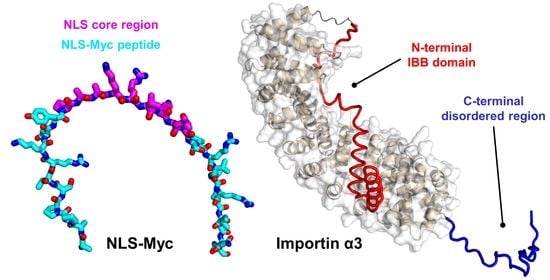
Deciphering the Binding of the Nuclear Localization Sequence of Myc Protein to the Nuclear Carrier Importin α3
Abstract
1. Introduction
2. Results
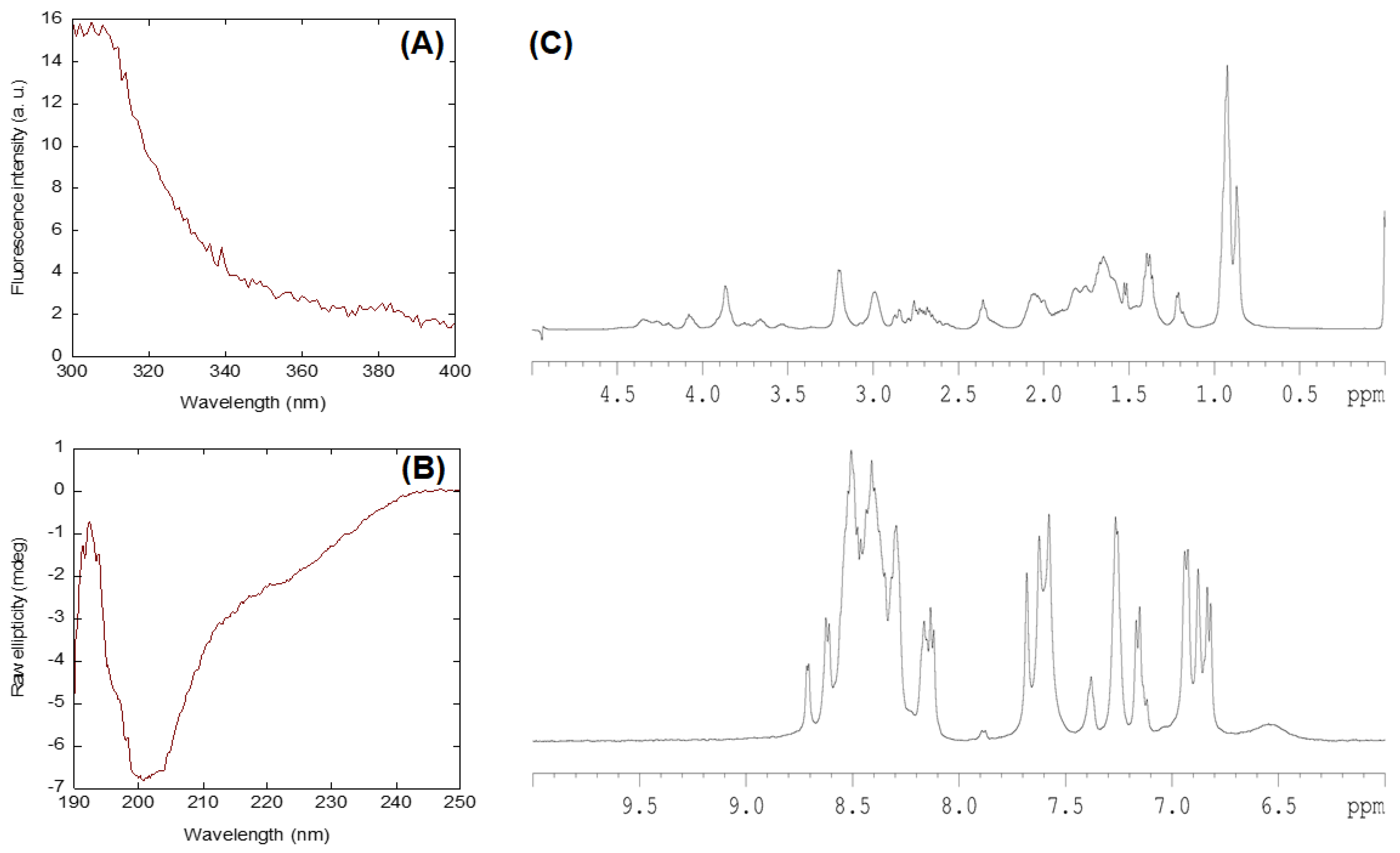
2.1. Conformational Features of Isolated NLS-Myc
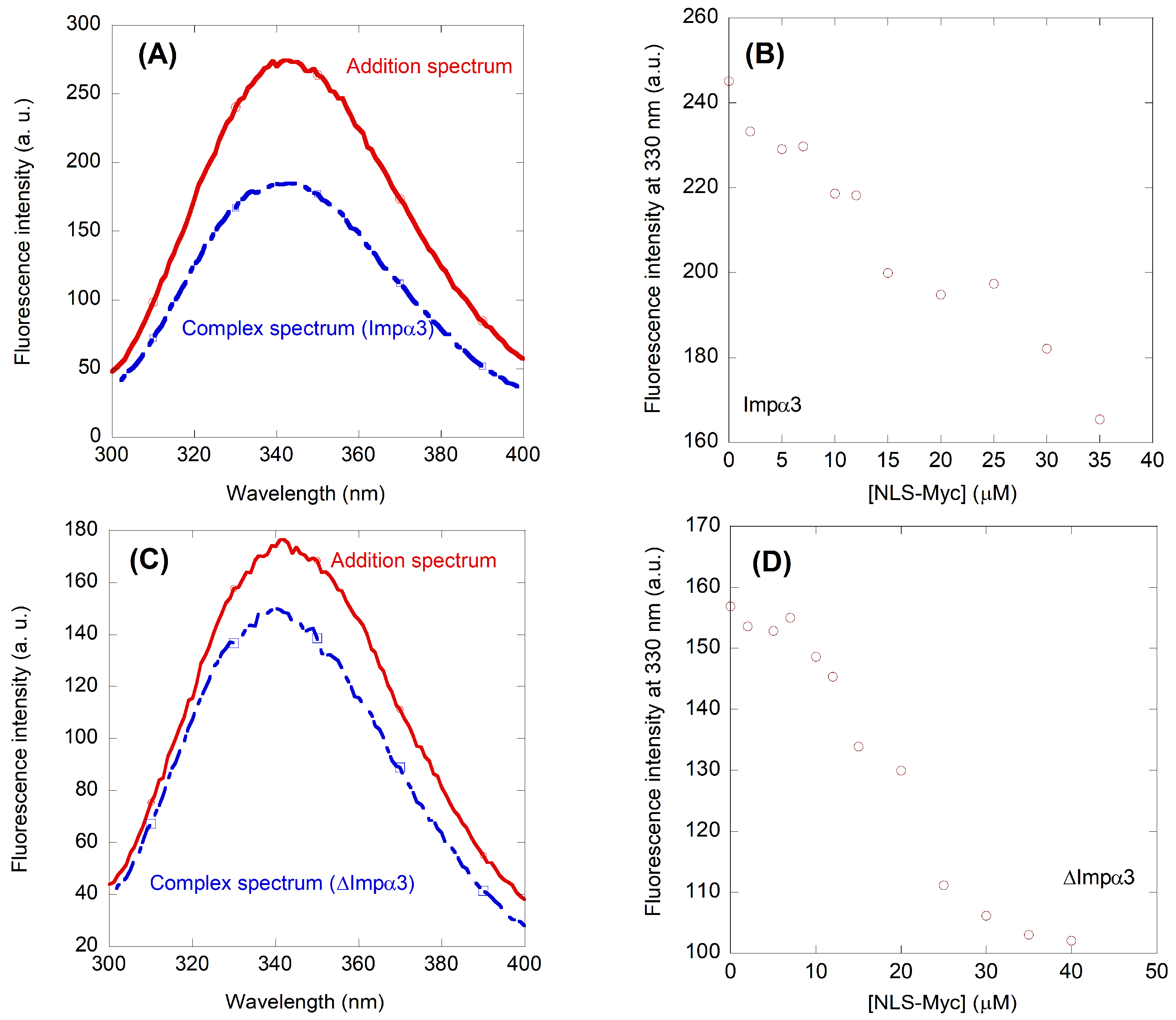
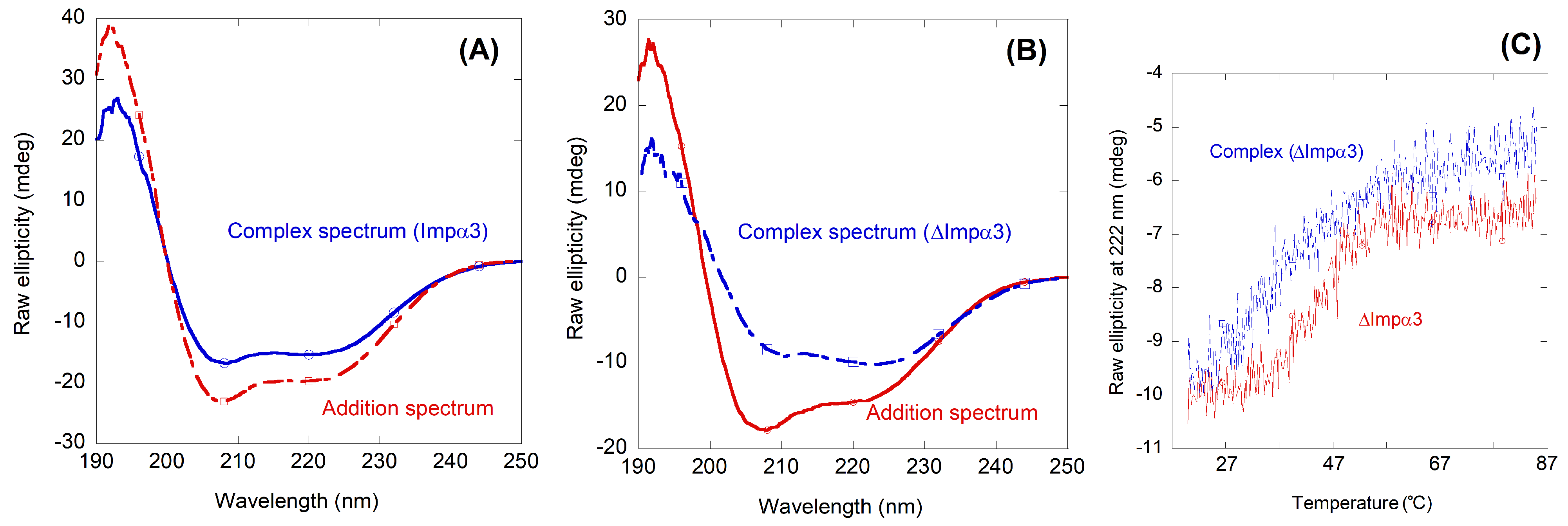
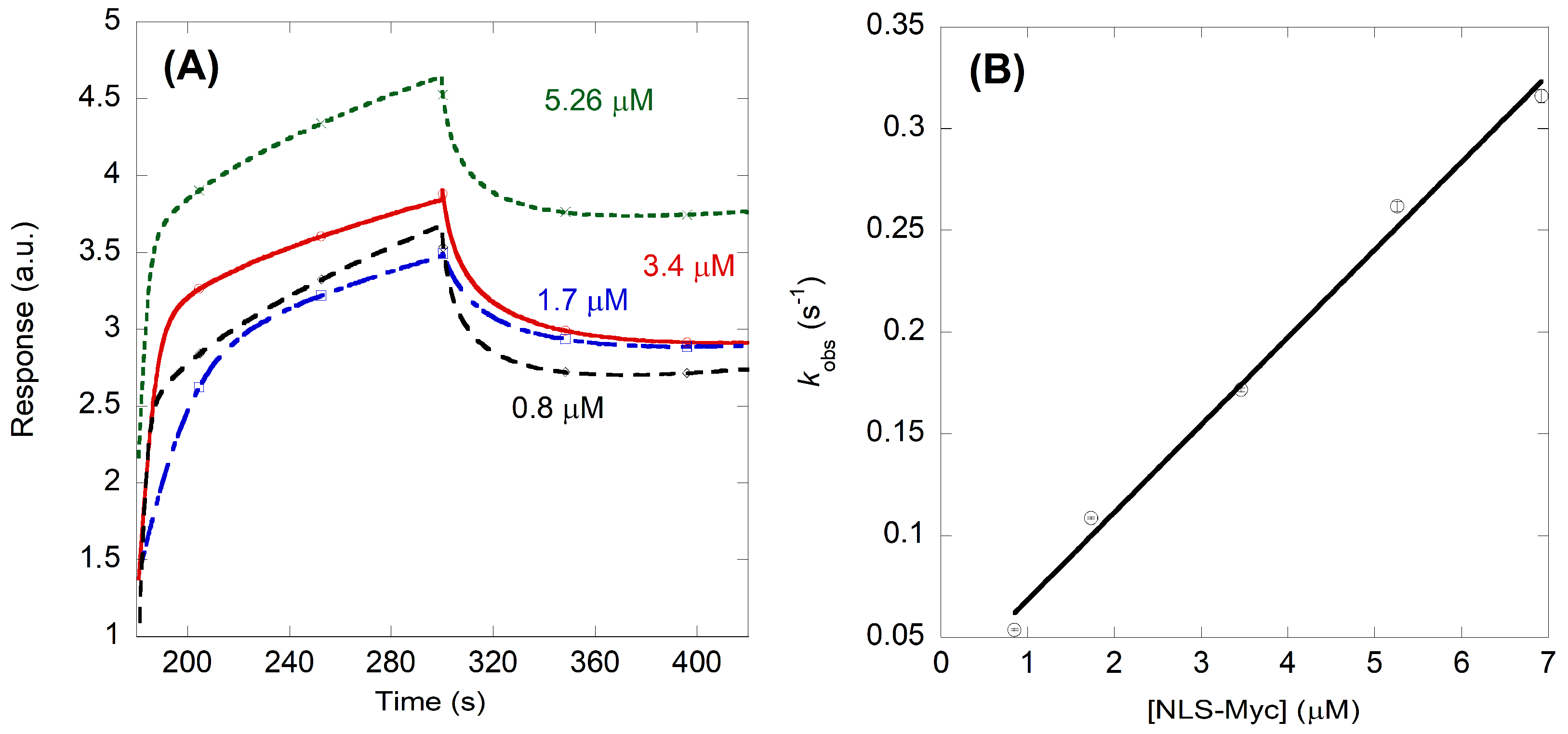
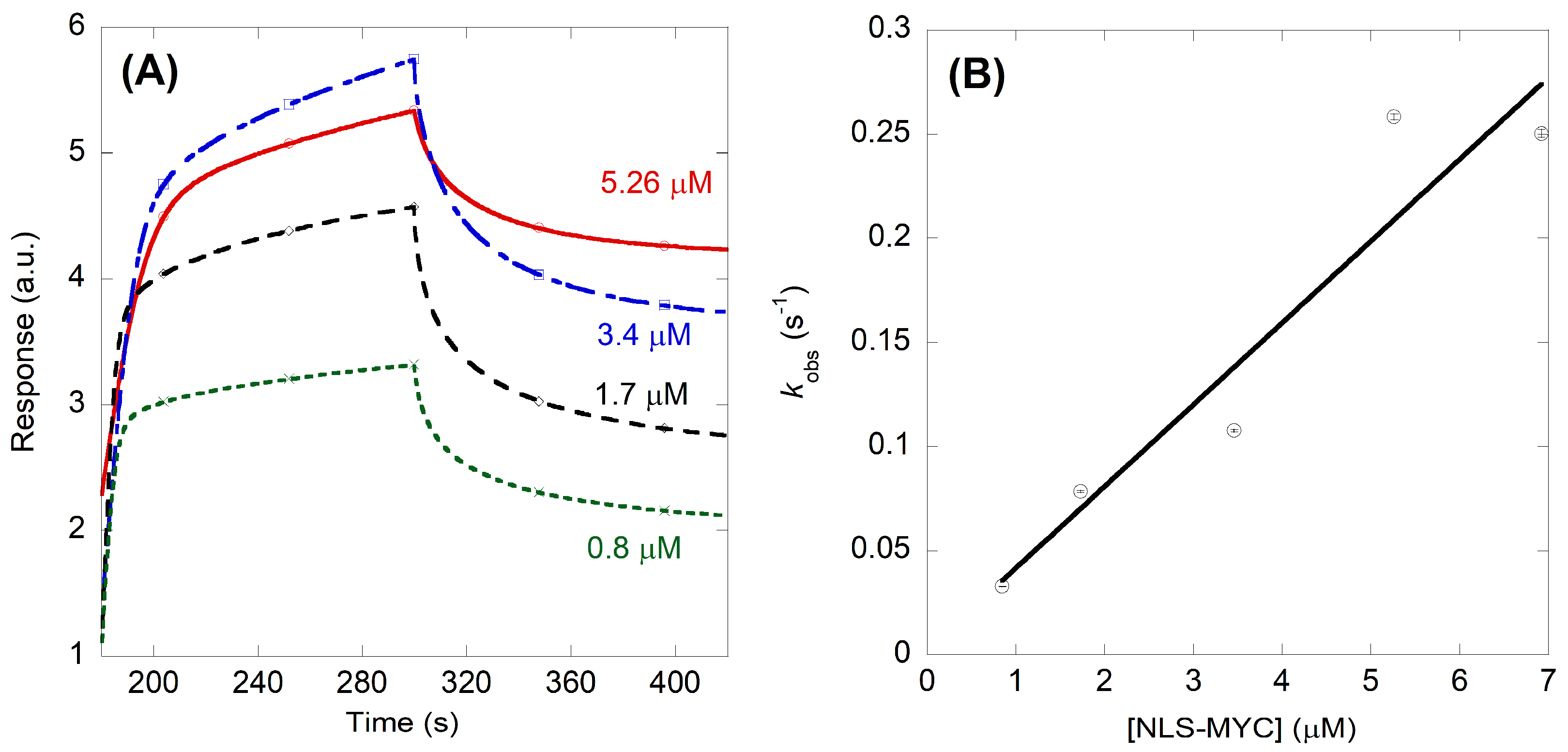
2.2. NLS-Myc Could Bind to Both Impα3 and ΔImpα3
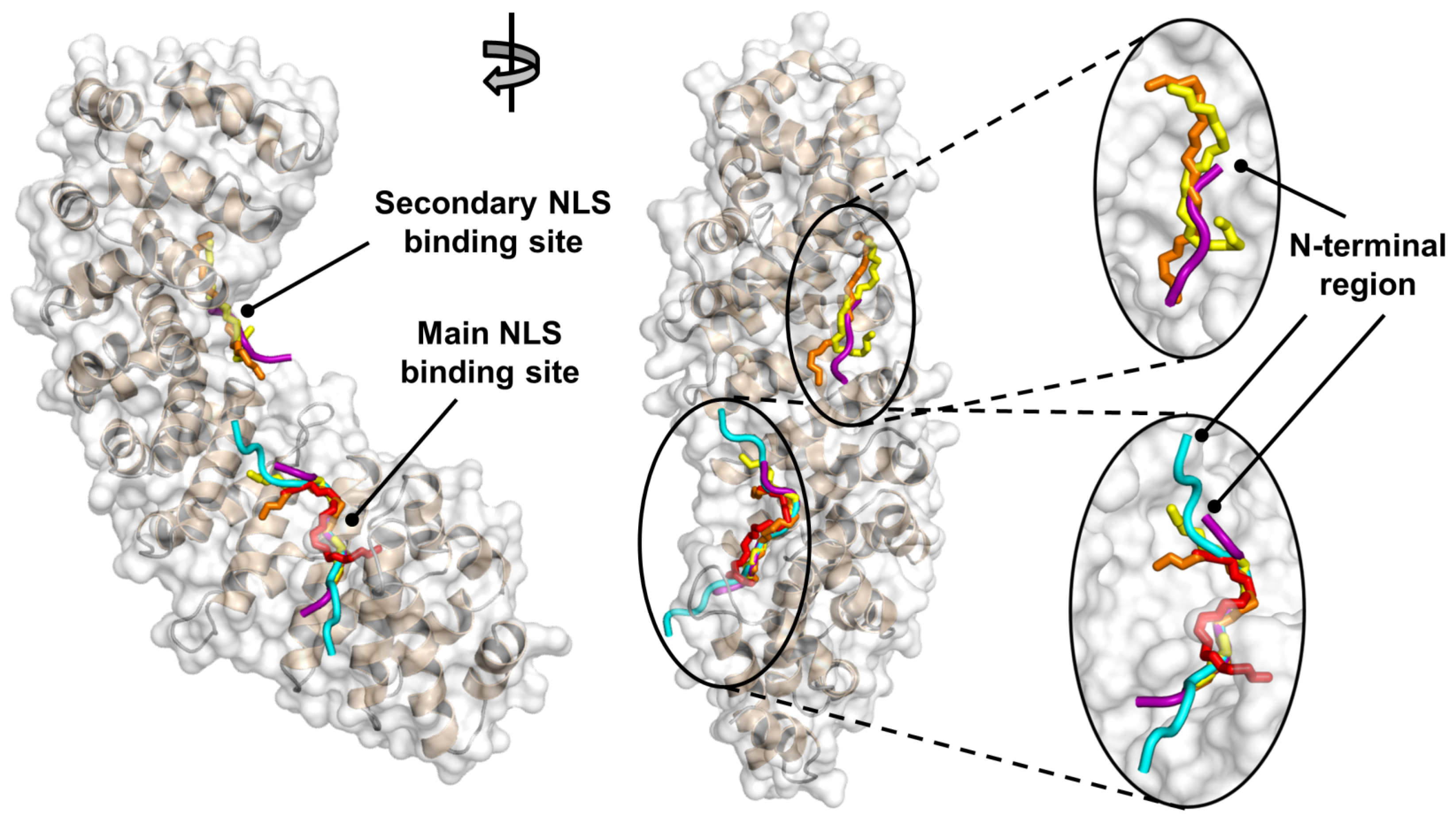
2.3. NLS-Myc Was Bound to the Major NLS Binding Site of Impα3
3. Discussion
4. Materials and Methods
4.1. Materials
4.1.1. Chemicals
4.1.2. Protein Expression and Purification
4.1.3. Synthesis of NLS-Myc
4.2. Fluorescence
4.2.1. Steady-State Fluorescence
4.2.2. Binding Fluorescence Experiments on NLS-Myc
4.3. Circular Dichroism (CD)
4.4. Nuclear Magnetic Resonance (NMR) Spectroscopy
4.4.1. 1D-1H-NMR Spectra
4.4.2. Translational Diffusion NMR (DOSY)
4.4.3. 2D-1H-NMR Spectra
4.5. Biolayer Interferometry (BLI)
4.5.1. Experimental Design of BLI Experiments
4.5.2. Fitting of the BLI Sensorgrams
4.6. Molecular Docking
4.6.1. Structure of the Docking Receptor Impα3
4.6.2. Binding of the Docking Guest NLS-Myc
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BLI | biolayer interferometry |
| CD | circular dichroism |
| DIPSI | decoupling in the presence of scalar interactions |
| DOSY | diffusion ordered spectroscopy |
| GFP | green fluorescent protein |
| IBB | importin β-binding domain |
| Impα3 | human importin α3 isoform (residues 1–521) |
| ΔImpα3 | IBB-depleted species of importin α3 (residues 64–521 of the intact protein) |
| NLS | nuclear localization sequence |
| NLS-Myc | peptide comprising the canonical nuclear localization sequence of Myc protein (residues 310–337 of the intact protein) |
| NMR | nuclear magnetic resonance |
| NOESY | nuclear Overhauser effect spectroscopy |
| NPC | nuclear pore complex |
| PDB | Protein Data Bank |
| TOCSY | total correlation spectroscopy |
| TSP | 3-(trimethylsilyl) propionic acid-2,2,3,3-2H4-sodium salt |
| UV | ultraviolet |
References
- Gabay, M.; Li, Y.; Felsher, D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Kalkat, M.; De Melo, J.; Hickamn, K.A.; Lourenco, C.; Redel, C.; Resetca, D.; Tamachi, A.; Tu, W.B.; Penn, L.Z. MYC deregulation in primary human cancers. Genes 2017, 8, 151. [Google Scholar] [CrossRef]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef]
- Dang, C.V.; Resar, L.M.; Emison, E.; Kim, S.; Li, Q.; Prescott, J.E.; Wonsey, D.; Zeller, K. Function of the c-Myc oncogenic transcription factor. Exp. Cell Res. 1999, 253, 63–77. [Google Scholar] [CrossRef]
- Sears, R.C. The life cycle of c-Myc: From synthesis to degradation. Cell Cycle 2004, 3, 1133–1137. [Google Scholar] [CrossRef]
- Conacci-Sorrell, M.; McFerrin, L.; Eisenman, R.N. An overview of MYC and its interactome. Cold Spring Harbor Perspect. Med. 2014, 4, a014357. [Google Scholar] [CrossRef]
- Felsher, D.W. MYC inactivation elicits oncogene addiction through both tumor cell-intrinsic and host-dependent mechanisms. Genes Cancer 2010, 1, 597–604. [Google Scholar] [CrossRef]
- Dang, C.V.; Lee, W.M. Identification of the human c-myc protein nuclear translocation signal. Mol. Cell Biol. 1988, 8, 4048–4054. [Google Scholar] [CrossRef]
- Christie, M.; Chang, C.-W.; Róna, G.; Smith, K.M.; Stewart, A.G.; Takeda, A.A.S.; Fontes, M.R.M.; Stewart, M.; Vértessy, B.G.; Foorwood, J.K.; et al. Structural biology and regulation of protein import into the nucleus. J. Mol. Biol. 2016, 428, 2060–2090. [Google Scholar] [CrossRef]
- Stewart, M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007, 8, 195–208. [Google Scholar] [CrossRef]
- Wing, C.E.; Fung, H.Y.J.; Chook, Y.M. Karyopherin-mediated nucleocytoplasmic transport. Nat. Rev. Mol. Cell Biol. 2022, 23, 307–328. [Google Scholar] [CrossRef]
- Conti, E.; Kuriyan, J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin α. Structure 2000, 8, 329–338. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucl. Acid Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Kobe, B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat. Struct. Biol. 1999, 6, 388–397. [Google Scholar] [CrossRef]
- Smith, K.M.; Tsimbalyuk, S.; Edwards, M.G.; Cross, E.M.; Batra, J.; Soares da Costa, T.P.; Aragao, D.; Basler, C.F.; Forwood, J.K. Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses. Nat. Commun. 2018, 9, 3703. [Google Scholar] [CrossRef]
- Thiele, S.; Stanelle-Bertram, S.; Beck, S.; Kouassi, N.M.; Zickler, M.; Müller, M.; Tuku, B.; Resa-Infante, P.; Van Riel, D.; Alawi, M.; et al. Cellular Importin-α3 expression dynamics in the lung regulate antiviral response pathways against influenza A virus infection. Cell Rep. 2020, 31, 107549. [Google Scholar] [CrossRef]
- Neira, J.L.; Rizzuti, B.; Jiménez-Alesanco, A.; Abián, O.; Velázquez-Campoy, A.; Iovanna, J.L. The paralogue of the intrinsically disordered Nuclear Protein 1 has a nuclear localization sequence that binds to human importin α3. Int. J. Mol. Sci. 2020, 21, 7428. [Google Scholar] [CrossRef]
- Neira, J.L.; Rizzuti, B.; Jiménez-Alesanco, A.; Palomino-Schätzlein, M.; Abián, O.; Velázquez-Campoy, A.; Iovanna, J.L. A phosphorylation-induced switch in the nuclear localization sequence of the intrinsically disordered NUPR1 hampers binding to importin. Biomolecules 2020, 10, 1313. [Google Scholar] [CrossRef]
- Neira, J.L.; Jiménez-Alesanco, A.; Rizzuti, B.; Velázquez-Campoy, A. The nuclear localization sequence of the epigenetic factor RYBP binds to human importin α3. Biochim. Biophys. Acta Proteins Proteom. 2021, 1869, 140670. [Google Scholar] [CrossRef]
- Benz, C.; Ali, M.; Krystkowiak, I.; Simonetti, L.; Sayadi, A.; Mihalic, F.; Kliche, J.; Andersson, E.; Jemth, P.; Davey, N.E.; et al. Proteome-scale mapping of binding sites in the unstructured regions of the human proteome. Mol. Syst. Biol. 2022, 18, e10584. [Google Scholar] [CrossRef]
- Pantoja-Uceda, D.; Neira, J.L.; Contreras, L.M.; Manton, C.A.; Welch, D.R.; Rizzuti, B. The isolated C-terminal nuclear localization sequence of the breast cancer metastasis supressor is disordered. Arch. Biochem. Biophys. 2019, 664, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.; Santofimia-Castaño, P.; Swayden, M.; Xia, Y.; Zhou, Z.; Audebert, S.; Camoin, L.; Huang, C.; Peng, L.; Jiménez-Alesanco, A.; et al. ZZW-115-dependent inhibition of NUPR1 nuclear translocation sensitizes cancer cells to genotoxic agents. JCI Insight 2020, 5, e138117. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Woody, R.W.; Kallenbach, N.R. Is poly-proline II a major backbone conformation in unfolded proteins? Adv. Prot. Chem. 2002, 62, 163–240. [Google Scholar] [CrossRef]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Wüthrich, K. NMR of Proteins and Nucleic Acids; John Wiley and Sons: New York, NY, USA, 1986. [Google Scholar]
- Kjaergaard, M.; Brander, S.; Poulsen, F.M. Random coil chemical shifts for intrinsically disordered proteins: Effects of temperature and pH. J. Biomol. NMR 2011, 49, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Kjaergaard, M.; Poulsen, F.M. Sequence correction of random coil chemical shifts: Correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR 2011, 50, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Danielsson, J.; Jarvet, J.; Damberg, P.; Gräslund, A. Translational diffusion measured by PFG-NMR on full length and fragments of the Alzheimer Aβ(1–40) peptide. Determination of hydrodynamic radii of random coil peptides of varying length. Magn. Reson. Chem. 2002, 40, S89–S97. [Google Scholar] [CrossRef]
- Diaz-Garcia, C.; Hornos, F.; Giudici, A.M.; Camara-Artigas, A.; Luque-Ortega, J.R.; Arbe, A.; Rizzuti, B.; Alfonso, C.; Forwood, J.K.; Iovanna, J.L.; et al. Human importin α3 and its N-terminal truncated form, without the importin-β-binding domain, are oligomeric species with a low conformational stability in solution. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129609. [Google Scholar] [CrossRef]
- Koyama, M.; Matsuura, Y. Crystal structure of importin-α3 bound to the nuclear localization signal of Ran-binding protein 3. Biochem. Biophys. Res. Commun. 2017, 491, 609–613. [Google Scholar] [CrossRef]
- Saphire, A.C.; Bark, S.J.; Gerace, L. All four homochiral enantiomers of a nuclear localization sequence derived from c-Myc serve as functional import signals. J. Biol. Chem. 1998, 273, 29764–29769. [Google Scholar] [CrossRef]
- Neira, J.L.; Rizzuti, B.; Abián, O.; Araujo-Abad, S.; Velázquez-Campoy, A.; de Juan Romero, C. Human enzyme PADI4 binds to the nuclear carrier Importin α3. Cells 2022, 11, 2166. [Google Scholar] [CrossRef]
- Hodel, M.R.; Corbett, A.H.; Hodel, A.E. Dissection of a nuclear localization signal. J. Biol. Chem. 2000, 276, 1317–1325. [Google Scholar] [CrossRef]
- Llombart, V.; Mansur, M.R. Therapeutic targeting of “undruggable” MYC. eBioMedicine 2022, 75, 103756. [Google Scholar] [CrossRef]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta Proteins Proteom. 2005, 1751, 119–139. [Google Scholar] [CrossRef]
- Kelly, S.M.; Price, N.C. The use of circular dichroism in the investigation of protein structure and function. Curr. Prot. Pept. Sci. 2000, 1, 349–384. [Google Scholar] [CrossRef]
- Woody, R.W. Circular dichroism. Methods Enzymol. 1995, 246, 34–71. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 2004, 383, 318–351. [Google Scholar] [CrossRef]
- Miyatake, H.; Sanjoh, A.; Unzai, S.; Matsuda, G.; Tatsumi, Y.; Miyamoto, Y.; Dohmae, N.; Aida, Y. Crystal structure of human Importin α1 (Rch1) revealing a potential autoinhibition mode involving homodimerization. PLoS ONE 2015, 10, e0115995. [Google Scholar] [CrossRef]
- Cautain, B.; Hill, R.; de Pedro, N.; Link, W. Components and regulation of nuclear transport processes. FEBS J. 2015, 282, 445–462. [Google Scholar] [CrossRef]
- Pumroy, R.A.; Cingolani, G. Diversification of importin-α isoforms in cellular trafficking and disease states. Biochem. J. 2015, 466, 13–28. [Google Scholar] [CrossRef]
- Gill, S.C.; von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989, 182, 319–326. [Google Scholar] [CrossRef]
- Neira, J.L.; Hornos, F.; Bacarizo, J.; Cámara-Artigas, A.; Gómez, J. The monomeric species of the regulatory domain of tyrosine hydroxylase has a low conformational stability. Biochemistry 2017, 55, 3418–3431. [Google Scholar] [CrossRef]
- Birdsall, B.; King, R.W.; Wheeler, M.R.; Lewis, C.A., Jr.; Goode, S.; Dunlap, R.B.; Roberts, G.C. Correction for light absorption in fluorescence studies of protein-ligand interactions. Anal. Biochem. 1983, 132, 353–361. [Google Scholar] [CrossRef]
- Beckett, D. Measurement and analysis of equilibrium binding titrations: A beginner’s guide. Methods Enzymol. 2011, 488, 1–16. [Google Scholar] [CrossRef]
- Royer, C.A.; Scarlata, S.F. Fluorescence approaches to quantifying biomolecular interactions. Methods Enzymol. 2008, 450, 79–106. [Google Scholar] [CrossRef]
- Hornos, F.; Fen, H.-Z.; Rizzuti, B.; Palomino-Schätzlein, M.; Wieczorek, D.; Neira, J.L.; Jin, J.-P. The muscle-relaxing C-terminal peptide from troponin I populates a nascent helix, facilitating binding to tropomyosin with a potent therapeutic effect. J. Biol. Chem. 2021, 296, 100228. [Google Scholar] [CrossRef]
- Cavanagh, J.; Fairbrother, W.J.; Palmer, A.G.; Skelton, N.J. Protein NMR Spectroscopy: Principles and Practice; Academic Press: New York, NY, USA, 1996. [Google Scholar]
- Piotto, M.; Saudek, V.; Sklenár, V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 1992, 2, 661–675. [Google Scholar] [CrossRef]
- Wilkins, D.K.; Grimshaw, S.B.; Receveur, V.; Dobson, C.M.; Jones, J.A.; Smith, L.J. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR technique. Biochemistry 1999, 38, 16424–16431. [Google Scholar] [CrossRef]
- Marion, D.; Wüthrich, K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 1983, 11, 967–975. [Google Scholar] [CrossRef]
- Bax, A.; Davis, D.G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985, 65, 355–360. [Google Scholar] [CrossRef]
- Kumar, A.; Ernst, R.R.; Wüthrich, K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 1980, 95, 1–6. [Google Scholar] [CrossRef]
- Cavanagh, J.; Rance, M. Suppression of cross-relaxation effects in TOCSY spectra via a modified DIPSI-2 mixing sequence. J. Magn. Reson. 1992, 96, 660–678. [Google Scholar] [CrossRef]
- Frenzel, D.; Willbold, D. Kinetic titration series with biolayer interferometry. PLoS ONE 2014, 9, e106882. [Google Scholar] [CrossRef] [PubMed]
- Pantoja-Uceda, D.; Neira, J.L.; Saelices, L.; Robles-Rengel, R.; Florencio, F.J.; Muro-Pastor, M.I.; Santoro, J. Dissecting the binding between glutamine synthetase and its two natively unfolded protein inhibitors. Biochemistry 2016, 55, 3370–3382. [Google Scholar] [CrossRef]
- Santofimia-Castaño, P.; Rizzuti, B.; Pey, A.L.; Soubeyran, P.; Vidal, M.; Urrutia, R.; Iovanna, J.L.; Neira, J.L. Intrinsically disordered chromatin protein NUPR1 binds to the C-terminal region of Polycomb RING1B. Proc. Natl. Acad. Sci. USA 2017, 114, E6332–E6341. [Google Scholar] [CrossRef]
- Santofimia-Castaño, P.; Rizzuti, B.; Abián, O.; Velázquez-Campoy, A.; Iovanna, J.L.; Neira, J.L. Amphipathic helical peptides hamper protein-protein interactions of the intrinsically disordered chromatin nuclear protein 1 (NUPR1). Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 1283–1295. [Google Scholar] [CrossRef]
- Santofimia-Castaño, P.; Xia, Y.; Lan, W.; Zhou, Z.; Huang, C.; Peng, L.; Soubeyran, P.; Velázquez-Campoy, A.; Abián, O.; Rizzuti, B.; et al. Ligand-based design identifies a potent NUPR1 inhibitor exerting anticancer activity via necroptosis. J. Clin. Investig. 2019, 129, 2500–2513. [Google Scholar] [CrossRef]
- Rizzuti, B.; Lan, W.; Santofimia-Castaño, P.; Zhou, Z.; Velázquez-Campoy, A.; Abián, O.; Peng, L.; Neira, J.L.; Xia, Y.; Iovanna, J.L. Design of inhibitors of the intrinsically disordered protein NUPR1: Balance between drug affinity and target function. Biomolecules 2021, 11, 1453. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]

| Fragment | Sequence | Number of Rotatable Bonds | Binding Affinity (kcal/mol) |
|---|---|---|---|
| F1 | A310APPST315 | 20 | −6.3 |
| F2 | P312PSTRK317 | 26 | −6.5 |
| F3 | S314TRKDY319 | 30 | −6.5 |
| F4 | R316KDYPA321 | 28 | −5.9 |
| F5 | D318YPAAK323 | 24 | −6.4 |
| F6 | P320AAKRV325 | 21 | −6.9 |
| F7 | A322KRVKL327 | 29 | −6.5 |
| F8 | R324VKLDS329 | 28 | −6.7 |
| F9 | K326LDSVR331 | 28 | −6.3 |
| F10 | D328SVRVL333 | 24 | −6.6 |
| F11 | V330RVLRQ335 | 27 | −6.7 |
| F12 | V332LRQIS337 | 26 | −6.4 |
| F13 | R334QISNN339 | 27 | −6.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rizzuti, B.; Iovanna, J.L.; Neira, J.L. Deciphering the Binding of the Nuclear Localization Sequence of Myc Protein to the Nuclear Carrier Importin α3. Int. J. Mol. Sci. 2022, 23, 15333. https://doi.org/10.3390/ijms232315333
Rizzuti B, Iovanna JL, Neira JL. Deciphering the Binding of the Nuclear Localization Sequence of Myc Protein to the Nuclear Carrier Importin α3. International Journal of Molecular Sciences. 2022; 23(23):15333. https://doi.org/10.3390/ijms232315333
Chicago/Turabian StyleRizzuti, Bruno, Juan L. Iovanna, and José L. Neira. 2022. "Deciphering the Binding of the Nuclear Localization Sequence of Myc Protein to the Nuclear Carrier Importin α3" International Journal of Molecular Sciences 23, no. 23: 15333. https://doi.org/10.3390/ijms232315333
APA StyleRizzuti, B., Iovanna, J. L., & Neira, J. L. (2022). Deciphering the Binding of the Nuclear Localization Sequence of Myc Protein to the Nuclear Carrier Importin α3. International Journal of Molecular Sciences, 23(23), 15333. https://doi.org/10.3390/ijms232315333