Novel Genes Involved in Hypertrophic Cardiomyopathy: Data of Transcriptome and Methylome Profiling
Abstract
:1. Introduction
2. Results
2.1. Clinical Characteristics of the Studied Individuals
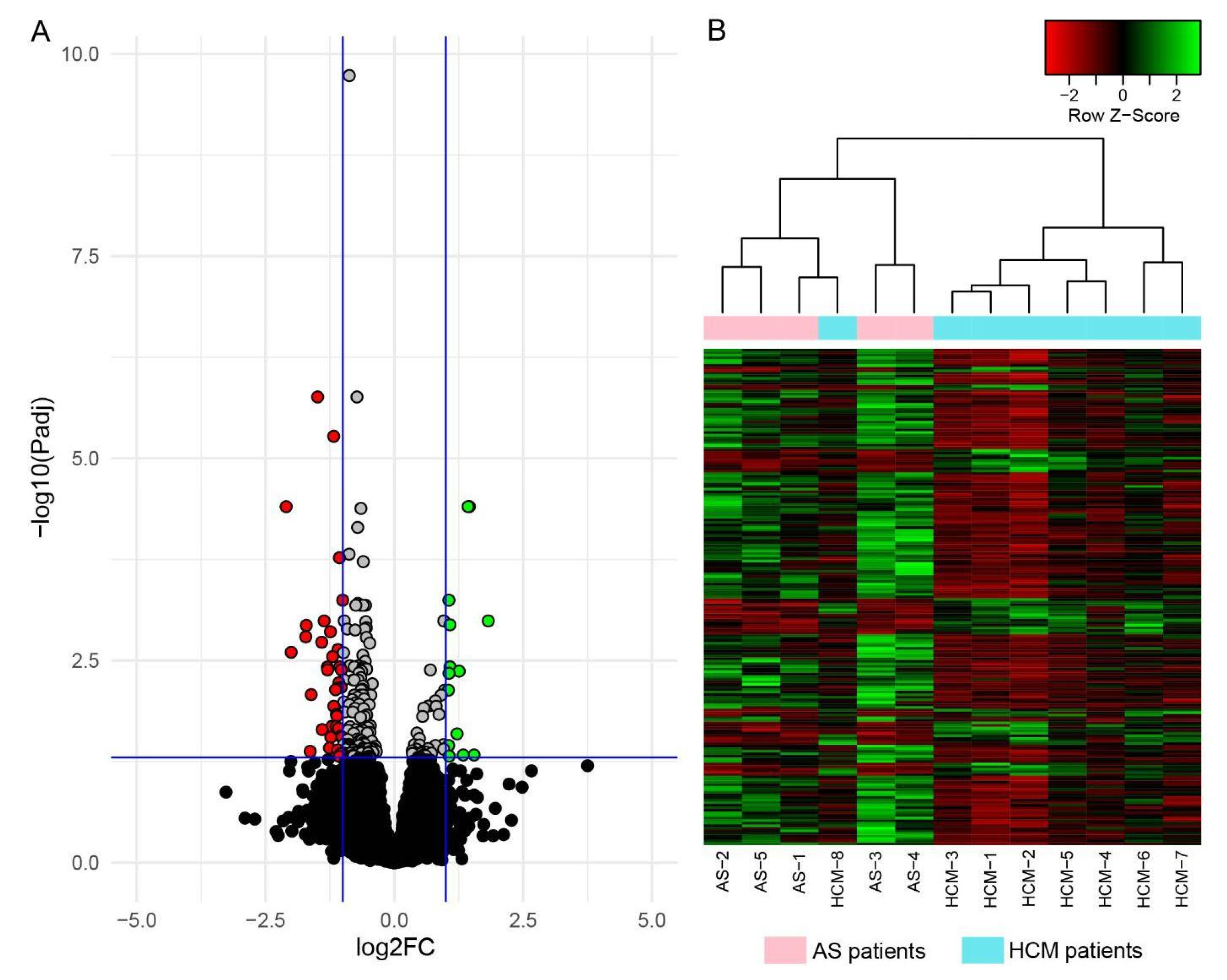
2.2. Analysis of Gene Differential Expression Using RNA Sequencing
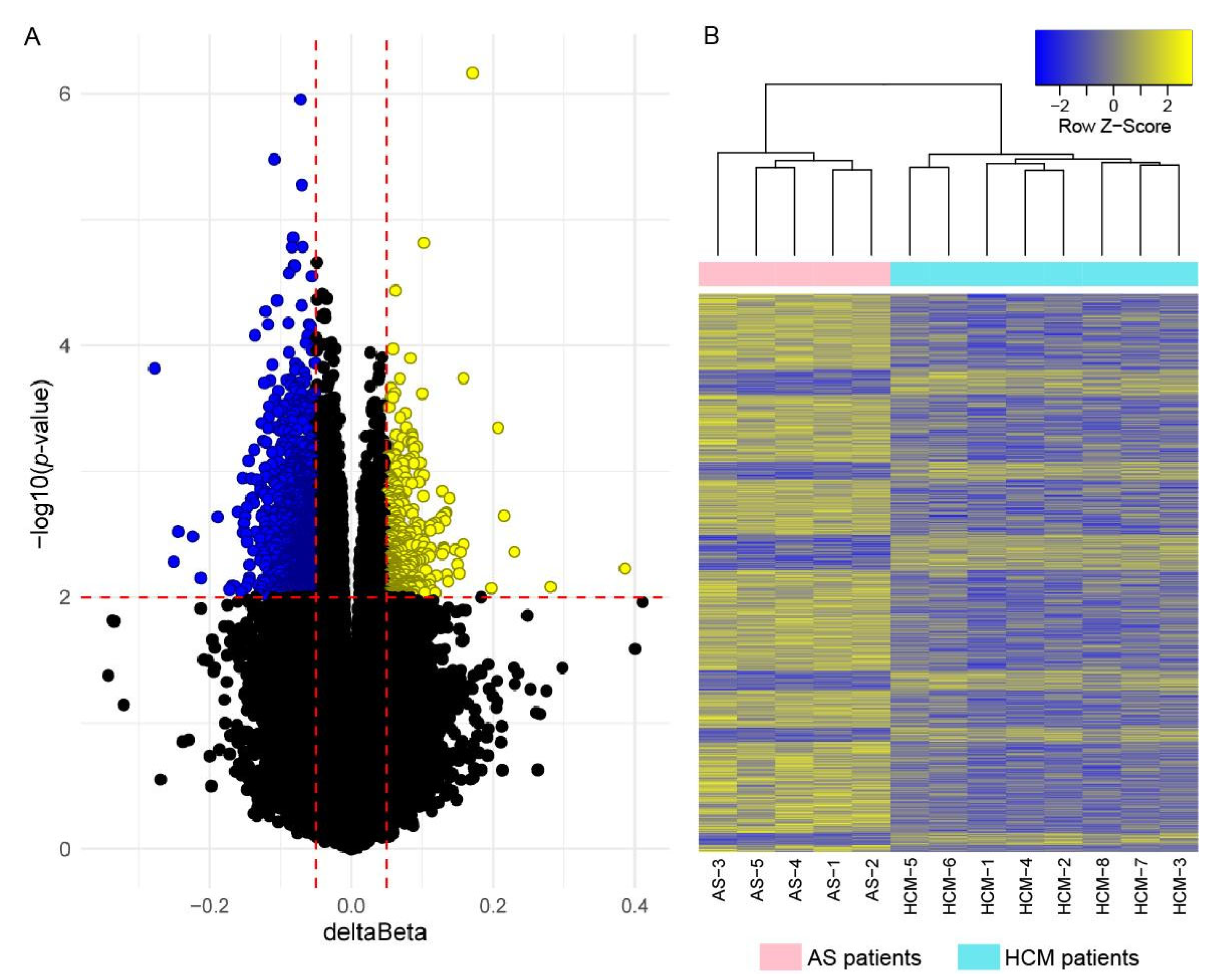
2.3. Genome-Wide DNA Methylation Analysis
2.4. Correlation of Gene Expression and DNA Methylation Data
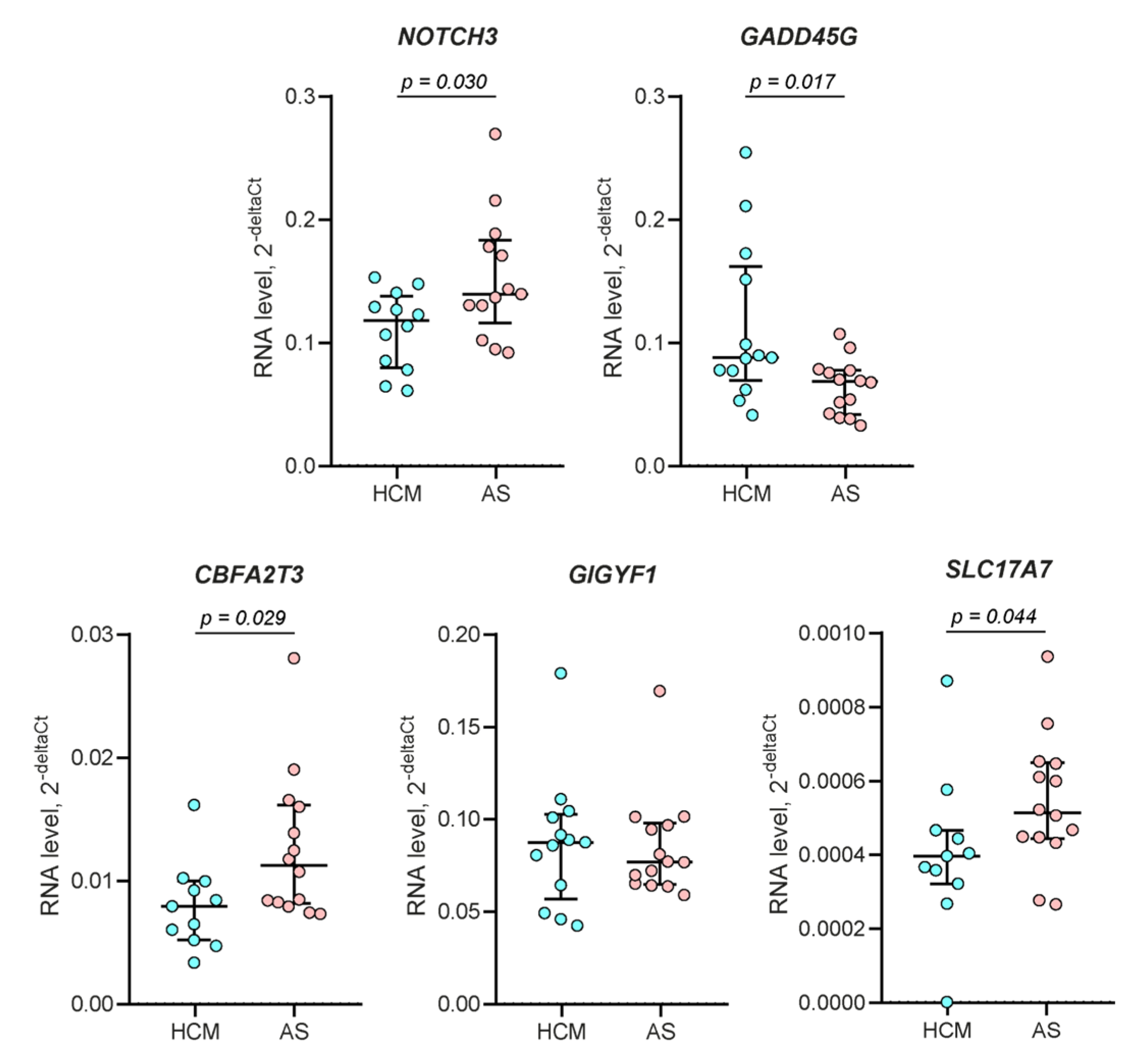
2.5. RT-qPCR Validation
2.6. Gene Ontology Enrichment Analysis
3. Discussion
4. Materials and Methods
4.1. Patients and Controls
4.2. Sample Processing
4.3. RNA-Seq Analysis
4.4. Genome-Wide DNA Methylation Analysis
4.5. RT-qPCR
4.6. Gene Set Enrichment Analysis and Data Visualization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maron, B.J.; Desai, M.Y.; Nishimura, R.A.; Spirito, P.; Rakowski, H.; Towbin, J.A.; Dearani, J.A.; Rowin, E.J.; Maron, M.S.; Sherrid, V.M. Management of Hypertrophic Cardiomyopathy: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2022, 79, 390–414. [Google Scholar] [CrossRef] [PubMed]
- Almeida, P.C.; Lopes, V.; Ferreira, L.A.; Moreira, N.; Marto, C.M.; Gonçalves, L.; Donato, P. Role of Cardiac Magnetic Resonance in the Diagnosis of Infiltrative, Hypertrophic, and Arrhythmogenic Cardiomyopathies. Front. Biosci. 2022, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Oliva-Sandoval, M.J.; Ruiz-Espejo, F.; Monserrat, L.; Hermida-Prieto, M.; Sabater, M.; García-Molina, E.; Ortiz, M.; Rodríguez-García, M.I.; Núñez, L.; Gimeno, J.R.; et al. Insights into genotype-phenotype correlation in hypertrophic cardiomyopathy. Findings from 18 Spanish families with a single mutation in MYBPC3. Heart 2010, 96, 1980–1984. [Google Scholar] [CrossRef] [PubMed]
- Jansweijer, J.A.; van Spaendonck-Zwarts, K.Y.; Tanck, M.W.T.; van Tintelen, J.P.; Christiaans, I.; van der Smagt, J.; Vermeer, A.; Bos, J.M.; Moss, A.J.; Swan, H.; et al. Heritability in genetic heart disease: The role of genetic background. Open Heart 2019, 6, e000929. [Google Scholar] [CrossRef] [PubMed]
- Cerrone, M.; Remme, C.A.; Tadros, R.; Bezzina, C.R.; Delmar, M. Beyond the One Gene-One Disease Paradigm: Complex Genetics and Pleiotropy in Inheritable Cardiac Disorders. Circulation 2019, 140, 595–610. [Google Scholar] [CrossRef]
- Parvari, R.; Levitas, A. The mutations associated with dilated cardiomyopathy. Biochem. Res. Int. 2012, 2012, 639250. [Google Scholar] [CrossRef]
- Maron, B.J.; Maron, M.S.; Maron, B.A.; Loscalzo, J. Moving Beyond the Sarcomere to Explain Heterogeneity in Hypertrophic Cardiomyopathy: JACC Review Topic of the Week. J. Am. Coll. Cardiol. 2019, 73, 1978–1986. [Google Scholar] [CrossRef]
- Tadros, R.; Francis, C.; Xu, X.; Vermeer, A.M.C.; Harper, A.R.; Huurman, R.; Bisabu, K.K.; Walsh, R.; Hoorntje, E.T.; Rijdt, W.P.T.; et al. Shared genetic pathways contribute to risk of hypertrophic and dilated cardiomyopathies with opposite directions of effect. Nat. Genet. 2021, 53, 128–134. [Google Scholar] [CrossRef]
- Harper, A.R.; Goel, A.; Grace, C.; Thomson, K.L.; Petersen, S.E.; Xu, X.; Waring, A.; Ormondroyd, E.; Kramer, C.M.; Ho, C.Y.; et al. Common genetic variants and modifiable risk factors underpin hypertrophic cardiomyopathy susceptibility and expressivity. Nat. Genet. 2021, 53, 135–142. [Google Scholar] [CrossRef]
- Vakrou, S.; Liu, Y.; Zhu, L.; Greenland, G.V.; Simsek, B.; Hebl, V.B.; Guan, Y.; Woldemichael, K.; Talbot, C.C.; Aon, M.A.; et al. Differences in molecular phenotype in mouse and human hypertrophic cardiomyopathy. Sci. Rep. 2021, 11, 13163. [Google Scholar] [CrossRef]
- Liu, C.-F.; Ni, Y.; Thachil, V.; Morley, M.; Moravec, C.S.; Tang, W.H.W. Differential expression of members of SOX family of transcription factors in failing human hearts. Transl. Res. 2022, 242, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Bos, J.M.; Hebl, V.B.; Oberg, A.L.; Sun, Z.; Herman, D.S.; Teekakirikul, P.; Seidman, J.G.; Seidman, C.E.; Remedios, C.G.D.; Maleszewski, J.J.; et al. Marked Up-Regulation of ACE2 in Hearts of Patients With Obstructive Hypertrophic Cardiomyopathy: Implications for SARS-CoV-2-Mediated COVID-19. Mayo Clin. Proc. 2020, 95, 1354–1368. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.-W.; Liu, J.-J.; Li, J.-H.; Li, J.-W.; Dai, J.; Lai, Y.-Q. RNA-seq profiling of mRNA associated with hypertrophic cardiomyopathy. Mol. Med. Rep. 2016, 14, 5573–5586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, J.; Schuldt, M.; Nagyova, E.; Gu, Z.; el Bouhaddani, S.; Yiangou, L.; Jansen, M.; Calis, J.J.A.; Dorsch, L.M.; Blok, C.S.; et al. Multi-omics integration identifies key upstream regulators of pathomechanisms in hypertrophic cardiomyopathy due to truncating MYBPC3 mutations. Clin. Epigenetics 2021, 13, 61. [Google Scholar] [CrossRef]
- Maron, B.A.; Wang, R.-S.; Shevtsov, S.; Drakos, S.G.; Arons, E.; Wever-Pinzon, O.; Huggins, G.S.; Samokhin, A.O.; Oldham, W.M.; Aguib, Y.; et al. Individualized interactomes for network-based precision medicine in hypertrophic cardiomyopathy with implications for other clinical pathophenotypes. Nat. Commun. 2021, 12, 873. [Google Scholar] [CrossRef]
- Ranjbarvaziri, S.; Kooiker, K.B.; Ellenberger, M.; Fajardo, G.; Zhao, M.; Roest, A.S.V.; Woldeyes, R.A.; Koyano, T.T.; Fong, R.; Ma, N.; et al. Altered Cardiac Energetics and Mitochondrial Dysfunction in Hypertrophic Cardiomyopathy. Circulation 2021, 144, 1714–1731. [Google Scholar] [CrossRef]
- Mahmoud, S.A.; Poizat, C. Epigenetics and chromatin remodeling in adult cardiomyopathy. J. Pathol. 2013, 231, 147–157. [Google Scholar] [CrossRef] [Green Version]
- Movassagh, M.; Choy, M.-K.; Knowles, D.A.; Cordeddu, L.; Haider, S.; Down, T.; Siggens, L.; Vujic, A.; Simeoni, I.; Penkett, C.; et al. Distinct epigenomic features in end-stage failing human hearts. Circulation 2011, 124, 2411–2422. [Google Scholar] [CrossRef] [Green Version]
- Glezeva, N.; Moran, B.; Collier, P.; Moravec, C.S.; Phelan, D.; Donnellan, E.; Russell-Hallinan, A.; O’Connor, D.P.; Gallagher, W.M.; Gallagher, J.; et al. Targeted DNA Methylation Profiling of Human Cardiac Tissue Reveals Novel Epigenetic Traits and Gene Deregulation Across Different Heart Failure Patient Subtypes. Circ. Heart Fail. 2019, 12, e005765. [Google Scholar] [CrossRef] [Green Version]
- Single Cell Type—C4B—The Human Protein Atlas, (n.d.). Available online: https://www.proteinatlas.org/ENSG00000224389-C4B/single+cell+type (accessed on 24 June 2022).
- Rehulkova, H.; Rehulka, P.; Fucikova, A.M.; Stulik, J.; Pudi, R. Identification of novel biomarker candidates for hypertrophic cardiomyopathy and other cardiovascular diseases leading to heart failure. Physiol. Res. 2016, 65, 751–762. [Google Scholar] [CrossRef]
- Zhang, M.; Pan, X.; Zou, Q.; Xia, Y.; Chen, J.; Hao, Q.; Wang, H.; Sun, D. Notch3 Ameliorates Cardiac Fibrosis After Myocardial Infarction by Inhibiting the TGF-β1/Smad3 Pathway. Cardiovasc. Toxicol. 2016, 16, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Cao, D.; Zhang, D.; Zhang, Y.; Yue, Y. MicroRNA-1 facilitates hypoxia-induced injury by targeting NOTCH3. J. Cell. Biochem. 2020, 12, 4458–4469. [Google Scholar] [CrossRef] [PubMed]
- Elmadhun, N.Y.; Sabe, A.A.; Lassaletta, A.D.; Chu, L.M.; Kondra, K.; Sturek, M.; Sellke, F.W. Metabolic syndrome impairs notch signaling and promotes apoptosis in chronically ischemic myocardium. J. Thorac. Cardiovasc. Surg. 2014, 148, 1048–1055; discussion 1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwahana, T.; Okada, S.; Kanda, M.; Oshima, M.; Iwama, A.; Matsumiya, G.; Kobayashi, Y. Novel myocardial markers GADD45G and NDUFS5 identified by RNA-sequencing predicts left ventricular reverse remodeling in advanced non-ischemic heart failure: A retrospective cohort study. BMC Cardiovasc. Disord. 2020, 20, 116. [Google Scholar] [CrossRef] [PubMed]
- Biel, A.; Castanza, A.S.; Rutherford, R.; Fair, S.R.; Chifamba, L.; Wester, J.C.; Hester, M.E.; Hevner, R.F. AUTS2 Syndrome: Molecular Mechanisms and Model Systems. Front. Mol. Neurosci. 2022, 15, 858582. [Google Scholar] [CrossRef] [PubMed]
- Kishi, M.; Pan, Y.A.; Crump, J.G.; Sanes, J.R. Mammalian SAD kinases are required for neuronal polarization. Science 2005, 307, 929–932. [Google Scholar] [CrossRef]
- Plambeck, K.E.; He, C.-W.; Navarro, H.H.; Díaz, E. Mutually Dependent Clustering of SynDIG4/PRRT1 and AMPA Receptor Subunits GluA1 and GluA2 in Heterologous Cells and Primary Neurons. Front. Mol. Neurosci. 2022, 15, 788620. [Google Scholar] [CrossRef] [PubMed]
- Upmanyu, N.; Jin, J.; von der Emde, H.; Ganzella, M.; Bösche, L.; Malviya, V.N.; Zhuleku, E.; Politi, A.Z.; Ninov, M.; Silbern, I.; et al. Colocalization of different neurotransmitter transporters on synaptic vesicles is sparse except for VGLUT1 and ZnT3. Neuron 2022, 110, 1483–1497.e7. [Google Scholar] [CrossRef]
- Jjingo, D.; Conley, A.B.; Yi, S.V.; Lunyak, V.V.; Jordan, I.K. On the presence and role of human gene-body DNA methylation. Oncotarget 2012, 3, 462–474. [Google Scholar] [CrossRef] [Green Version]
- Ganassi, M.; Zammit, P.S. Involvement of muscle satellite cell dysfunction in neuromuscular disorders: Expanding the portfolio of satellite cell-opathies. Eur. J. Transl. Myol. 2022, 32, 10064. [Google Scholar] [CrossRef]
- Tampakakis, E.; Mahmoud, A.I. The role of hormones and neurons in cardiomyocyte maturation. Semin. Cell Dev. Biol. 2021, 118, 136–143. [Google Scholar] [CrossRef] [PubMed]
- loras, J.S. Sympathetic nervous system activation in human heart failure: Clinical implications of an updated model. J. Am. Coll. Cardiol. 2009, 54, 375–385. [Google Scholar] [CrossRef] [Green Version]
- Binkley, P.F.; Nunziata, E.; Haas, G.J.; Nelson, S.D.; Cody, R.J. Parasympathetic withdrawal is an integral component of autonomic imbalance in congestive heart failure: Demonstration in human subjects and verification in a paced canine model of ventricular failure. J. Am. Coll. Cardiol. 1991, 18, 464–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knuuti, J.; Sipola, P. Is it time for cardiac innervation imaging? Q. J. Nucl. Med. Mol. Imaging 2005, 49, 97–105. [Google Scholar] [PubMed]
- Elliott, P.M.; Anastasakis, A.; Borger, M.A.; Borggrefe, M.; Cecchi, F.; Charron, P.; Hagege, A.A.; Lafont, A.; Limongelli, G.; Mahrholdt, H.; et al. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: The Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). Eur. Heart J. 2014, 35, 2733–2779. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Klipper-Aurbach, Y.; Wasserman, M.; Braunspiegel-Weintrob, N.; Borstein, D.; Peleg, S.; Assa, S.; Karp, M.; Benjamini, Y.; Hochberg, Y.; Laron, Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med. Hypotheses 1995, 45, 486–490. [Google Scholar] [CrossRef]
- Tian, Y.; Morris, T.J.; Webster, A.P.; Yang, Z.; Beck, S.; Feber, A.; Teschendorff, A.E. ChAMP: Updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 2017, 33, 3982–3984. [Google Scholar] [CrossRef] [Green Version]
- Teschendorff, A.E.; Marabita, F.; Lechner, M.; Bartlett, T.; Tegner, J.; Gomez-Cabrero, D.; Beck, S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013, 29, 189–196. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D. RStudio, ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. 2022. Available online: https://CRAN.R-project.org/package=ggplot2 (accessed on 24 June 2022).
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. Gplots: Various R Programming Tools for Plotting Data. 2022. Available online: https://CRAN.R-project.org/package=gplots (accessed on 24 June 2022).
- Tennekes, M.; Ellis, P. Treemap: Treemap Visualization. 2021. Available online: https://CRAN.R-project.org/package=treemap (accessed on 24 June 2022).


| Characteristics | Overall Sample | Discovery Sample | ||
|---|---|---|---|---|
| HCM, N = 13 | AS, N = 14 | HCM, N = 8 | AS, N = 5 | |
| Age, years | 56.5 ± 12.0 | 60.4 ± 9.0 | 55.0 ± 12.7 | 64.0 ± 9.5 |
| Female, n (%) | 6 (46.2) | 5 (35.7) | 4 (50.0) | 1 (20.0) |
| BMI, kg/m2 | 28.7 ± 4.3 | 28.1 ± 4.0 | 30.6 ± 3.5 | 29.2 ± 5.4 |
| Atrial fibrillation, n (%) | 3 (23.1) | 2 (14.3) | 2 (25.0) | 1 (20.0) |
| Ventricular tachycardia, n (%) | 5 (38.5) | 4 (28.6) | 2 (25.0) | 1 (20.0) |
| Arterial hypertension, n (%) | 11 (84.6) | 12 (85.7) | 6 (75.0) | 5 (100.0) |
| Coronary heart disease, n (%) | 5 (38.5) | 4 (28.6) | 4 (50.0) | 4 (80.0) |
| Diabetes mellitus, n (%) | 2 (15.4) | 4 (28.6) | 2 (25.0) | 1 (20.0) |
| Data of instrumental examination and laboratory tests | ||||
| Maximal LV wall thickness, mm | 22.5 ± 5.0 | 16.6 ± 3.3 | 23.5 ± 5.6 | 15.0 ± 2.0 |
| LA diameter, mm | 44.5 ± 3.7 | 40.7 ± 4.6 | 44.1 ± 3.5 | 41.4 ± 2.3 |
| LA end-systolic volume index, mL/m2 | 49.9 ± 11.3 | 40.8 ± 9.6 | 49.3 ± 11.1 | 38.8 ± 9.5 |
| Maximal LV outflow tract pressure gradient, mmHg | 114.6 ± 27.6 | 100.8 ± 28.5 | 122.1 ± 28.4 | 90.0 ± 35.8 |
| LV ejection fraction, % | 61.4 ± 6.9 | 55.8 ± 7.4 | 62.3 ± 8.3 | 58.0 ± 5.1 |
| Severe mitral regurgitation, n (%) | 5 (38.5) | 0 (0.0) | 4 (50.0) | 0 (0.0) |
| Giant T-wave inversions, n (%) | 4 (30.8) | 0 (0.0) | 2 (20.0) | 0 (0.0) |
| Sokolow-Lyon index, mm | 39.5 ± 13.5 | 29.6 ± 10.6 | 38.3 ± 15.4 | 27.2 ± 10.8 |
| eGFR, mL/min | 99.0 ± 40.6 | 99.0 ± 33.1 | 108.8 ± 41.0 | 87.1 ± 37.3 |
| NT-proBNP, pg/ml | 1646.6 2± 1497.2 | 728.6 ± 1462.6 | 1901.6 ± 1757.8 | 348.4 ± 356.5 |
| Drug administration | ||||
| Beta-blockers, n (%) | 12 (92.3) | 9 (64.3) | 7 (87.5) | 5 (100.0) |
| ACE inhibitors or ARBs, n (%) | 10 (76.9) | 8 (57.1) | 6 (75.0) | 4 (80.0) |
| Loop diuretics, n (%) | 6 (46.2) | 7 (50.0) | 4 (50.0) | 4 (80.0) |
| MRAs, n (%) | 3 (23.1) | 3 (21.4) | 2 (25.0) | 2 (40.0) |
| No. | Gene | Genomic Location | Log2 FC | padj-Value | No. | Gene | Genomic Location | Log2 FC | padj-Value |
|---|---|---|---|---|---|---|---|---|---|
| Genes downregulated in HCM | |||||||||
| 1 | C4B | 6p21.33 | −1.49 | 1.74 × 10−6 | 20 | PTGIR | 19q13.32 | −1.10 | 0.0070 |
| 2 | NOTCH3 | 19p13.12 | −1.18 | 5.33 × 10−6 | 21 | BGN | Xq28 | −1.14 | 0.0073 |
| 3 | IGF2 | 11p15.5 | −2.10 | 3.95 × 10−5 | 22 | GRAMD1C | 3q13.31 | −1.61 | 0.0084 |
| 4 | LAMA5 | 20q13.33 | −1.06 | 0.00017 | 23 | NACA | 12q13.3 | −1.17 | 0.012 |
| 5 | LTBP4 | 19q13.2 | −1.00 | 0.00057 | 24 | ENSG00000272789.1 | 2q14.3 | −1.12 | 0.015 |
| 6 | C4A | 6p21.33 | −1.36 | 0.0010 | 25 | SSPOP | 7q36.1 | −1.11 | 0.015 |
| 7 | LMX1B | 9q33.3 | −1.71 | 0.0012 | 26 | CCDC80 | 3q13.2 | −1.21 | 0.021 |
| 8 | PKD1P4 | 16p12.3 | −1.24 | 0.0014 | 27 | MN1 | 22q12.1 | −1.13 | 0.021 |
| 9 | SLC35F2 | 11q22.3 | −1.72 | 0.0016 | 28 | MRC2 | 17q23.2 | −1.08 | 0.022 |
| 10 | SPOCK1 | 5q31.2 | −1.41 | 0.0019 | 29 | GPC6 | 13q31.3 | −1.40 | 0.023 |
| 11 | ITGA11 | 15q23 | −1.09 | 0.0023 | 30 | NR1D1 | 17q21.1 | −1.04 | 0.027 |
| 12 | UCKL1-AS1 | 20q13.33 | −2.00 | 0.0025 | 31 | SLC6A9 | 1p34.1 | −1.23 | 0.028 |
| 13 | KCNC3 | 19q13.33 | −1.20 | 0.0028 | 32 | PSD4 | 2q14.1 | −1.01 | 0.028 |
| 14 | NECTIN1 | 11q23.3 | −1.06 | 0.0038 | 33 | ADAMTS5 | 21q21.3 | −1.26 | 0.038 |
| 15 | BRSK2 | 11p15.5 | −1.30 | 0.0038 | 34 | TPTEP1 | 22q11.1 | −1.10 | 0.040 |
| 16 | CHGB | 20p12.3 | −1.29 | 0.0041 | 35 | NDUFA13 | 19p13.11 | −1.63 | 0.042 |
| 17 | KCNT1 | 9q34.3 | −1.03 | 0.0041 | 36 | MYH11 | 16p13.11 | −1.00 | 0.045 |
| 18 | CCN3 | 8q24.12 | −1.08 | 0.0060 | 37 | PTK7 | 6p21.1 | −1.04 | 0.045 |
| 19 | NCOR2 | 12q24.31 | −1.03 | 0.0068 | 38 | DPT | 1q24.2 | −1.07 | 0.049 |
| Genes upregulated in HCM | |||||||||
| 1 | EIF4EBP3 | 5q31.3 | 1.45 | 3.95 × 10−5 | 8 | ENSG00000279041.1 | 8p12 | 1.06 | 0.0046 |
| 2 | CTXND1 | 15q25.1 | 1.43 | 3.95 × 10−5 | 9 | APOD | 3q29 | 1.05 | 0.0074 |
| 3 | GADD45G | 9q22.2 | 1.06 | 0.00057 | 10 | ENSG00000287047.1 | 10q25.1 | 1.22 | 0.026 |
| 4 | ATRNL1 | 10q25.3 | 1.82 | 0.0010 | 11 | SLC26A4 | 7q22.3 | 1.05 | 0.036 |
| 5 | ST8SIA5 | 18q21.1 | 1.08 | 0.0011 | 12 | ENSG00000286401.1 | 10q11.23 | 1.34 | 0.047 |
| 6 | SOCS2-AS1 | 12q22 | 1.08 | 0.0038 | 13 | C2CD6 | 2q33.1 | 1.55 | 0.047 |
| 7 | SH3GL2 | 9p22.2 | 1.26 | 0.0043 | 14 | APOA1 | 11q23.3 | 1.06 | 0.048 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiselev, I.; Kozin, M.; Baulina, N.; Pisklova, M.; Danilova, L.; Zotov, A.; Chumakova, O.; Zateyshchikov, D.; Favorova, O. Novel Genes Involved in Hypertrophic Cardiomyopathy: Data of Transcriptome and Methylome Profiling. Int. J. Mol. Sci. 2022, 23, 15280. https://doi.org/10.3390/ijms232315280
Kiselev I, Kozin M, Baulina N, Pisklova M, Danilova L, Zotov A, Chumakova O, Zateyshchikov D, Favorova O. Novel Genes Involved in Hypertrophic Cardiomyopathy: Data of Transcriptome and Methylome Profiling. International Journal of Molecular Sciences. 2022; 23(23):15280. https://doi.org/10.3390/ijms232315280
Chicago/Turabian StyleKiselev, Ivan, Maxim Kozin, Natalia Baulina, Maria Pisklova, Ludmila Danilova, Alexandr Zotov, Olga Chumakova, Dmitry Zateyshchikov, and Olga Favorova. 2022. "Novel Genes Involved in Hypertrophic Cardiomyopathy: Data of Transcriptome and Methylome Profiling" International Journal of Molecular Sciences 23, no. 23: 15280. https://doi.org/10.3390/ijms232315280
APA StyleKiselev, I., Kozin, M., Baulina, N., Pisklova, M., Danilova, L., Zotov, A., Chumakova, O., Zateyshchikov, D., & Favorova, O. (2022). Novel Genes Involved in Hypertrophic Cardiomyopathy: Data of Transcriptome and Methylome Profiling. International Journal of Molecular Sciences, 23(23), 15280. https://doi.org/10.3390/ijms232315280







