α |
| PERK | Protein kinase RNA-like endoplasmic reticulum kinase |
| Pol I | RNA Polymerase I |
| Pt | Platinum |
| Pt | Platinum |
| ROS | Reactive oxygen species |
| rRNA | Ribosomal RNA |
| ssDNA | Single strand DNA breaks |
| SSRP1 | structure specific recognition protein 1 |
| TFIIH | Transcription factor II H |
| TLR4 | Toll-like receptor 4 |
| UBF | Upstream binding factor |
| XPC | Xeroderma pigmentosum |
References
- Pénzváltó, Z.; Lánczky, A.; Lénárt, J.; Meggyesházi, N.; Krenács, T.; Szoboszlai, N.; Denkert, C.; Pete, I.; Győrffy, B. MEK1 is associated with carboplatin resistance and is a prognostic biomarker in epithelial ovarian cancer. BMC Cancer 2014, 14, 837. [Google Scholar] [CrossRef]
- DeVita, V.T., Jr.; Chu, E. A History of Cancer Chemotherapy. Cancer Res. 2008, 68, 8643–8653. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, C.; Gao, X.; Yao, Q. Platinum-based drugs for cancer therapy and anti-tumor strategies. Theranostics 2022, 12, 2115–2132. [Google Scholar] [CrossRef]
- Ndagi, U.; Mhlongo, N.; Soliman, M.E. Metal complexes in cancer therapy—An update from drug design perspective. Drug. Des. Devel. Ther. 2017, 11, 599–616. [Google Scholar] [CrossRef]
- Famurewa, A.C.; Mukherjee, A.G.; Wanjari, U.R.; Sukumar, A.; Murali, R.; Renu, K.; Vellingiri, B.; Dey, A.; Gopalakrishnan, A.V. Repurposing FDA-approved drugs against the toxicity of platinum-based anticancer drugs. Life Sci. 2022, 305. [Google Scholar] [CrossRef]
- Dilruba, S.; Kalayda, G.V. Platinum-based drugs: Past, present and future. Cancer Chemotherapy and Pharmacology 2016, 77, 1103–1124. [Google Scholar] [CrossRef]
- Kelland, L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Rottenberg, S.; Disler, C.; Perego, P. The rediscovery of platinum-based cancer therapy. Nat. Rev. Cancer 2020, 21, 37–50. [Google Scholar] [CrossRef]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Karbownik, A.; Szałek, E.; Urjasz, H.; Głęboka, A.; Mierzwa, E.; Grzeskowiak, E. The physical and chemical stability of cisplatin (Teva) in concentrate and diluted in sodium chloride 0.9%. Contemp. Oncol. 2012, 5, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ye, Z.-W.; Tew, K.D.; Townsend, D.M. Cisplatin chemotherapy and renal function. Adv. Cancer Res. 2021, 152, 305–327. [Google Scholar] [CrossRef] [PubMed]
- Hayati, F.; Hossainzadeh, M.; Shayanpour, S.; Abedi-Gheshlaghi, Z.; Mousavi, S.S.B. Prevention of cisplatin nephrotoxicity. J. Nephropharmacol. 2015, 5, 57–60. [Google Scholar]
- Hastings, J.; Owen, G.; Dekker, A.; Ennis, M.; Kale, N.; Muthukrishnan, V.; Turner, S.; Swainston, N.; Mendes, P.; Steinbeck, C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2015, 44, D1214–D1219. [Google Scholar] [CrossRef]
- Peyrone, M. Ueber die Einwirkung des Ammoniaks auf Platinchlorür. Eur. J. Org. Chem. 1844, 51, 1–29. [Google Scholar] [CrossRef]
- Rosenberg, B.; VAN Camp, L.; Krigas, T. Inhibition of Cell Division in Escherichia coli by Electrolysis Products from a Platinum Electrode. Nature 1965, 205, 698–699. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, B.; Vancamp, L.; Trosko, J.E.; Mansour, V.H. Platinum Compounds: A New Class of Potent Antitumour Agents. Nature 1969, 222, 385–386. [Google Scholar] [CrossRef] [PubMed]
- Barry, N.P.E.; Sadler, P.J. 100 years of metal coordination chemistry: From Alfred Werner to anticancer metallodrugs. Pure Appl. Chem. 2014, 86, 1897–1910. [Google Scholar] [CrossRef]
- Higby, D.J.; Wallace, H.J.; Albert, D.J.; Holland, J.F. Diaminodichloroplatinum: A phase I study showing responses in testicular and other tumors. Cancer 1974, 33, 1219–1225. [Google Scholar] [CrossRef]
- Higby, D.J.; Wallace, H.J., Jr.; Albert, D.; Holland, J.F. Diamminodichloroplatinum in the Chemotherapy of Testicular Tumors. J. Urol. 1974, 112, 100–104. [Google Scholar] [CrossRef]
- Cersosimo, R.J. Cisplatin neurotoxicity. Cancer Treat. Rev. 1989, 16, 195–211. [Google Scholar] [CrossRef]
- Wiltshaw, E. Cisplatin in the treatment of cancer. Platinum Met. Rev. 1979, 23, 90–98. [Google Scholar]
- Daugaard, G.; Abildgaard, U. Cisplatin nephrotoxicity. Cancer Chemother. Pharmacol. 1989, 25, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, E.R.; Lippard, S.J. Structure, Recognition, and Processing of Cisplatin−DNA Adducts. Chem. Rev. 1999, 99, 2467–2498. [Google Scholar] [CrossRef] [PubMed]
- Einhorn, L.H. Treatment of testicular cancer: A new and improved model. J. Clin. Oncol. 1990, 8, 1777–1781. [Google Scholar] [CrossRef]
- Fung, C.; Dinh, P.C.; Fossa, S.D.; Travis, L.B. Testicular Cancer Survivorship. J. Natl. Compr. Cancer Netw. 2019, 17, 1557–1568. [Google Scholar] [CrossRef] [PubMed]
- Boulikas, T.; Vougiouka, M. Recent clinical trials using cisplatin, carboplatin and their combination chemotherapy drugs (review). Oncol. Rep. 2004, 11, 559–595. [Google Scholar] [CrossRef]
- DeConti, R.C.; Toftness, B.R.; Lange, R.C.; A Creasey, W. Clinical and pharmacological studies with cis-diamminedichloroplatinum (II). Cancer Res. 1973, 33, 1310–1315. [Google Scholar]
- Gale, G.R.; Morris, C.R.; Atkins, L.M.; Smith, A.B. Binding of an antitumor platinum compound to cells as influenced by physical factors and pharmacologically active agents. Cancer Res. 1973, 33, 813–818. [Google Scholar]
- Eljack, N.D.; Ma, H.-Y.M.; Drucker, J.; Shen, C.; Hambley, T.W.; New, E.J.; Friedrich, T.; Clarke, R.J. Mechanisms of cell uptake and toxicity of the anticancer drug cisplatin. Metallomics 2014, 6, 2126–2133. [Google Scholar] [CrossRef]
- Binks, S.P.; Dobrota, M. Kinetics and mechanism of uptake of platinum-based pharmaceuticals by the rat small intestine. Biochem. Pharmacol. 1990, 40, 1329–1336. [Google Scholar] [CrossRef]
- Andrews, P.A.; Mann, S.C.; Velury, S.; Howell, S.B. Cisplatin Uptake Mediated Cisplatin-Resistance in Human Ovarian Carcinoma Cells. In Platinum and Other Metal Coordination Compounds in Cancer Chemotherapy: Proceedings of the Fifth International Symposium on Platinum and Other Metal Coordination Compounds in Cancer Chemotherapy Abano, Padua, Italy, 29 June–2 July 1987; Nicolini, M., Ed.; Springer: Boston, MA, USA, 1988; pp. 248–254. [Google Scholar]
- Andrews, P.A.; Albright, K.D. Role of Membrane Ion Transport in Cisplatin Accumulation. In Platinum and Other Metal Coordination Compounds in Cancer Chemotherapy; Howell, S.B., Ed.; Springer: Boston, MA, USA, 1991; pp. 151–159. [Google Scholar]
- Ivy, K.D.; Kaplan, J.H. A Re-Evaluation of the Role of hCTR1, the Human High-Affinity Copper Transporter, in Platinum-Drug Entry into Human Cells. Mol. Pharmacol. 2013, 83, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Mann, S.C.; Andrews, P.A.; Howell, S.B. Short-term cis-diamminedichloroplatinum(II) accumulation in sensitive and resistant human ovarian carcinoma cells. Cancer Chemother Pharmacol. 1990, 25, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Ishida, S.; Lee, J.; Thiele, D.J.; Herskowitz, I. Uptake of the anticancer drug cisplatin mediated by the copper transporter Ctr1 in yeast and mammals. Proc. Natl. Acad. Sci. USA 2002, 99, 14298–14302. [Google Scholar] [CrossRef] [PubMed]
- Katano, K.; Kondo, A.; Safaei, R.; Holzer, A.; Samimi, G.; Mishima, M.; Kuo, Y.-M.; Rochdi, M.; Howell, S.B. Acquisition of resistance to cisplatin is accompanied by changes in the cellular pharmacology of copper. Cancer Res. 2002, 62, 6559–6565. [Google Scholar] [PubMed]
- Holzer, A.K.; Samimi, G.; Katano, K.; Naerdemann, W.; Lin, X.; Safaei, R.; Howell, S.B. The Copper Influx Transporter Human Copper Transport Protein 1 Regulates the Uptake of Cisplatin in Human Ovarian Carcinoma Cells. Mol. Pharmacol. 2004, 66, 817–823. [Google Scholar] [CrossRef]
- Holzer, A.K.; Katano, K.; Klomp, L.W.J.; Howell, S.B. Cisplatin Rapidly Down-regulates Its Own Influx Transporter hCTR1 in Cultured Human Ovarian Carcinoma Cells. Clin. Cancer Res. 2004, 10, 6744–6749. [Google Scholar] [CrossRef]
- Petris, M.J.; Smith, K.; Lee, J.; Thiele, D.J. Copper-stimulated Endocytosis and Degradation of the Human Copper Transporter, hCtr1. J. Biol. Chem. 2003, 278, 9639–9646. [Google Scholar] [CrossRef]
- Kalayda, G.V.; Wagner, C.H.; Jaehde, U. Relevance of copper transporter 1 for cisplatin resistance in human ovarian carcinoma cells. J. Inorg. Biochem. 2012, 116, 1–10. [Google Scholar] [CrossRef]
- Sinani, D.; Adle, D.J.; Kim, H.; Lee, J. Distinct Mechanisms for Ctr1-mediated Copper and Cisplatin Transport. J. Biol. Chem. 2007, 282, 26775–26785. [Google Scholar] [CrossRef]
- Beretta, G.L.; Gatti, L.; Tinelli, S.; Corna, E.; Colangelo, D.; Zunino, F.; Perego, P. Cellular pharmacology of cisplatin in relation to the expression of human copper transporter CTR1 in different pairs of cisplatin-sensitive and -resistant cells. Biochem. Pharmacol. 2004, 68, 283–291. [Google Scholar] [CrossRef]
- Hall, M.D.; Okabe, M.; Shen, D.-W.; Liang, X.-J.; Gottesman, M.M. The Role of Cellular Accumulation in Determining Sensitivity to Platinum-Based Chemotherapy. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 495–535. [Google Scholar] [CrossRef] [PubMed]
- Song, I.S.; Savaraj, N.; Siddik, Z.H.; Liu, P.; Wei, Y.; Wu, C.J.; Kuo, M.T. Role of human copper transporter Ctr1 in the transport of platinum-based antitumor agents in cisplatin-sensitive and cisplatin-resistant cells. Mol. Cancer Ther. 2004, 3, 1543–1549. [Google Scholar] [CrossRef] [PubMed]
- A Andrews, P.; Mann, S.C.; Huynh, H.H.; Albright, K.D. Role of the Na+, K(+)-adenosine triphosphatase in the accumulation of cis-diamminedichloroplatinum(II) in human ovarian carcinoma cells. Cancer Res. 1991, 51, 3677–3681. [Google Scholar]
- Kishimoto, S.; Kawazoe, Y.; Ikeno, M.; Saitoh, M.; Nakano, Y.; Nishi, Y.; Fukushima, S.; Takeuchi, Y. Role of Na+, K+-ATPase α1 subunit in the intracellular accumulation of cisplatin. Cancer Chemother. Pharmacol. 2006, 57, 84–90. [Google Scholar] [CrossRef] [PubMed]
- A Andrews, P.; Albright, K.D. Mitochondrial defects in cis-diamminedichloroplatinum(II)-resistant human ovarian carcinoma cells. Cancer Res. 1992, 52, 1895–1901. [Google Scholar]
- Walker, W.F.; Johnston, I.D.A. 2—Water and Electrolyte Metabolism. In The Metabolic Basis of Surgical Care; Walker, W.F., Johnston, I.D.A., Eds.; Butterworth-Heinemann: Oxford, UK, 1971. [Google Scholar]
- El-Khateeb, M.; Appleton, T.G.; Gahan, L.R.; Charles, B.G.; Berners-Price, S.J.; Bolton, A.-M. Reactions of cisplatin hydrolytes with methionine, cysteine, and plasma ultrafiltrate studied by a combination of HPLC and NMR techniques. J. Inorg. Biochem. 1999, 77, 13–21. [Google Scholar] [CrossRef]
- Pinto, A.L.; Lippard, S.J. Binding of the antitumor drug cis-diamminedichloroplatinum(II) (cisplatin) to DNA. Biochim. Biophys. Acta 1985, 780, 167–180. [Google Scholar] [CrossRef]
- Miller, S.E.; House, D.A. The hydrolysis products of cis-diamminedichloroplatinum(II) 5. The anation kinetics of cis-Pt(X)(NH3)2(OH2)+ (X-Cl, OH) with glycine, monohydrogen malonate and chloride. Inorganica Chim. Acta 1991, 187, 125–132. [Google Scholar] [CrossRef]
- Kelland, L.R. New platinum antitumor complexes. Critical Rev. Oncol. /Hematol. 1993, 15, 191–219. [Google Scholar] [CrossRef]
- Eastman, A. Characterization of the adducts produced in DNA by cis-diamminedichloroplatinum(II) and cis-dichloro(ethylenediamine)platinum(II). Biochemistry 1983, 22, 3927–3933. [Google Scholar] [CrossRef]
- Plooy, A.C.; Fichtinger-Schepman, A.M.J.; Schutte, H.H.; Van Dijk, M.; Lohman, P.H. The quantitative detection of various Pt-DNA-adducts in Chinese hamster ovary cells treated with cisplatin: Application of immunochemical techniques. Carcinogenesis 1985, 6, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Eastman, A. The formation, isolation and characterization of DNA adducts produced by anticancer platinum complexes. Pharmacol. Ther. 1987, 34, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Giraud-Panis, M.-J.; Malinge, J.-M.; Sedletska, Y. Cisplatin Is a DNA-Damaging Antitumour Compound Triggering Multifactorial Biochemical Responses in Cancer Cells: Importance of Apoptotic Pathways. Curr. Med. Chem. Agents 2005, 5, 251–265. [Google Scholar] [CrossRef]
- Beck, D.J.; Brubaker, R.R. Effect of cis-platinum(II)diamminodichloride on wild type and deoxyribonucleic acid repair deficient mutants of Escherichia coli. J. Bacteriol. 1973, 116, 1247–1252. [Google Scholar] [CrossRef]
- Fraval, H.; Rawlings, C.; Roberts, J. Increased sensitivity of UV-repair-deficient human cells to DNA bound platinum products which unlike thymine dimers are not recognized by an endonuclease extracted from Micrococcus luteus. Mutat. Res. Mol. Mech. Mutagen. 1978, 51, 121–132. [Google Scholar] [CrossRef]
- Harder, H.C.; Smith, R.G.; Leroy, A.F. Template primer inactivation by cis- and trans-dichlorodiammine platinum for human DNA polymerase alpha, beta, and Rauscher murine leukemia virus reverse transcriptase, as a mechanism of cytotoxicity. Cancer Res. 1976, 36, 3821–3829. [Google Scholar]
- Pinto, A.L.; Lippard, S.J. Sequence-dependent termination of in vitro DNA synthesis by cis- and trans-diamminedichloroplatinum (II). Proc. Natl. Acad. Sci. USA 1985, 82, 4616–4619. [Google Scholar] [CrossRef]
- Sorenson, C.M.; Eastman, A. Mechanism of cis-diamminedichloroplatinum(II)-induced cytotoxicity: Role of G2 arrest and DNA double-strand breaks. Cancer Res. 1988, 48, 4484–4488. [Google Scholar]
- Sorenson, C.M.; Eastman, A. Influence of cis-diamminedichloroplatinum(II) on DNA synthesis and cell cycle progression in excision repair proficient and deficient Chinese hamster ovary cells. Cancer Res. 1988, 48, 6703–6707. [Google Scholar]
- Sorenson, C.M.; Barry, M.A.; Eastman, A. Analysis of Events Associated With Cell Cycle Arrest at G2 Phase and Cell Death Induced by Cisplatin. Gynecol. Oncol. 1990, 82, 749–755. [Google Scholar] [CrossRef]
- Jung, Y.; Lippard, S.J. Multiple States of Stalled T7 RNA Polymerase at DNA Lesions Generated by Platinum Anticancer Agents. J. Biol. Chem. 2003, 278, 52084–52092. [Google Scholar] [CrossRef]
- Tornaletti, S.; Patrick, S.M.; Turchi, J.J.; Hanawalt, P.C. Behavior of T7 RNA Polymerase and Mammalian RNA Polymerase II at Site-specific Cisplatin Adducts in the Template DNA. J. Biol. Chem. 2003, 278, 35791–35797. [Google Scholar] [CrossRef] [PubMed]
- Tremeau-Bravard, A.; Riedl, T.; Egly, J.-M.; Dahmus, M.E. Fate of RNA Polymerase II Stalled at a Cisplatin Lesion. J. Biol. Chem. 2004, 279, 7751–7759. [Google Scholar] [CrossRef] [PubMed]
- Corda, Y.; Job, C.; Anin, M.F.; Leng, M.; Job, D. Transcription by eucaryotic and procaryotic RNA polymerases of DNA modified at a d(GG) or a d(AG) site by the antitumor drug cis-diamminedichloroplatinum(II). Biochemistry 1991, 30, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Corda, Y.; Anin, M.F.; Leng, M.; Job, D. RNA polymerases react differently at d(ApG) and d(GpG) adducts in DNA modified by cis-diamminedichloroplatinum(II). Biochemistry 1992, 31, 1904–1908. [Google Scholar] [CrossRef] [PubMed]
- Corda, Y.; Job, C.; Anin, M.F.; Leng, M.; Job, D. Spectrum of DNA-platinum adduct recognition by prokaryotic and eukaryotic DNA-dependent RNA polymerases. Biochemistry 1993, 32, 8582–8588. [Google Scholar] [CrossRef]
- Mello, J.A.; Lippard, S.J.; Essigmann, J.M. DNA Adducts of cis-Diamminedichloroplatinum(II) and Its Trans Isomer Inhibit RNA Polymerase II Differentially in Vivo. Biochemistry 1995, 34, 14783–14791. [Google Scholar] [CrossRef]
- Todd, R.C.; Lippard, S.J. Inhibition of transcription by platinum antitumor compounds. Metallomics 2009, 1, 280–291. [Google Scholar] [CrossRef]
- Vichi, P.; Coin, F.; Renaud, J.; Vermeulen, W.; Hoeijmakers, J.; Moras, D.; Egly, J. Cisplatin- and UV-damaged DNA lure the basal transcription factor TFIID/TBP. EMBO J. 1997, 16, 7444–7456. [Google Scholar] [CrossRef]
- Cullinane, C.; Mazur, S.J.; Essigmann, J.M.; Phillips, A.D.R.; Bohr, V.A. Inhibition of RNA Polymerase II Transcription in Human Cell Extracts by Cisplatin DNA Damage. Biochemistry 1999, 38, 6204–6212. [Google Scholar] [CrossRef]
- Treiber, D.K.; Zhai, X.; Jantzen, H.M.; Essigmann, J.M. Cisplatin-DNA adducts are molecular decoys for the ribosomal RNA transcription factor hUBF (human upstream binding factor). Proc. Natl. Acad. Sci. USA 1994, 91, 5672–5676. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Beckmann, H.; Jantzen, H.-M.; Essigmann, J.M. Cisplatin−DNA Adducts Inhibit Ribosomal RNA Synthesis by Hijacking the Transcription Factor Human Upstream Binding Factor. Biochemistry 1998, 37, 16307–16315. [Google Scholar] [CrossRef] [PubMed]
- Ise, T.; Nagatani, G.; Imamura, T.; Kato, K.; Takano, H.; Nomoto, M.; Izumi, H.; Ohmori, H.; Okamoto, T.; Ohga, T.; et al. Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res. 1999, 59, 342–346. [Google Scholar] [PubMed]
- Yarnell, A.T.; Oh, S.; Reinberg, D.; Lippard, S.J. Interaction of FACT, SSRP1, and the High Mobility Group (HMG) Domain of SSRP1 with DNA Damaged by the Anticancer Drug Cisplatin *. J. Biol. Chem. 2001, 276, 25736–25741. [Google Scholar] [CrossRef] [PubMed]
- Dunham, S.U.; Lippard, S.J. DNA Sequence Context and Protein Composition Modulate HMG-Domain Protein Recognition of Cisplatin-Modified DNA. Biochemistry 1997, 36, 11428–11436. [Google Scholar] [CrossRef]
- Mymryk, J.S.; Zaniewski, E.; Archer, T.K. Cisplatin Inhibits Chromatin Remodeling, Transcription Factor Binding, and Transcription from the Mouse Mammary Tumor Virus Promoter in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 2076–2080. [Google Scholar] [CrossRef]
- E Damsma, G.; Alt, A.; Brueckner, F.; Carell, T.; Cramer, P. Mechanism of transcriptional stalling at cisplatin-damaged DNA. Nat. Struct. Mol. Biol. 2007, 14, 1127–1133. [Google Scholar] [CrossRef]
- Strauss, B.S. The ‘A rule’ of mutagen specificity: A consequence of DNA polymerase bypass of non-instructional lesions? BioEssays 1991, 13, 79–84. [Google Scholar] [CrossRef]
- Wu, C. Chromatin Remodeling and the Control of Gene Expression. J. Biol. Chem. 1997, 272, 28171–28174. [Google Scholar] [CrossRef]
- Li, B.; Carey, M.; Workman, J.L. The Role of Chromatin during Transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef]
- Ober, M.; Lippard, S.J. Cisplatin Damage Overrides the Predefined Rotational Setting of Positioned Nucleosomes. J. Am. Chem. Soc. 2007, 129, 6278–6286. [Google Scholar] [CrossRef][Green Version]
- Ober, M.; Lippard, S.J. A 1,2-d(GpG) cisplatin intrastrand cross-link influences the rotational and translational setting of DNA in nucleosomes. J. Am. Chem. Soc. 2008, 130, 2851–2861. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Todd, R.C.; Lippard, S.J. Consequences of Cisplatin Binding on Nucleosome Structure and Dynamics. Chem. Biol. 2010, 17, 1334–1343. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.-M.; Park, J.-S.; Lee, I.-B.; Kang, Y.-I.; Jung, H.J.; An, D.; Shin, Y.; Kim, M.J.; I Kim, H.; Song, J.-J.; et al. Cisplatin fastens chromatin irreversibly even at a high chloride concentration. Nucleic Acids Res. 2021, 49, 12035–12047. [Google Scholar] [CrossRef]
- Charlier, C.; Kintz, P.; Dubois, N.; Plomteux, G. Fatal Overdosage with Cisplatin. J. Anal. Toxicol. 2004, 28, 138–140. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Song, L.; Lippard, S.J. Visualizing Inhibition of Nucleosome Mobility and Transcription by Cisplatin–DNA Interstrand Crosslinks in Live Mammalian Cells. Cancer Res. 2013, 73, 4451–4460. [Google Scholar] [CrossRef] [PubMed]
- Pucci, B.; Kasten, M.; Giordano, A. Cell cycle and apoptosis. Neoplasia 2000, 2, 291–299. [Google Scholar] [CrossRef]
- Donahue, B.A.; Augot, M.; Bellon, S.F.; Treiber, D.K.; Toney, J.H.; Lippard, S.J.; Essigmann, J.M. Characterization of a DNA damage-recognition protein from mammalian cells that binds specifically to intrastrand d(GpG) and d(ApG) DNA adducts of the anticancer drug cisplatin. Biochemistry 1990, 29, 5872–5880. [Google Scholar] [CrossRef]
- Fink, D.; Aebi, S.; Howell, S.B. The role of DNA mismatch repair in drug resistance. Clin. Cancer Res. 1998, 4, 1–6. [Google Scholar]
- Chaney, S.G.; Vaisman, A. Specificity of platinum–DNA adduct repair. J. Inorg. Biochem. 1999, 77, 71–81. [Google Scholar] [CrossRef]
- Appella, E.; Anderson, C.W. Post-translational modifications and activation of p53 by genotoxic stresses. JBIC J. Biol. Inorg. Chem. 2001, 268, 2764–2772. [Google Scholar] [CrossRef] [PubMed]
- Persons, D.L.; Yazlovitskaya, E.M.; Pelling, J.C. Effect of Extracellular Signal-regulated Kinase on p53 Accumulation in Response to Cisplatin. J. Biol. Chem. 2000, 275, 35778–35785. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Chaudhry, P.; Fabi, F.; Asselin, E. Cisplatin-induced caspase activation mediates PTEN cleavage in ovarian cancer cells: A potential mechanism of chemoresistance. BMC Cancer 2013, 13, 233–239. [Google Scholar] [CrossRef]
- Cummings, B.S.; Schnellmann, R.G. Cisplatin-Induced Renal Cell Apoptosis: Caspase 3-Dependent and -Independent Pathways. J. Pharmacol. Exp. Ther. 2002, 302, 8–17. [Google Scholar] [CrossRef]
- Jones, E.V.; Dickman, M.; Whitmarsh, A.J. Regulation of p73-mediated apoptosis by c-Jun N-terminal kinase. Biochem. J. 2007, 405, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Paul, I.; Chacko, A.D.; Stasik, I.; Busacca, S.; Crawford, N.; McCoy, F.; McTavish, N.; Wilson, B.; Barr, M.; O’Byrne, K.J.; et al. Acquired differential regulation of caspase-8 in cisplatin-resistant non-small-cell lung cancer. Cell Death Dis. 2012, 3, e449. [Google Scholar] [CrossRef]
- Mansouri, A.; Ridgway, L.D.; Korapati, A.L.; Zhang, Q.; Tian, L.; Wang, Y.; Siddik, Z.H.; Mills, G.B.; Claret, F.X. Sustained Activation of JNK/p38 MAPK Pathways in Response to Cisplatin Leads to Fas Ligand Induction and Cell Death in Ovarian Carcinoma Cells. J. Biol. Chem. 2003, 278, 19245–19256. [Google Scholar] [CrossRef]
- Yang, Z.; Schumaker, L.M.; Egorin, M.J.; Zuhowski, E.G.; Guo, Z.; Cullen, K.J. Cisplatin Preferentially Binds Mitochondrial DNA and Voltage-Dependent Anion Channel Protein in the Mitochondrial Membrane of Head and Neck Squamous Cell Carcinoma: Possible Role in Apoptosis. Clin. Cancer Res. 2006, 12, 5817–5825. [Google Scholar] [CrossRef]
- Mandic, A.; Hansson, J.; Linder, S.; Shoshan, M.C. Cisplatin Induces Endoplasmic Reticulum Stress and Nucleus-independent Apoptotic Signaling. J. Biol. Chem. 2003, 278, 9100–9106. [Google Scholar] [CrossRef]
- Yu, F.; Megyesi, J.; Price, P.M. Cytoplasmic initiation of cisplatin cytotoxicity. Am. J. Physiol. -Renal Physiol. 2008, 295, F44–F52. [Google Scholar] [CrossRef]
- Gutekunst, M.; Oren, M.; Weilbacher, A.; Dengler, M.A.; Markwardt, C.; Thomale, J.; Aulitzky, W.E.; Van Der Kuip, H. p53 Hypersensitivity Is the Predominant Mechanism of the Unique Responsiveness of Testicular Germ Cell Tumor (TGCT) Cells to Cisplatin. PLoS ONE 2011, 6, e19198. [Google Scholar] [CrossRef]
- Olivero, O.A.; Chang, P.K.; Lopez-Larraza, D.M.; Semino-Mora, M.C.; Poirier, M.C. Preferential formation and decreased removal of cisplatin–DNA adducts in Chinese hamster ovary cell mitochondrial DNA as compared to nuclear DNA. Mutat. Res. Toxicol. Environ. Mutagen. 1997, 391, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Montopoli, M.; Bellanda, M.; Lonardoni, F.; Ragazzi, E.; Dorigo, P.; Froldi, G.; Mammi, S.; Caparrotta, L. “Metabolic reprogramming” in ovarian cancer cells resistant to cisplatin. Curr. Cancer Drug Targets 2011, 11, 226–235. [Google Scholar] [CrossRef]
- Martins, N.M.; Santos, N.A.G.; Curti, C.; Bianchi, M.L.P.; Santos, A.C. Cisplatin induces mitochondrial oxidative stress with resultant energetic metabolism impairment, membrane rigidification and apoptosis in rat liver. J. Appl. Toxicol. 2007, 28, 337–344. [Google Scholar] [CrossRef]
- Santandreu, F.M.; Roca, P.; Oliver, J. Uncoupling protein-2 knockdown mediates the cytotoxic effects of cisplatin. Free Radic. Biol. Med. 2010, 49, 658–666. [Google Scholar] [CrossRef]
- Li-ping, X.; Skrezek, C.; Wand, H.; Reibe, F. Mitochondrial dysfunction at the early stage of cisplatin-induced acute renal failure in rats. J. Zhejiang Univ. -Sci. 2000, 1, 91–96. [Google Scholar]
- Marullo, R.; Werner, E.; Degtyareva, N.; Moore, B.; Altavilla, G.; Ramalingam, S.S.; Doetsch, P.W. Cisplatin Induces a Mitochondrial-ROS Response That Contributes to Cytotoxicity Depending on Mitochondrial Redox Status and Bioenergetic Functions. PLoS ONE 2013, 8, e81162. [Google Scholar] [CrossRef]
- Choi, Y.-M.; Kim, H.; Shim, W.; Anwar, M.A.; Kwon, J.-W.; Kwon, H.-K.; Kim, H.J.; Jeong, H.; Kim, H.M.; Hwang, D.; et al. Mechanism of Cisplatin-Induced Cytotoxicity Is Correlated to Impaired Metabolism Due to Mitochondrial ROS Generation. PLoS ONE 2015, 10, e0135083. [Google Scholar] [CrossRef]
- Garrido, N.; Pérez-Martos, A.; Faro, M.; Lou-Bonafonte, J.M.; Fernández-Silva, P.; López-Pérez, M.J.; Montoya, J.; Enríquez, J.A. Cisplatin-mediated impairment of mitochondrial DNA metabolism inversely correlates with glutathione levels. Biochem. J. 2008, 414, 93–102. [Google Scholar] [CrossRef]
- Redza-Dutordoir, M.; Averill-Bates, D.A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2016, 1863, 2977–2992. [Google Scholar] [CrossRef]
- Fleury, C.; Mignotte, B.; Vayssière, J.-L. Mitochondrial reactive oxygen species in cell death signaling. Biochimie 2002, 84, 131–141. [Google Scholar] [CrossRef]
- Wu, C.-C.; Bratton, S.B. Regulation of the Intrinsic Apoptosis Pathway by Reactive Oxygen Species. Antioxidants Redox Signal. 2013, 19, 546–558. [Google Scholar] [CrossRef]
- Halestrap, A.P.; Woodfield, K.-Y.; Connern, C.P. Oxidative Stress, Thiol Reagents, and Membrane Potential Modulate the Mitochondrial Permeability Transition by Affecting Nucleotide Binding to the Adenine Nucleotide Translocase *. J. Biol. Chem. 1997, 272, 3346–3354. [Google Scholar] [CrossRef] [PubMed]
- Kagan, V.E.; Borisenko, G.G.; Tyurina, Y.Y.; Tyurin, V.; Jiang, J.; Potapovich, A.I.; Kini, V.; Amoscato, A.A.; Fujii, Y. Oxidative lipidomics of apoptosis: Redox catalytic interactions of cytochrome c with cardiolipin and phosphatidylserine. Free. Radic. Biol. Med. 2004, 37, 1963–1985. [Google Scholar] [CrossRef]
- Kleih, M.; Böpple, K.; Dong, M.; Gaißler, A.; Heine, S.; Olayioye, M.A.; Aulitzky, W.E.; Essmann, F. Direct impact of cisplatin on mitochondria induces ROS production that dictates cell fate of ovarian cancer cells. Cell Death Dis. 2019, 10, 851. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Blower, M.D. The endoplasmic reticulum: Structure, function and response to cellular signaling. Cell. Mol. Life Sci. 2016, 73, 79–94. [Google Scholar] [CrossRef]
- Avril, T.; Vauléon, E.; Chevet, E. Endoplasmic reticulum stress signaling and chemotherapy resistance in solid cancers. Oncogenesis 2017, 6, e373. [Google Scholar] [CrossRef]
- Hetz, C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012, 13, 89–102. [Google Scholar] [CrossRef]
- Lee, A.S. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods 2005, 35, 373–381. [Google Scholar] [CrossRef]
- Costa-Mattioli, M.; Walter, P. The integrated stress response: From mechanism to disease. Science 2020, 368. [Google Scholar] [CrossRef]
- Tabas, I.; Ron, D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 2011, 13, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Tian, M.; Ding, C.; Yu, S. The C/EBP Homologous Protein (CHOP) Transcription Factor Functions in Endoplasmic Reticulum Stress-Induced Apoptosis and Microbial Infection. Front. Immunol. 2019, 9, 3083. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, T.; Imaizumi, K.; Oono, K.; Yui, D.; Gomi, F.; Katayama, T.; Tohyama, M. Activation of Caspase-12, an Endoplastic Reticulum (ER) Resident Caspase, through Tumor Necrosis Factor Receptor-associated Factor 2-dependent Mechanism in Response to the ER Stress *. J. Biol. Chem. 2001, 276, 13935–13940. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Zhu, H.; Morishima, N.; Li, E.; Xu, J.; Yankner, B.A.; Yuan, J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature 2000, 403, 98–103. [Google Scholar] [CrossRef]
- Yang, H.; Niemeijer, M.; van de Water, B.; Beltman, J.B. ATF6 Is a Critical Determinant of CHOP Dynamics during the Unfolded Protein Response. iScience 2020, 23, 100860. [Google Scholar] [CrossRef]
- Xu, Y.; Yu, H.; Qin, H.; Kang, J.; Yu, C.; Zhong, J.; Su, J.; Li, H.; Sun, L. Inhibition of autophagy enhances cisplatin cytotoxicity through endoplasmic reticulum stress in human cervical cancer cells. Cancer Lett. 2012, 314, 232–243. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, C.; Su, J.; Xie, Q.; Ma, L.; Zeng, L.; Yu, Y.; Liu, S.; Li, S.; Li, Z.; et al. Tolerance to endoplasmic reticulum stress mediates cisplatin resistance in human ovarian cancer cells by maintaining endoplasmic reticulum and mitochondrial homeostasis. Oncol. Rep. 2015, 34, 3051–3060. [Google Scholar] [CrossRef]
- Liu, H.; Baliga, R. Endoplasmic Reticulum Stress–Associated Caspase 12 Mediates Cisplatin-Induced LLC-PK1 Cell Apoptosis. J. Am. Soc. Nephrol. 2005, 16, 1985–1992. [Google Scholar] [CrossRef]
- Shen, L.; Wen, N.; Xia, M.; Zhang, Y.; Liu, W.; Xu, Y.; Sun, L. Calcium efflux from the endoplasmic reticulum regulates cisplatin-induced apoptosis in human cervical cancer HeLa cells. Oncol. Lett. 2016, 11, 2411–2419. [Google Scholar] [CrossRef]
- Li, C.; Wei, J.; Li, Y.; He, X.; Zhou, Q.; Yan, J.; Zhang, J.; Liu, Y.; Shu, H.-B. Transmembrane Protein 214 (TMEM214) Mediates Endoplasmic Reticulum Stress-induced Caspase 4 Enzyme Activation and Apoptosis. J. Biol. Chem. 2013, 288, 17908–17917. [Google Scholar] [CrossRef]
- Harwood, S.M.; Yaqoob, M.M.; A Allen, D. Caspase and calpain function in cell death: Bridging the gap between apoptosis and necrosis. Ann. Clin. Biochem. Int. J. Biochem. Lab. Med. 2005, 42, 415–431. [Google Scholar] [CrossRef]
- Danese, A.; Leo, S.; Rimessi, A.; Wieckowski, M.R.; Fiorica, F.; Giorgi, C.; Pinton, P. Cell death as a result of calcium signaling modulation: A cancer-centric prospective. Biochim. et Biophys. Acta (BBA) Mol. Cell Res. 2021, 1868, 119061. [Google Scholar] [CrossRef] [PubMed]
- Giorgi, C.; Marchi, S.; Pinton, P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat. Rev. Mol. Cell Biol. 2018, 19, 713–730. [Google Scholar] [CrossRef] [PubMed]
- Akman, M.; Belisario, D.C.; Salaroglio, I.C.; Kopecka, J.; Donadelli, M.; De Smaele, E.; Riganti, C. Hypoxia, endoplasmic reticulum stress and chemoresistance: Dangerous liaisons. J. Exp. Clin. Cancer Res. 2021, 40, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Liu, R.; Qu, Q. Role of endoplasmic reticulum stress on cisplatin resistance in ovarian carcinoma. Oncol. Lett. 2017, 13, 1437–1443. [Google Scholar] [CrossRef] [PubMed]
- Rashid, H.-O.; Yadav, R.K.; Kim, H.-R.; Chae, H.-J. ER stress: Autophagy induction, inhibition and selection. Autophagy 2015, 11, 1956–1977. [Google Scholar] [CrossRef] [PubMed]
- Burger, K.N.; Staffhorst, R.W.; De Kruijff, B. Interaction of the anti-cancer drug cisplatin with phosphatidylserine in intact and semi-intact cells. Biochim. Biophys. Acta (BBA) -Biomembr. 1999, 1419, 43–54. [Google Scholar] [CrossRef][Green Version]
- Speelmans, G.; Sips, W.H.; Grisel, R.J.; Staffhorst, R.W.; Fichtinger-Schepman, A.M.J.; Reedijk, J.; de Kruijff, B. The interaction of the anti-cancer drug cisplatin with phospholipids is specific for negatively charged phospholipids and takes place at low chloride ion concentration. Biochim. Biophys. Acta (BBA) -Biomembr. 1996, 1283, 60–66. [Google Scholar] [CrossRef][Green Version]
- Suwalsky, M.; Hernández, P.; Villena, F.; Sotomayor, C.P. The Anticancer Drug Cisplatin Interacts with the Human Erythrocyte Membrane. Z. Naturforschung C 2000, 55, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Quist, A.P.; Kumar, S.; Lal, R. Cisplatin Nanoliposomes for Cancer Therapy: AFM and Fluorescence Imaging of Cisplatin Encapsulation, Stability, Cellular Uptake, and Toxicity. Langmuir 2006, 22, 8156–8162. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Lu, J.; Li, R. The events that occur when cisplatin encounters cells. Coord. Chem. Rev. 1996, 151, 53–88. [Google Scholar] [CrossRef]
- Lacour, S.; Hammann, A.; Grazide, S.; Lagadic-Gossmann, D.; Athias, A.; Sergent, O.; Laurent, G.; Gambert, P.; Solary, E.; Dimanche-Boitrel, M.-T. Cisplatin-Induced CD95 Redistribution into Membrane Lipid Rafts of HT29 Human Colon Cancer Cells. Cancer Res. 2004, 64, 3593–3598. [Google Scholar] [CrossRef] [PubMed]
- Rebillard, A.; Tekpli, X.; Meurette, O.; Sergent, O.; LeMoigne-Muller, G.; Vernhet, L.; Gorria, M.; Chevanne, M.; Christmann, M.; Kaina, B.; et al. Cisplatin-Induced Apoptosis Involves Membrane Fluidification via Inhibition of NHE1 in Human Colon Cancer Cells. Cancer Res. 2007, 67, 7865–7874. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, W.; Jia, F.; Ye, J.; Zhao, Y.; Luo, Q.; Zhu, Z.; Wang, F. Cisplatin-induced alteration on membrane composition of A549 cells revealed by ToF-SIMS. Surf. Interface Anal. 2020, 52, 256–263. [Google Scholar] [CrossRef]
- Nganga, R.; Oleinik, N.; Ogretmen, B. Chapter One—Mechanisms of Ceramide-Dependent Cancer Cell Death. In Advances in Cancer Research; Chalfant, C.E., Fisher, P.B., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 140, pp. 1–25. [Google Scholar]
- Fife, C.M.; A McCarroll, J.; Kavallaris, M. Movers and shakers: Cell cytoskeleton in cancer metastasis. Br. J. Pharmacol. 2014, 171, 5507–5523. [Google Scholar] [CrossRef]
- Köpf-Maier, P.; Mühlhausen, S. Changes in the cytoskeleton pattern of tumor cells by cisplatin in vitro. Chem. Interact. 1992, 82, 295–316. [Google Scholar] [CrossRef]
- Zeidan, Y.H.; Jenkins, R.W.; Hannun, Y.A. Remodeling of cellular cytoskeleton by the acid sphingomyelinase/ceramide pathway. J. Cell Biol. 2008, 181, 335–350. [Google Scholar] [CrossRef]
- Min, Y.J.; Poruchynsky, M.S.; Sackett, D.L.; Murphy, B.; Fojo, T. Cisplatin markedly enhances microtubule depolymerization in A549 cell line compared with oxaliplatin. Cancer Res. 2008, 68 (Suppl. 9), 3335. [Google Scholar]
- Tulub, A.A.; Stefanov, V.E. Cisplatin stops tubulin assembly into microtubules. A new insight into the mechanism of antitumor activity of platinum complexes. Int. J. Biol. Macromol. 2001, 28, 191–198. [Google Scholar] [CrossRef]
- Raudenska, M.; Kratochvilova, M.; Vicar, T.; Gumulec, J.; Balvan, J.; Polanska, H.; Pribyl, J.; Masarik, M. Cisplatin enhances cell stiffness and decreases invasiveness rate in prostate cancer cells by actin accumulation. Sci. Rep. 2019, 9, 1660. [Google Scholar] [CrossRef]
- Pierson-Marchandise, M.; Gras, V.; Moragny, J.; Micallef, J.; Gaboriau, L.; Picard, S.; Choukroun, G.; Masmoudi, K.; Liabeuf, S.; The French National Network of Pharmacovigilance Centres. The drugs that mostly frequently induce acute kidney injury: A case − noncase study of a pharmacovigilance database. Br. J. Clin. Pharmacol. 2017, 83, 1341–1349. [Google Scholar] [CrossRef]
- Hoek, J.; Bloemendal, K.M.; van der Velden, L.A.; van Diessen, J.N.; van Werkhoven, E.; Klop, W.M.; Tesselaar, M.E. Nephrotoxicity as a Dose-Limiting Factor in a High-Dose Cisplatin-Based Chemoradiotherapy Regimen for Head and Neck Carcinomas. Cancers 2016, 8, 21. [Google Scholar] [CrossRef]
- Hanigan, M.H.; Devarajan, P. Cisplatin nephrotoxicity: Molecular mechanisms. Cancer Ther. 2003, 1, 47–61. [Google Scholar]
- Gonzalez-Vitale, J.C.; Hayes, D.M.; Cvitkovic, E.; Sternberg, S.S. The renal pathology in clinical trials of Cis-platinum (II) diamminedichloride. Cancer 1977, 39, 1362–1371. [Google Scholar] [CrossRef]
- Arany, I.; Safirstein, R.L. Cisplatin nephrotoxicity. Semin. Nephrol. 2003, 23, 460–464. [Google Scholar] [CrossRef]
- Megyesi, J.; Safirstein, R.L.; Price, P.M. Induction of p21WAF1/CIP1/SDI1 in kidney tubule cells affects the course of cisplatin-induced acute renal failure. J. Clin. Investig. 1998, 101, 777–782. [Google Scholar] [CrossRef]
- Tsuruya, K.; Ninomiya, T.; Tokumoto, M.; Hirakawa, M.; Masutani, K.; Taniguchi, M.; Fukuda, K.; Kanai, H.; Kishihara, K.; Hirakata, H.; et al. Direct involvement of the receptor-mediated apoptotic pathways in cisplatin-induced renal tubular cell death. Kidney Int. 2003, 63, 72–82. [Google Scholar] [CrossRef]
- Baliga, R.; Ueda, N.; Walker, P.D.; Shah, S.V. Oxidant Mechanisms in Toxic Acute Renal Failure*. Drug Metab. Rev. 1999, 31, 971–997. [Google Scholar] [CrossRef]
- Ramesh, G.; Reeves, W.B. Cisplatin Increases TNF-α mRNA Stability in Kidney Proximal Tubule Cells. Ren. Fail. 2006, 28, 583–592. [Google Scholar] [CrossRef]
- Ramesh, G.; Reeves, W.B. TNF-α mediates chemokine and cytokine expression and renal injury in cisplatin nephrotoxicity. J. Clin. Investig. 2002, 110, 835–842. [Google Scholar] [CrossRef]
- Ramesh, G.; Reeves, W. TNFR2-mediated apoptosis and necrosis in cisplatin-induced acute renal failure. Am. J. Physiol. Physiol. 2003, 285, F610–F618. [Google Scholar] [CrossRef]
- Winston, J.A.; Safirstein, R. Reduced renal blood flow in early cisplatin-induced acute renal failure in the rat. Am. J. Physiol. Physiol. 1985, 249, F490–F496. [Google Scholar] [CrossRef]
- Offerman, J.J.G.; Meijer, S.; Sleijfer, D.T.; Mulder, N.H.; Donker, A.J.M.; Koops, H.S.; Van Der Hem, G.K. Acute effects of cis-diamminedichloroplatinum (CDDP) on renal function. Cancer Chemother. Pharmacol. 1984, 12, 36–38. [Google Scholar] [CrossRef]
- Callejo, A.; Sedó-Cabezón, L.; Juan, I.D.; Llorens, J. Cisplatin-Induced Ototoxicity: Effects, Mechanisms and Protection Strategies. Toxics 2015, 3, 268–293. [Google Scholar] [CrossRef]
- Knight, K.R.G.; Kraemer, D.F.; Neuwelt, E.A. Ototoxicity in Children Receiving Platinum Chemotherapy: Underestimating a Commonly Occurring Toxicity That May Influence Academic and Social Development. J. Clin. Oncol. 2005, 23, 8588–8596. [Google Scholar] [CrossRef]
- Kushner, B.H.; Budnick, A.; Kramer, K.; Modak, S.; Cheung, N.-K.V. Ototoxicity from high-dose use of platinum compounds in patients with neuroblastoma. Cancer 2006, 107, 417–422. [Google Scholar] [CrossRef]
- Kopke, R.; Allen, K.A.; Henderson, D.; Hoffer, M.; Frenz, D.; VAN DE Water, T. A Radical Demise: Toxins and Trauma Share Common Pathways in Hair Cell Death. Ann. N. Y. Acad. Sci. 1999, 884, 171–191. [Google Scholar] [CrossRef]
- Breglio, A.M.; Rusheen, A.E.; Shide, E.D.; Fernandez, K.A.; Spielbauer, K.K.; McLachlin, K.M.; Hall, M.D.; Amable, L.; Cunningham, L.L. Cisplatin is retained in the cochlea indefinitely following chemotherapy. Nat. Commun. 2017, 8, 1654. [Google Scholar] [CrossRef]
- Filipski, K.K.; Mathijssen, R.H.; Mikkelsen, T.S.; Schinkel, A.H.; Sparreboom, A. Contribution of Organic Cation Transporter 2 (OCT2) to Cisplatin-Induced Nephrotoxicity. Clin. Pharmacol. Ther. 2009, 86, 396–402. [Google Scholar] [CrossRef]
- Ciarimboli, G.; Deuster, D.; Knief, A.; Sperling, M.; Holtkamp, M.; Edemir, B.; Pavenstädt, H.; Lanvers-Kaminsky, C.; am Zehnhoff-Dinnesen, A.; Schinkel, A.H.; et al. Organic cation transporter 2 mediates cisplatin-induced oto- and nephrotoxicity and is a target for protective interventions. Am. J. Pathol. 2010, 176, 1169–1180. [Google Scholar] [CrossRef]
- Gregg, R.W.; Molepo, J.M.; Monpetit, V.J.; Mikael, N.Z.; Redmond, D.; Gadia, M.; Stewart, D.J. Cisplatin neurotoxicity: The relationship between dosage, time, and platinum concentration in neurologic tissues, and morphologic evidence of toxicity. J. Clin. Oncol. 1992, 10, 795–803. [Google Scholar] [CrossRef]
- Starobova, H.; Vetter, I. Pathophysiology of Chemotherapy-Induced Peripheral Neuropathy. Front. Mol. Neurosci. 2017, 10, 174. [Google Scholar] [CrossRef]
- Lomonaco, M.; Milone, M.; Batocchi, A.P.; Padua, L.; Restuccia, D.; Tonali, P. Cisplatin neuropathy: Clinical course and neurophysiological findings. J. Neurol. 1992, 239, 199–204. [Google Scholar] [CrossRef]
- Siegal, T.; Haim, N. Cisplatin-induced peripheral neuropathy. Frequent off-therapy deterioration, demyelinating syndromes, and muscle cramps. Cancer 1990, 66, 1117–1123. [Google Scholar] [CrossRef]
- Kerckhove, N.; Collin, A.; Condé, S.; Chaleteix, C.; Pezet, D.; Balayssac, D. Long-Term Effects, Pathophysiological Mechanisms, and Risk Factors of Chemotherapy-Induced Peripheral Neuropathies: A Comprehensive Literature Review. Front. Pharmacol. 2017, 8, 86. [Google Scholar] [CrossRef]
- Cavalli, F.; Tschopp, L.; Sonntag, R.W.; Zimmermann, A. A case of liver toxicity following cis-dichlorodiammineplatinum(II) treatment. Cancer Treat. Rep. 1978, 62, 2125–2126. [Google Scholar]
- Hu, Y.; Sun, B.; Zhao, B.; Mei, D.; Gu, Q.; Tian, Z. Cisplatin-induced cardiotoxicity with midrange ejection fraction: A case report and review of the literature. Medicine 2018, 97, e13807. [Google Scholar] [CrossRef]
- Astolfi, L.; Ghiselli, S.; Guaran, V.; Chicca, M.; Simoni, E.; Olivetto, E.; Lelli, G.; Martini, A. Correlation of adverse effects of cisplatin administration in patients affected by solid tumours: A retrospective evaluation. Oncol. Rep. 2013, 29, 1285–1292. [Google Scholar] [CrossRef]
- Batchelor, D. Hair and cancer chemotherapy: Consequences and nursing care—A literature study. Eur. J. Cancer Care 2001, 10, 147–163. [Google Scholar] [CrossRef]
- Kartalou, M.; Essigmann, J.M. Mechanisms of resistance to cisplatin. Mutation Res. /Fundam. Molecular Mech. Mutagen. 2001, 478, 23–43. [Google Scholar] [CrossRef] [PubMed]
- Kilari, D.; Guancial, E.; Kim, E.S. Role of copper transporters in platinum resistance. World J. Clin. Oncol. 2016, 7, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Öhrvik, H.; Logeman, B.; Turk, B.; Reinheckel, T.; Thiele, D.J. Cathepsin Protease Controls Copper and Cisplatin Accumulation via Cleavage of the Ctr1 Metal-binding Ectodomain. J. Biol. Chem. 2016, 291, 13905–13916. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Matsumoto, H.; Yamada, S.; Kawai, K.; Suemizu, H.; Gika, M.; Takanami, I.; Iwazaki, M.; Nakamura, M. Association of ATP7A expression and in vitro sensitivity to cisplatin in non-small cell lung cancer. Oncol. Lett. 2010, 1, 837–840. [Google Scholar] [CrossRef] [PubMed]
- Samimi, G.; Varki, N.M.; Wilczynski, S.; Safaei, R.; Alberts, D.S.; Howell, S.B. Increase in expression of the copper transporter ATP7A during platinum drug-based treatment is associated with poor survival in ovarian cancer patients. Clin. Cancer Res. 2003, 9, 5853–5859. [Google Scholar] [PubMed]
- Yang, T.; Chen, M.; Chen, T.; Thakur, A. Expression of the copper transporters hCtr1, ATP7A and ATP7B is associated with the response to chemotherapy and survival time in patients with resected non-small cell lung cancer. Oncol. Lett. 2015, 10, 2584–2590. [Google Scholar] [CrossRef]
- Song, L.; Li, Y.; Li, W.; Wu, S.; Li, Z. miR-495 Enhances the Sensitivity of Non-Small Cell Lung Cancer Cells to Platinum by Modulation of Copper-Transporting P-type Adenosine Triphosphatase A (ATP7A). J. Cell. Biochem. 2013, 115, 1234–1242. [Google Scholar] [CrossRef]
- Yu, Z.; Cao, W.; Ren, Y.; Zhang, Q.; Liu, J. ATPase copper transporter A, negatively regulated by miR-148a-3p, contributes to cisplatin resistance in breast cancer cells. Clin. Transl. Med. 2020, 10, 57–73. [Google Scholar] [CrossRef]
- Hinoshita, E.; Uchiumi, T.; Taguchi, K.; Kinukawa, N.; Tsuneyoshi, M.; Maehara, Y.; Sugimachi, K.; Kuwano, M. Increased expression of an ATP-binding cassette superfamily transporter, multidrug resistance protein 2, in human colorectal carcinomas. Clin. Cancer Res. 2000, 6, 2401–2407. [Google Scholar]
- Yamasaki, M.; Makino, T.; Masuzawa, T.; Kurokawa, Y.; Miyata, H.; Takiguchi, S.; Nakajima, K.; Fujiwara, Y.; Matsuura, N.; Mori, M.; et al. Role of multidrug resistance protein 2 (MRP2) in chemoresistance and clinical outcome in oesophageal squamous cell carcinoma. Br. J. Cancer 2011, 104, 707–713. [Google Scholar] [CrossRef]
- Amable, L. Cisplatin resistance and opportunities for precision medicine. Pharmacol. Res. 2016, 106, 27–36. [Google Scholar] [CrossRef]
- Rudin, C.M.; Yang, Z.; Schumaker, L.M.; VanderWeele, D.; Newkirk, K.; Egorin, M.J.; Zuhowski, E.G.; Cullen, K.J. Inhibition of glutathione synthesis reverses Bcl-2-mediated cisplatin resistance. Cancer Res. 2003, 63, 312–318. [Google Scholar]
- Chen, H.H.W.; Kuo, M.T. Role of Glutathione in the Regulation of Cisplatin Resistance in Cancer Chemotherapy. Met. Based Drugs 2010, 2010, 430939. [Google Scholar] [CrossRef]
- Byun, S.-S.; Kim, S.W.; Choi, H.; Lee, C.; Lee, E. Augmentation of cisplatin sensitivity in cisplatin-resistant human bladder cancer cells by modulating glutathione concentrations and glutathione-related enzyme activities. BJU Int. 2005, 95, 1086–1090. [Google Scholar] [CrossRef]
- Rocha, C.R.R.; Garcia, C.C.M.; Vieira, D.B.; Quinet, A.; de Andrade-Lima, L.C.; Munford, V.; Belizário, J.E.; Menck, C.F.M. Glutathione depletion sensitizes cisplatin- and temozolomide-resistant glioma cells in vitro and in vivo. Cell Death Dis. 2014, 5, e1505. [Google Scholar] [CrossRef]
- Hayden, A.; Douglas, J.; Sommerlad, M.; Andrews, L.; Gould, K.; Hussain, S.; Thomas, G.J.; Packham, G.; Crabb, S.J. The Nrf2 transcription factor contributes to resistance to cisplatin in bladder cancer. Urol. Oncol. Semin. Orig. Investig. 2014, 32, 806–814. [Google Scholar] [CrossRef]
- Li, D.; Hong, X.; Zhao, F.; Ci, X.; Zhang, S. Targeting Nrf2 may reverse the drug resistance in ovarian cancer. Cancer Cell Int. 2021, 21, 1–10. [Google Scholar] [CrossRef]
- Wangy, X.-J.; Suny, Z.; Villeneuve, N.F.; Zhang, S.; Zhao, F.; Li, Y.; Chen, W.; Yi, X.; Zheng, W.; Wondrak, G.T.; et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinog. 2008, 29, 1235–1243. [Google Scholar] [CrossRef]
- Jiang, T.; Harder, B.; Rojo de la Vega, M.; Wong, P.K.; Chapman, E.; Zhang, D.D. p62 links autophagy and Nrf2 signaling. Free Radic. Biol. Med. 2015, 88 Pt B, 199–204. [Google Scholar] [CrossRef]
- Yu, H.; Su, J.; Xu, Y.; Kang, J.; Li, H.; Zhang, L.; Yi, H.; Xiang, X.; Liu, F.; Sun, L. p62/SQSTM1 involved in cisplatin resistance in human ovarian cancer cells by clearing ubiquitinated proteins. Eur. J. Cancer 2011, 47, 1585–1594. [Google Scholar] [CrossRef]
- Bao, L.-J.; Jaramillo, M.C.; Zhang, Z.-B.; Zheng, Y.-X.; Yao, M.; Zhang, D.D.; Yi, X.-F. Nrf2 induces cisplatin resistance through activation of autophagy in ovarian carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 1502–1513. [Google Scholar]
- Rocha, C.R.R.; Silva, M.M.; Quinet, A.; Cabral-Neto, J.B.; Menck, C.F.M. DNA repair pathways and cisplatin resistance: An intimate relationship. Clinics 2018, 73, e478s. [Google Scholar] [CrossRef]
- Schärer, O.D. Nucleotide Excision Repair in Eukaryotes. Cold Spring Harb. Perspect. Biol. 2013, 5, a012609. [Google Scholar] [CrossRef]
- Duan, M.; Ulibarri, J.; Liu, K.J.; Mao, P. Role of Nucleotide Excision Repair in Cisplatin Resistance. Int. J. Mol. Sci. 2020, 21, 9248. [Google Scholar] [CrossRef]
- Ng, J.M.; Vermeulen, W.; van der Horst, G.T.; Bergink, S.; Sugasawa, K.; Vrieling, H.; Hoeijmakers, J.H. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003, 17, 1630–1645. [Google Scholar] [CrossRef]
- Nocentini, S.; Coin, F.; Saijo, M.; Tanaka, K.; Egly, J.-M. DNA Damage Recognition by XPA Protein Promotes Efficient Recruitment of Transcription Factor II H. J. Biol. Chem. 1997, 272, 22991–22994. [Google Scholar] [CrossRef]
- Yokoi, M.; Masutani, C.; Maekawa, T.; Sugasawa, K.; Ohkuma, Y.; Hanaoka, F. The Xeroderma Pigmentosum Group C Protein Complex XPC-HR23B Plays an Important Role in the Recruitment of Transcription Factor IIH to Damaged DNA. J. Biol. Chem. 2000, 275, 9870–9875. [Google Scholar] [CrossRef]
- Oksenych, V.; de Jesus, B.B.; Zhovmer, A.; Egly, J.-M.; Coin, F. Molecular insights into the recruitment of TFIIH to sites of DNA damage. EMBO J. 2009, 28, 2971–2980. [Google Scholar] [CrossRef]
- Coin, F.; Oksenych, V.; Egly, J.-M. Distinct Roles for the XPB/p52 and XPD/p44 Subcomplexes of TFIIH in Damaged DNA Opening during Nucleotide Excision Repair. Mol. Cell 2007, 26, 245–256. [Google Scholar] [CrossRef]
- Houtsmuller, A.B.; Rademakers, S.; Nigg, A.L.; Hoogstraten, D.; Hoeijmakers, J.H.; Vermeulen, W. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science 1999, 284, 958–961. [Google Scholar] [CrossRef]
- Ito, S.; Kuraoka, I.; Chymkowitch, P.; Compe, E.; Takedachi, A.; Ishigami, C.; Coin, F.; Egly, J.-M.; Tanaka, K. XPG Stabilizes TFIIH, Allowing Transactivation of Nuclear Receptors: Implications for Cockayne Syndrome in XP-G/CS Patients. Mol. Cell 2007, 26, 231–243. [Google Scholar] [CrossRef]
- Kemp, M.G.; Reardon, J.T.; Lindsey-Boltz, L.; Sancar, A. Mechanism of Release and Fate of Excised Oligonucleotides during Nucleotide Excision Repair. J. Biol. Chem. 2012, 287, 22889–22899. [Google Scholar] [CrossRef]
- Ogi, T.; Limsirichaikul, S.; Overmeer, R.M.; Volker, M.; Takenaka, K.; Cloney, R.; Nakazawa, Y.; Niimi, A.; Miki, Y.; Jaspers, N.G.; et al. Three DNA Polymerases, Recruited by Different Mechanisms, Carry Out NER Repair Synthesis in Human Cells. Mol. Cell 2010, 37, 714–727. [Google Scholar] [CrossRef] [PubMed]
- Ferry, K.V.; Hamilton, T.C.; Johnson, S.W. Increased nucleotide excision repair in cisplatin-resistant ovarian cancer cells: Role of ercc1–xpf. Biochem. Pharmacol. 2000, 60, 1305–1313. [Google Scholar] [CrossRef]
- Prakash, R.; Zhang, Y.; Feng, W.; Jasin, M. Homologous Recombination and Human Health: The Roles of BRCA1, BRCA2, and Associated Proteins. Cold Spring Harb. Perspect. Biol. 2015, 7, a016600. [Google Scholar] [CrossRef]
- Toh, M.; Ngeow, J. Homologous Recombination Deficiency: Cancer Predispositions and Treatment Implications. Oncologist 2021, 26, e1526–e1537. [Google Scholar] [CrossRef]
- Sakai, W.; Swisher, E.M.; Karlan, B.Y.; Agarwal, M.K.; Higgins, J.; Friedman, C.; Villegas, E.; Jacquemont, C.; Farrugia, D.J.; Couch, F.J.; et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 2008, 451, 1116–1120. [Google Scholar] [CrossRef]
- Swisher, E.M.; Sakai, W.; Karlan, B.Y.; Wurz, K.; Urban, N.; Taniguchi, T. Secondary BRCA1 Mutations in BRCA1-Mutated Ovarian Carcinomas with Platinum Resistance. Cancer Res. 2008, 68, 2581–2586. [Google Scholar] [CrossRef]
- Sawant, A.; Kothandapani, A.; Zhitkovich, A.; Sobol, R.W.; Patrick, S.M. Role of mismatch repair proteins in the processing of cisplatin interstrand cross-links. DNA Repair 2015, 35, 126–136. [Google Scholar] [CrossRef]
- Yamada, M.; O’Regan, E.; Brown, R.; Karran, P. Selective recognition of a cisplatin-DNA adduct by human mismatch repair proteins. Nucleic Acids Res. 1997, 25, 491–496. [Google Scholar] [CrossRef]
- Stojic, L.; Brun, R.; Jiricny, J. Mismatch repair and DNA damage signalling. DNA Repair 2004, 3, 1091–1101. [Google Scholar] [CrossRef]
- Topping, R.P.; Wilkinson, J.C.; Scarpinato, K.D. Mismatch Repair Protein Deficiency Compromises Cisplatin-induced Apoptotic Signaling. J. Biol. Chem. 2009, 284, 14029–14039. [Google Scholar] [CrossRef] [PubMed]
- Aebi, S.; Kurdi-Haidar, B.; Gordon, R.; Cenni, B.; Zheng, H.; Fink, D.; Christen, R.D.; Boland, C.R.; Koi, M.; Fishel, R.; et al. Loss of DNA mismatch repair in acquired resistance to cisplatin. Cancer Res. 1996, 56, 3087–3090. [Google Scholar] [PubMed]
- Fink, D.; Zheng, H.; Nebel, S.; Norris, P.S.; Aebi, S.; Lin, T.P.; Nehmé, A.; Christen, R.D.; Haas, M.; MacLeod, C.L.; et al. In vitro and in vivo resistance to cisplatin in cells that have lost DNA mismatch repair. Cancer Res. 1997, 57, 1841–1845. [Google Scholar] [PubMed]
- Fink, D.; Nebel, S.; Norris, P.S.; Baergen, R.N.; Wilczynski, S.P.; Costa, M.J.; Haas, M.; Cannistra, S.A.; Howell, S.B. Enrichment for DNA mismatch repair-deficient cells during treatment with cisplatin. Int. J. Cancer 1998, 77, 741–746. [Google Scholar] [CrossRef]
- Monneret, C. Platinum anticancer drugs. From serendipity to rational design. Annales Pharmaceutiques Françaises 2011, 69, 286–295. [Google Scholar] [CrossRef]
- Schoch, S.; Gajewski, S.; Rothfuß, J.; Hartwig, A.; Köberle, B. Comparative Study of the Mode of Action of Clinically Approved Platinum-Based Chemotherapeutics. Int. J. Mol. Sci. 2020, 21, 6928. [Google Scholar] [CrossRef] [PubMed]
- van der Vijgh, W.J. Clinical pharmacokinetics of carboplatin. Clin. Pharmacokinet 1991, 21, 242–261. [Google Scholar] [CrossRef]
- Vasey, P.A.; Jayson, G.C.; Gordon, A.; Gabra, H.; Coleman, R.; Atkinson, R.; Parkin, D.; Paul, J.; Hay, A.; Kaye, S.B. Phase III Randomized Trial of Docetaxel-Carboplatin Versus Paclitaxel-Carboplatin as First-line Chemotherapy for Ovarian Carcinoma. JNCI 2004, 96, 1682–1691. [Google Scholar] [CrossRef]
- Swenerton, K.; Jeffrey, J.; Stuart, G.; Roy, M.; Krepart, G.; Carmichael, J.; Drouin, P.; Stanimir, R.; O’Connell, G.; MacLean, G.; et al. Cisplatin-cyclophosphamide versus carboplatin-cyclophosphamide in advanced ovarian cancer: A randomized phase III study of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 1992, 10, 718–726. [Google Scholar] [CrossRef]
- Doz, F.; Neuenschwander, S.; Plantaz, D.; Courbon, B.; Gentet, J.C.; Bouffet, E.; Mosseri, V.; Vannier, J.P.; Mechinaud, F.; Desjardins, L.; et al. Etoposide and carboplatin in extraocular retinoblastoma: A study by the Societe Francaise d'Oncologie Pediatrique. J. Clin. Oncol. 1995, 13, 902–909. [Google Scholar] [CrossRef]
- Robert, N.; Leyland-Jones, B.; Asmar, L.; Belt, R.; Ilegbodu, D.; Loesch, D.; Raju, R.; Valentine, E.; Sayre, R.; Cobleigh, M.; et al. Randomized phase III study of trastuzumab, paclitaxel, and carboplatin compared with trastuzumab and paclitaxel in women with HER-2-overexpressing metastatic breast cancer. J. Clin. Oncol. 2006, 24, 2786–2792. [Google Scholar] [CrossRef]
- Miller, D.S.; Filiaci, V.L.; Mannel, R.S.; Cohn, D.E.; Matsumoto, T.; Tewari, K.S.; DiSilvestro, P.; Pearl, M.L.; Argenta, P.A.; Powell, M.A.; et al. Carboplatin and Paclitaxel for Advanced Endometrial Cancer: Final Overall Survival and Adverse Event Analysis of a Phase III Trial (NRG Oncology/GOG0209). J. Clin. Oncol. 2020, 38, 3841–3850. [Google Scholar] [CrossRef]
- Martin, L.P.; Hamilton, T.C.; Schilder, R.J. Platinum Resistance: The Role of DNA Repair Pathways. Clin. Cancer Res. 2008, 14, 1291–1295. [Google Scholar] [CrossRef]
- Soori, H.; Rabbani-Chadegani, A.; Davoodi, J. Exploring binding affinity of oxaliplatin and carboplatin, to nucleoprotein structure of chromatin: Spectroscopic study and histone proteins as a target. Eur. J. Med. Chem. 2015, 89, 844–850. [Google Scholar] [CrossRef]
- Chaney, S.G.; Campbell, S.; Temple, B.; Bassett, E.; Wu, Y.; Faldu, M. Protein interactions with platinum–DNA adducts: From structure to function. J. Inorg. Biochem. 2004, 98, 1551–1559. [Google Scholar] [CrossRef]
- Teuben, J.-M.; Bauer, C.; Wang, A.H.J.; Reedijk, J. Solution Structure of a DNA Duplex Containing a cis-Diammineplatinum(II) 1,3-d(GTG) Intrastrand Cross-Link, a Major Adduct in Cells Treated with the Anticancer Drug Carboplatin. Biochemistry 1999, 38, 12305–12312. [Google Scholar] [CrossRef]
- Stefanou, D.T.; Souliotis, V.L.; Zakopoulou, R.; Liontos, M.; Bamias, A. DNA Damage Repair: Predictor of Platinum Efficacy in Ovarian Cancer? Biomedicines 2021, 10, 8. [Google Scholar] [CrossRef]
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005, 4, 307–320. [Google Scholar] [CrossRef]
- Rabik, C.A.; Dolan, M.E. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat. Rev. 2007, 33, 9–23. [Google Scholar] [CrossRef]
- Kilic, A.; Barlak, N.; Sanli, F.; Aytatli, A.; Capik, O.; Karatas, O.F. Mode of action of carboplatin via activating p53/miR-145 axis in head and neck cancers. Laryngoscope 2020, 130, 2818–2824. [Google Scholar] [CrossRef]
- Brozovic, A.; Vuković, L.; Polančac, D.S.; Arany, I.; Köberle, B.; Fritz, G.; Fiket, Ž.; Majhen, D.; Ambriović-Ristov, A.; Osmak, M. Endoplasmic Reticulum Stress Is Involved in the Response of Human Laryngeal Carcinoma Cells to Carboplatin but Is Absent in Carboplatin-Resistant Cells. PLoS ONE 2013, 8, e76397. [Google Scholar] [CrossRef][Green Version]
- Shen, B.; Mao, W.; Ahn, J.; Chung, P.; He, P. Mechanism of HN-3 cell apoptosis induced by carboplatin: Combination of mitochondrial pathway associated with Ca2+ and the nucleus pathways. Mol. Med. Rep. 2018, 18, 4978–4986. [Google Scholar] [CrossRef]
- Kashyap, D.; Garg, V.K.; Goel, N. Chapter Four—Intrinsic and extrinsic pathways of apoptosis: Role in cancer development and prognosis. In Advances in Protein Chemistry and Structural Biology; Donev, R., Ed.; Academic Press: Cambridge, MA, USA, 2021; Volume 125, pp. 73–120. [Google Scholar]
- David, K.K.; Andrabi, S.A.; Dawson, T.M.; Dawson, V.L. Parthanatos, a messenger of death. Front Biosci. 2009, 14, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- A Fatokun, A.; Dawson, V.L.; Dawson, T.M. Parthanatos: Mitochondrial-linked mechanisms and therapeutic opportunities. Br. J. Pharmacol. 2014, 171, 2000–2016. [Google Scholar] [CrossRef]
- Andrabi, S.A.; Kim, N.S.; Yu, S.-W.; Wang, H.; Koh, D.W.; Sasaki, M.; Klaus, J.A.; Otsuka, T.; Zhang, Z.; Koehler, R.C.; et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. USA 2006, 103, 18308–18313. [Google Scholar] [CrossRef]
- Cheng, Y.-J.; Wu, R.; Cheng, M.-L.; Du, J.; Hu, X.-W.; Yu, L.; Zhao, X.-K.; Yao, Y.-M.; Long, Q.-Z.; Zhu, L.-L.; et al. Carboplatin-induced hematotoxicity among patients with non-small cell lung cancer: Analysis on clinical adverse events and drug-gene interactions. Oncotarget 2016, 8, 32228–32236. [Google Scholar] [CrossRef][Green Version]
- Gore, M.; Fryatt, I.; Wiltshaw, E.; Dawson, T.; Robinson, B.; Calvert, A. Cisplatin/carboplatin cross-resistance in ovarian cancer. Br. J. Cancer 1989, 60, 767–769. [Google Scholar] [CrossRef]
- Pandey, A.; Bhosale, B.; Pandita, V.; Singh, A.; Ghosh, J.; Ghosh, J.; Bajpai, J. Carboplatin hypersensitivity in relapsed ovarian carcinoma: A therapeutic challenge. Indian J. Med. Paediatr. Oncol. 2014, 35, 17–20. [Google Scholar] [CrossRef]
- Stewart, D.J. Mechanisms of resistance to cisplatin and carboplatin. Crit. Rev. Oncol. 2007, 63, 12–31. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, G.F.; Wlodarczyk, S.R.; Monteiro, G. Carboplatin: Molecular mechanisms of action associated with chemoresistance. Braz. J. Pharm. Sci. 2014, 50, 693–701. [Google Scholar] [CrossRef]
- Gavande, N.S.; VanderVere-Carozza, P.S.; Hinshaw, H.D.; Jalal, S.I.; Sears, C.R.; Pawelczak, K.S.; Turchi, J.J. DNA repair targeted therapy: The past or future of cancer treatment? Pharmacol. Ther. 2016, 160, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, L. DNA crosslinking damage and cancer—A tale of friend and foe. Transl. Cancer Res. 2013, 2, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Wheate, N.J.; Walker, S.; Craig, G.E.; Oun, R. The status of platinum anticancer drugs in the clinic and in clinical trials. Dalton Trans. 2010, 39, 8113–8127. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.; Muhsin, M.; Kirkpatrick, P. Oxaliplatin. Nature Rev. Drug Discov. 2004, 3, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Bogliolo, S.; Cassani, C.; Gardella, B.; Musacchi, V.; Babilonti, L.; Venturini, P.-L.; Ferrero, S.; Spinillo, A. Oxaliplatin for the treatment of ovarian cancer. Expert Opin. Investig. Drugs 2015, 24, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Balibrea, E.; Martínez-Cardús, A.; Ginés, A.; de Porras, V.R.; Moutinho, C.; Layos, L.; Manzano, J.L.; Bugés, C.; Bystrup, S.; Esteller, M.; et al. Tumor-Related Molecular Mechanisms of Oxaliplatin Resistance. Mol. Cancer Ther. 2015, 14, 1767–1776. [Google Scholar] [CrossRef]
- Zhang, H.-Y.; Liu, Y.-R.; Ji, C.; Li, W.; Dou, S.-X.; Xie, P.; Wang, W.-C.; Zhang, L.-Y.; Wang, P.-Y. Oxaliplatin and Its Enantiomer Induce Different Condensation Dynamics of Single DNA Molecules. PLoS ONE 2013, 8, e71556. [Google Scholar] [CrossRef]
- Malina, J.; Novakova, O.; Vojtiskova, M.; Natile, G.; Brabec, V. Conformation of DNA GG Intrastrand Cross-Link of Antitumor Oxaliplatin and Its Enantiomeric Analog. Biophys. J. 2007, 93, 3950–3962. [Google Scholar] [CrossRef]
- Zhang, C.-M.; Lv, J.-F.; Gong, L.; Yu, L.-Y.; Chen, X.-P.; Zhou, H.-H.; Fan, L. Role of Deficient Mismatch Repair in the Personalized Management of Colorectal Cancer. Int. J. Environ. Res. Public Heal. 2016, 13, 892. [Google Scholar] [CrossRef]
- Ahmad, S. Platinum-DNA Interactions and Subsequent Cellular Processes Controlling Sensitivity to Anticancer Platinum Complexes. Chem. Biodivers. 2010, 7, 543–566. [Google Scholar] [CrossRef]
- Arnould, S.; Hennebelle, I.; Canal, P.; Bugat, R.; Guichard, S. Cellular determinants of oxaliplatin sensitivity in colon cancer cell lines. Eur. J. Cancer 2003, 39, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Devanabanda, B.; Kasi, A. StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Alcindor, T.; Beauger, N. Oxaliplatin: A Review in the Era of Molecularly Targeted Therapy. Curr. Oncol. 2011, 18, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Demols, A.; Peeters, M.; Polus, M.; Marechal, R.; Gay, F.; Monsaert, E.; Hendlisz, A.; Van Laethem, J.L. Gemcitabine and oxaliplatin (GEMOX) in gemcitabine refractory advanced pancreatic adenocarcinoma: A phase II study. Br. J. Cancer 2006, 94, 481–485. [Google Scholar] [CrossRef]
- Noordhuis, P.; Laan, A.C.; van de Born, K.; Honeywell, R.J.; Peters, G.J. Coexisting Molecular Determinants of Acquired Oxaliplatin Resistance in Human Colorectal and Ovarian Cancer Cell Lines. Int. J. Mol. Sci. 2019, 20, 3619. [Google Scholar] [CrossRef]
- Hah, S.S.; Sumbad, R.A.; White, R.W.D.V.; Turteltaub, K.W.; Henderson, P.T. Characterization of Oxaliplatin−DNA Adduct Formation in DNA and Differentiation of Cancer Cell Drug Sensitivity at Microdose Concentrations. Chem. Res. Toxicol. 2007, 20, 1745–1751. [Google Scholar] [CrossRef] [PubMed]
- Holzer, A.K.; Manorek, G.H.; Howell, S.B. Contribution of the Major Copper Influx Transporter CTR1 to the Cellular Accumulation of Cisplatin, Carboplatin, and Oxaliplatin. Mol. Pharmacol. 2006, 70, 1390–1394. [Google Scholar] [CrossRef] [PubMed]
- Larson, C.A.; Blair, B.G.; Safaei, R.; Howell, S.B. The Role of the Mammalian Copper Transporter 1 in the Cellular Accumulation of Platinum-Based Drugs. Mol. Pharmacol. 2008, 75, 324–330. [Google Scholar] [CrossRef]
- Yonezawa, A.; Masuda, S.; Yokoo, S.; Katsura, T.; Inui, K.-I. Cisplatin and Oxaliplatin, but Not Carboplatin and Nedaplatin, Are Substrates for Human Organic Cation Transporters (SLC22A1–3 and Multidrug and Toxin Extrusion Family). J. Pharmacol. Exp. Ther. 2006, 319, 879–886. [Google Scholar] [CrossRef]
- Fujita, S.; Hirota, T.; Sakiyama, R.; Baba, M.; Ieiri, I. Identification of drug transporters contributing to oxaliplatin-induced peripheral neuropathy. J. Neurochem. 2018, 148, 373–385. [Google Scholar] [CrossRef]
- Woynarowski, J.M.; Faivre, S.; Herzig, M.C.; Arnett, B.; Chapman, W.G.; Trevino, A.V.; Raymond, E.; Chaney, S.G.; Vaisman, A.; Varchenko, M.; et al. Oxaliplatin-Induced Damage of Cellular DNA. Mol. Pharmacol. 2000, 58, 920–927. [Google Scholar] [CrossRef]
- Woynarowski, J.M.; Chapman, W.G.; Napier, C.; Herzig, M.C.S.; Juniewicz, P. Sequence- and Region-Specificity of Oxaliplatin Adducts in Naked and Cellular DNA. Mol. Pharmacol. 1998, 54, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Faivre, S.; Chan, D.; Salinas, R.; Woynarowska, B.; Woynarowski, J.M. DNA strand breaks and apoptosis induced by oxaliplatin in cancer cells. Biochem. Pharmacol. 2003, 66, 225–237. [Google Scholar] [CrossRef]
- Gourdier, I.; Crabbe, L.; Andreau, K.; Pau, B.; Kroemer, G. Oxaliplatin-induced mitochondrial apoptotic response of colon carcinoma cells does not require nuclear DNA. Oncogene 2004, 23, 7449–7457. [Google Scholar] [CrossRef]
- Quenching of Fluorescence. Principles of Fluorescence Spectroscopy; Lakowicz, J.R., Ed.; Springer: Boston, MA, USA, 2006; pp. 277–330. [Google Scholar]
- Bruno, P.M.; Liu, Y.; Park, G.Y.; Murai, J.; Koch, C.E.; Eisen, T.J.; Pritchard, J.R.; Pommier, Y.; Lippard, S.J.; Hemann, M.T. A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat. Med. 2017, 23, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Sutton, E.C.; DeRose, V.J. Early nucleolar responses differentiate mechanisms of cell death induced by oxaliplatin and cisplatin. J. Biol. Chem. 2021, 296, 100633. [Google Scholar] [CrossRef]
- Yang, K.; Yang, J.; Yi, J. Nucleolar Stress: Hallmarks, sensing mechanism and diseases. Cell Stress 2018, 2, 125–140. [Google Scholar] [CrossRef]
- Sutton, E.C.; McDevitt, C.E.; Prochnau, J.Y.; Yglesias, M.V.; Mroz, A.M.; Yang, M.C.; Cunningham, R.M.; Hendon, C.H.; DeRose, V.J. Nucleolar Stress Induction by Oxaliplatin and Derivatives. J. Am. Chem. Soc. 2019, 141, 18411–18415. [Google Scholar] [CrossRef]
- Yuan, X.; Zhang, W.; He, Y.; Yuan, J.; Song, D.; Chen, H.; Qin, W.; Qian, X.; Yu, H.; Guo, Z. Proteomic analysis of cisplatin- and oxaliplatin-induced phosphorylation in proteins bound to Pt–DNA adducts. Metallomics 2020, 12, 1834–1840. [Google Scholar] [CrossRef]
- Gourdier, I.; Del Rio, M.; Crabbé, L.; Candeil, L.; Copois, V.; Ychou, M.; Auffray, C.; Martineau, P.; Mechti, N.; Pommier, Y.; et al. Drug specific resistance to oxaliplatin is associated with apoptosis defect in a cellular model of colon carcinoma. FEBS Lett. 2002, 529, 232–236. [Google Scholar] [CrossRef]
- Tesniere, A.; Schlemmer, F.; Boige, V.; Kepp, O.; Martins, I.; Ghiringhelli, F.; Aymeric, L.; Michaud, M.; Apetoh, L.; Barault, L.; et al. Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene 2009, 29, 482–491. [Google Scholar] [CrossRef]
- Zhu, H.; Shan, Y.; Ge, K.; Lu, J.; Kong, W.; Jia, C. Oxaliplatin induces immunogenic cell death in hepatocellular carcinoma cells and synergizes with immune checkpoint blockade therapy. Cell. Oncol. 2020, 43, 1203–1214. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Kepp, O.; Senovilla, L.; Menger, L.; Chaput, N.; Kroemer, G. Immunogenic Tumor Cell Death for Optimal Anticancer Therapy: The Calreticulin Exposure Pathway. Clin. Cancer Res. 2010, 16, 3100–3104. [Google Scholar] [CrossRef] [PubMed]
- Panaretakis, T.; Kepp, O.; Brockmeier, U.; Tesniere, A.; Bjorklund, A.-C.; Chapman, D.C.; Durchschlag, M.; Joza, N.; Pierron, G.; van Endert, P.; et al. Mechanisms of pre-apoptotic calreticulin exposure in immunogenic cell death. EMBO J. 2009, 28, 578–590. [Google Scholar] [CrossRef] [PubMed]
- Obeid, M.; Tesniere, A.; Ghiringhelli, F.; Fimia, G.M.; Apetoh, L.; Perfettini, J.-L.; Castedo, M.; Mignot, G.; Panaretakis, T.; Casares, N.; et al. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat. Med. 2006, 13, 54–61. [Google Scholar] [CrossRef]
- Stojanovska, V.; McQuade, R.M.; Fraser, S.; Prakash, M.; Gondalia, S.; Stavely, R.; Palombo, E.; Apostolopoulos, V.; Sakkal, S.; Nurgali, K. Oxaliplatin-induced changes in microbiota, TLR4+ cells and enhanced HMGB1 expression in the murine colon. PLoS ONE 2018, 13, e0198359. [Google Scholar] [CrossRef]
- Apetoh, L.; Ghiringhelli, F.; Tesniere, A.; Obeid, M.; Ortiz, C.; Criollo, A.; Mignot, G.; Maiuri, M.C.; Ullrich, E.; Saulnier, P.; et al. Toll-like receptor 4–dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat. Med. 2007, 13, 1050–1059. [Google Scholar] [CrossRef]
- Lim, S.-C.; Choi, J.E.; Kang, H.S.; SI, H. Ursodeoxycholic acid switches oxaliplatin-induced necrosis to apoptosis by inhibiting reactive oxygen species production and activating p53-caspase 8 pathway in HepG2 hepatocellular carcinoma. Int. J. Cancer 2010, 126, 1582–1595. [Google Scholar] [CrossRef]
- Rogers, B.B.; Cuddahy, T.; Briscella, C.; Ross, N.; Olszanski, A.; Denlinger, C.S. Oxaliplatin: Detection and Management of Hypersensitivity Reactions. Clin. J. Oncol. Nurs. 2019, 23, 68–75. [Google Scholar] [CrossRef]
- Zhu, C.; Ren, X.; Liu, D.; Zhang, C. Oxaliplatin-induced hepatic sinusoidal obstruction syndrome. Toxicology 2021, 460, 152882. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, B.; Gao, X.; Sun, J.; Ye, J.; Li, J.; Cao, P. Targeting strategies for oxaliplatin-induced peripheral neuropathy: Clinical syndrome, molecular basis, and drug development. J. Exp. Clin. Cancer Res. 2021, 40, 1–25. [Google Scholar] [CrossRef]
- Martinez-Balibrea, E.; Martínez-Cardús, A.; Musulen, E.; Ginés, A.; Manzano, J.L.; Aranda, E.; Plasencia, C.; Neamati, N.; Abad, A. Increased levels of copper efflux transporter ATP7B are associated with poor outcome in colorectal cancer patients receiving oxaliplatin-based chemotherapy. Int. J. Cancer 2009, 124, 2905–2910. [Google Scholar] [CrossRef] [PubMed]
- Beretta, G.L.; Benedetti, V.; Cossa, G.; Assaraf, Y.G.; Bram, E.; Gatti, L.; Corna, E.; Carenini, N.; Colangelo, D.; Howell, S.B.; et al. Increased levels and defective glycosylation of MRPs in ovarian carcinoma cells resistant to oxaliplatin. Biochem. Pharmacol. 2010, 79, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Tummala, R.; Wolle, D.; Barwe, S.P.; Sampson, V.B.; Rajasekaran, A.K.; Pendyala, L. Expression of Na,K-ATPase-beta(1) subunit increases uptake and sensitizes carcinoma cells to oxaliplatin. Cancer Chemother. Pharmacol. 2009, 64, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Trachootham, D.; Liu, J.; Chen, G.; Pelicano, H.; Garcia-Prieto, C.; Lu, W.; Burger, J.A.; Croce, C.M.; Plunkett, W.; et al. Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nature 2012, 14, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, J. Review of oxaliplatin: An active platinum agent in colorectal cancer. Int. J. Clin. Pract. 2000, 54, 399–402. [Google Scholar] [PubMed]
- Boyer, J.; McLean, E.G.; Aroori, S.; Wilson, P.; McCulla, A.; Carey, P.D.; Longley, D.B.; Johnston, P.G. Characterization of p53 Wild-Type and Null Isogenic Colorectal Cancer Cell Lines Resistant to 5-Fluorouracil, Oxaliplatin, and Irinotecan. Clin. Cancer Res. 2004, 10, 2158–2167. [Google Scholar] [CrossRef]
- Hatch, S.B.; Swift, L.P.; Caporali, S.; Carter, R.; Hill, E.J.; MacGregor, T.P.; D’Atri, S.; Middleton, M.R.; McHugh, P.J.; Sharma, R.A. XPF protein levels determine sensitivity of malignant melanoma cells to oxaliplatin chemotherapy: Suitability as a biomarker for patient selection. Int. J. Cancer 2013, 134, 1495–1503. [Google Scholar] [CrossRef]
- Bohanes, P.; LaBonte, M.J.; Lenz, H.-J. A Review of Excision Repair Cross-complementation Group 1 in Colorectal Cancer. Clin. Color. Cancer 2011, 10, 157–164. [Google Scholar] [CrossRef]
- Graf, N.; Ang, W.H.; Zhu, G.; Myint, M.; Lippard, S.J. Role of Endonucleases XPF and XPG in Nucleotide Excision Repair of Platinated DNA and Cisplatin/Oxaliplatin Cytotoxicity. ChemBioChem 2011, 12, 1115–1123. [Google Scholar] [CrossRef]
- Li, P.; Fang, Y.J.; Li, F.; Ou, Q.J.; Chen, G.; Ma, G. ERCC1, defective mismatch repair status as predictive biomarkers of survival for stage III colon cancer patients receiving oxaliplatin-based adjuvant chemotherapy. Br. J. Cancer 2013, 108, 1238–1244. [Google Scholar] [CrossRef]
- Yang, J.; Parsons, J.; Nicolay, N.H.; Caporali, S.; Harrington, C.F.; Singh, R.; Finch, D.; D’Atri, S.; Farmer, P.B.; Johnston, P.G.; et al. Cells deficient in the base excision repair protein, DNA polymerase beta, are hypersensitive to oxaliplatin chemotherapy. Oncogene 2009, 29, 463–468. [Google Scholar] [CrossRef]
- Teng, K.-Y.; Qiu, M.-Z.; Li, Z.-H.; Luo, H.-Y.; Zeng, Z.-L.; Luo, R.-Z.; Zhang, H.-Z.; Wang, Z.-Q.; Li, Y.-H.; Xu, R.-H. DNA polymeraseη protein expression predicts treatment response and survival of metastatic gastric adenocarcinoma patients treated with oxaliplatin-based chemotherapy. J. Transl. Med. 2010, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Wen, K.; Fu, Z.; Wu, X.; Feng, J.; Chen, W.; Qian, J. Oct-4 is required for an antiapoptotic behavior of chemoresistant colorectal cancer cells enriched for cancer stem cells: Effects associated with STAT3/Survivin. Cancer Lett. 2013, 333, 56–65. [Google Scholar] [CrossRef] [PubMed]
- van Houdt, W.J.; Emmink, B.L.; Pham, T.V.; Piersma, S.R.; Verheem, A.; Vries, R.G.; Fratantoni, S.A.; Pronk, A.; Clevers, H.; Rinkes, I.B.; et al. Comparative Proteomics of Colon Cancer Stem Cells and Differentiated Tumor Cells Identifies BIRC6 as a Potential Therapeutic Target. Mol. Cell. Proteom. 2011, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Almendro, V.; Ametller, E.; García-Recio, S.; Collazo, O.; Casas, I.; Augé, J.M.; Maurel, J.; Gascón, P. The Role of MMP7 and Its Cross-Talk with the FAS/FASL System during the Acquisition of Chemoresistance to Oxaliplatin. PLoS ONE 2009, 4, e4728. [Google Scholar] [CrossRef] [PubMed]

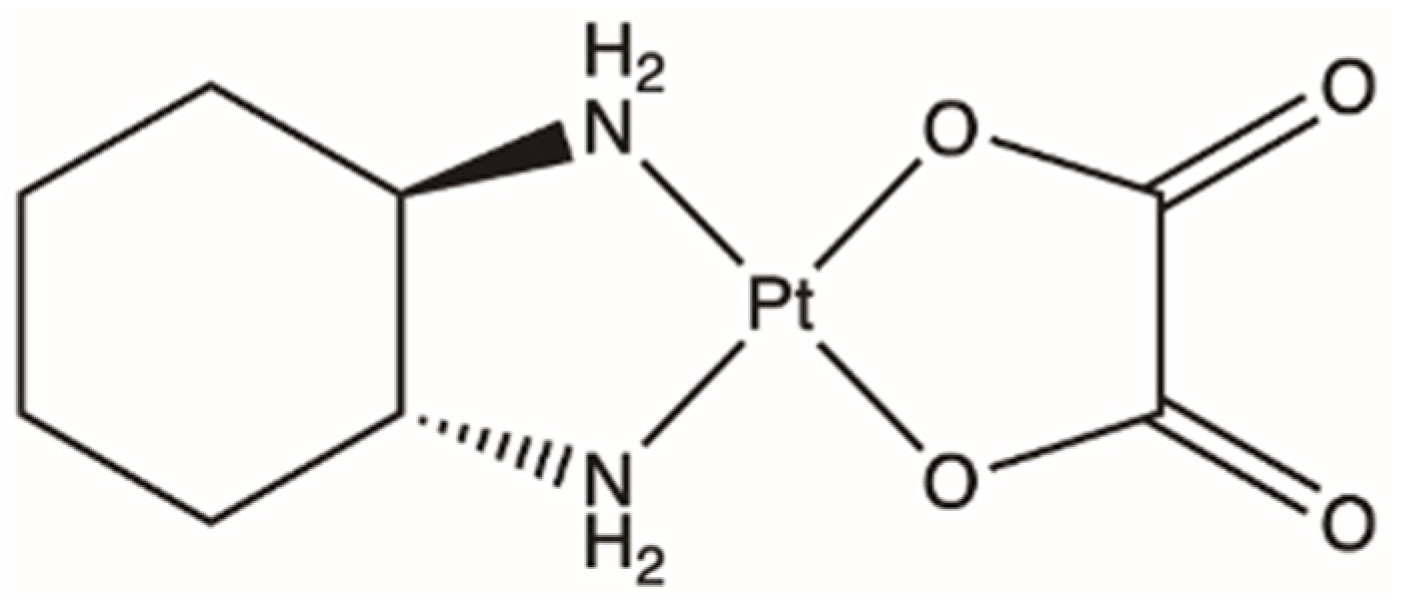

| Platinating Agent | Indication | Major Side Effect |
|---|---|---|
| Cisplatin | Breast, ovarian, testicular, head-and-neck, esophageal, lung, bladder, brain tumours | Nephrotoxicity, ototoxicity, neurotoxicity, hepatotoxicity, cardiotoxicity |
| Carboplatin | Ovarian, lung, certain head-and-neck | Myelosuppression (thrombocytopenia) |
| Oxaliplatin | Colorectal | Peripheral neurotoxicity, Hepatic Sinusoidal obstruction syndrome, myelosuppression |
| Platinating Agent | Influx | Efflux | Organelle Targets |
|---|---|---|---|
| Cisplatin | CTR1, passive diffusion | ATP7A, ATP7B, MRP2 | Nucleus, ER, Mitochondria, Cytoskeleton, Plasma Membrane |
| Carboplatin | CTR1, passive diffusion | Nucleus, ER, Mitochondria | |
| Oxaliplatin | OCT 1–3, CTR1 (minor), passive diffusion | Nucleus, Nucleolus, ER, Mitochondria |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forgie, B.N.; Prakash, R.; Telleria, C.M. Revisiting the Anti-Cancer Toxicity of Clinically Approved Platinating Derivatives. Int. J. Mol. Sci. 2022, 23, 15410. https://doi.org/10.3390/ijms232315410
Forgie BN, Prakash R, Telleria CM. Revisiting the Anti-Cancer Toxicity of Clinically Approved Platinating Derivatives. International Journal of Molecular Sciences. 2022; 23(23):15410. https://doi.org/10.3390/ijms232315410
Chicago/Turabian StyleForgie, Benjamin N., Rewati Prakash, and Carlos M. Telleria. 2022. "Revisiting the Anti-Cancer Toxicity of Clinically Approved Platinating Derivatives" International Journal of Molecular Sciences 23, no. 23: 15410. https://doi.org/10.3390/ijms232315410
APA StyleForgie, B. N., Prakash, R., & Telleria, C. M. (2022). Revisiting the Anti-Cancer Toxicity of Clinically Approved Platinating Derivatives. International Journal of Molecular Sciences, 23(23), 15410. https://doi.org/10.3390/ijms232315410








