Transcriptome Analysis Reveals the Molecular Regularity Mechanism Underlying Stem Bulblet Formation in Oriental Lily ‘Siberia’; Functional Characterization of the LoLOB18 Gene
Abstract
:1. Introduction
2. Results
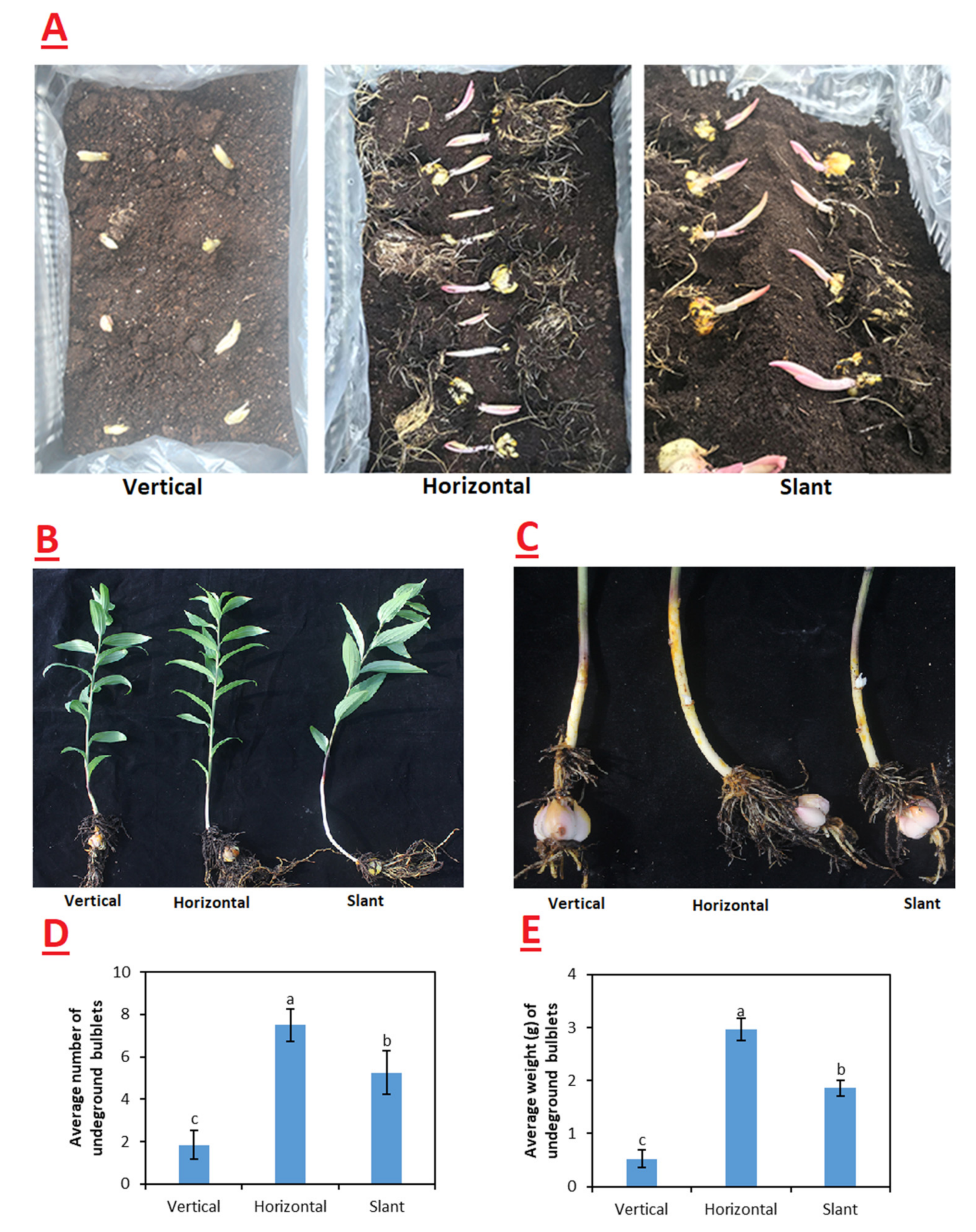
2.1. Phenotypic Analysis
2.2. Transcriptome Sequencing and Transcript Assembly
2.3. Functional Annotation and Classification of Unigenes
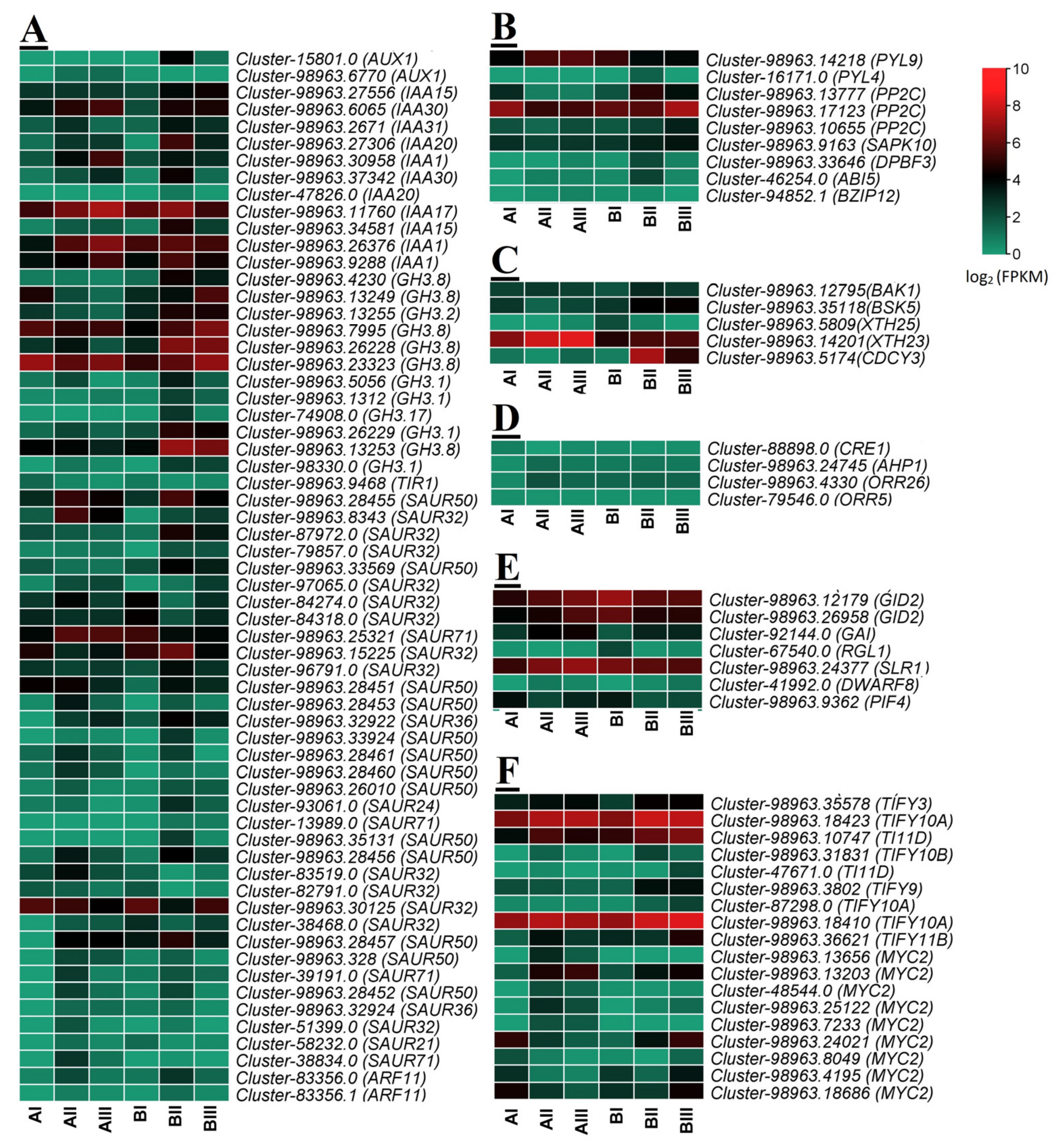
2.4. Analysis of DEGs and Candidate Gene Selection
2.5. Validation of Transcriptome Data with qRT-PCR Analysis
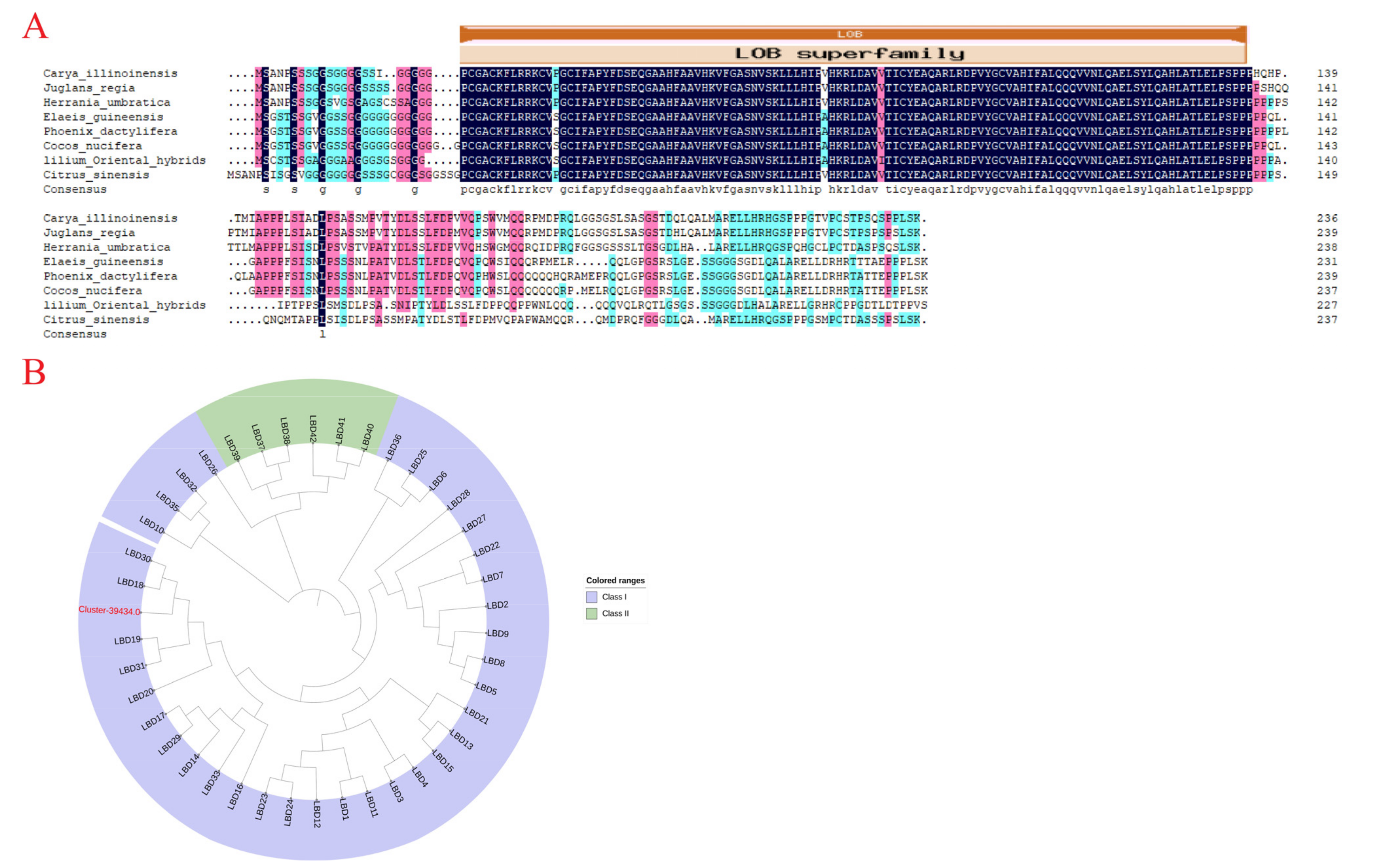
2.6. Candidate Gene Cloning and Characterization Analysis
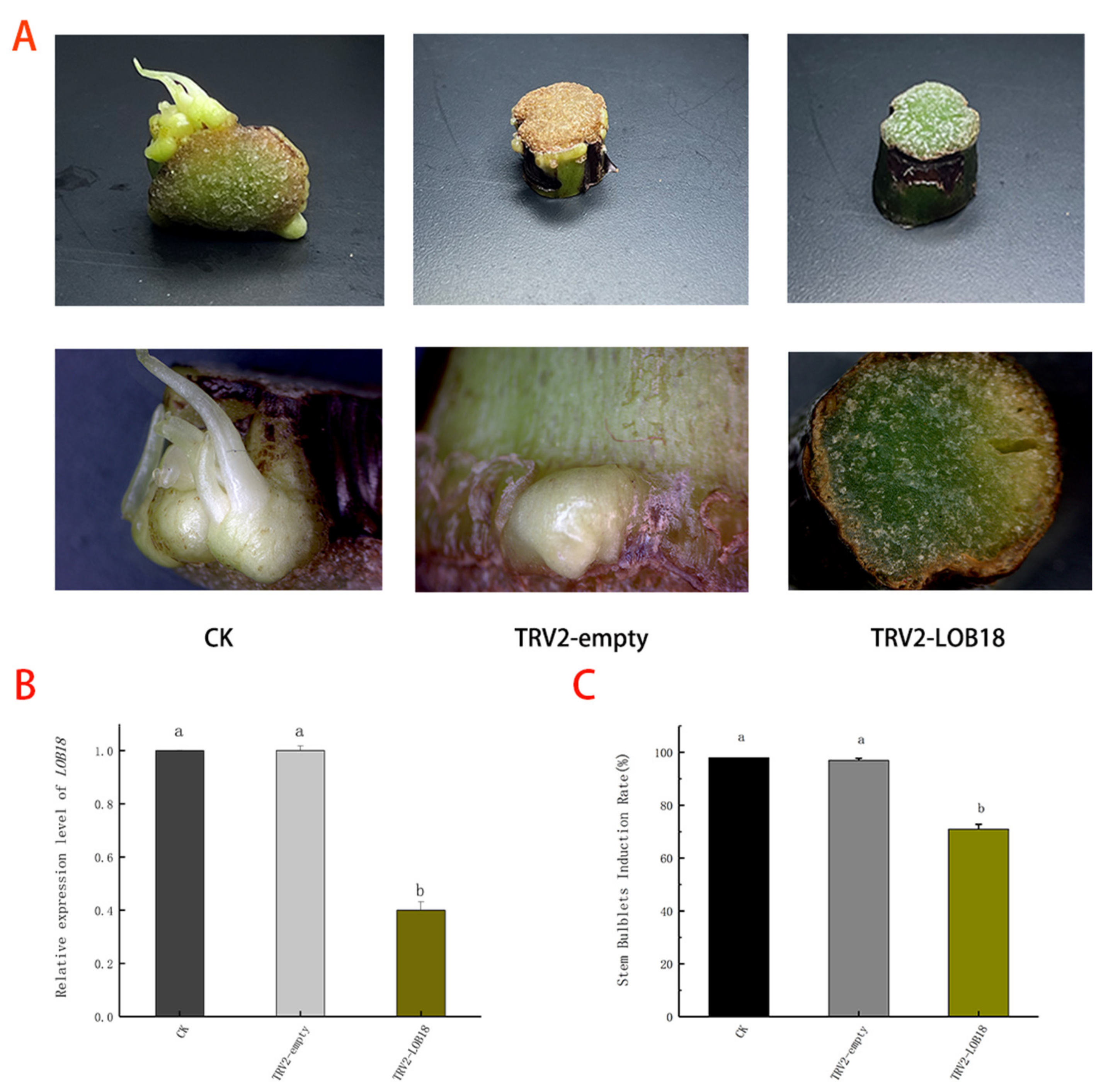
2.7. LoLOB18 Silencing Inhibits Stem Bulblet Formation
3. Discussion
3.1. Different Hormones Regulate Bulblet Formation
3.2. Starch Synthesis Contributes to Bulblet Formation
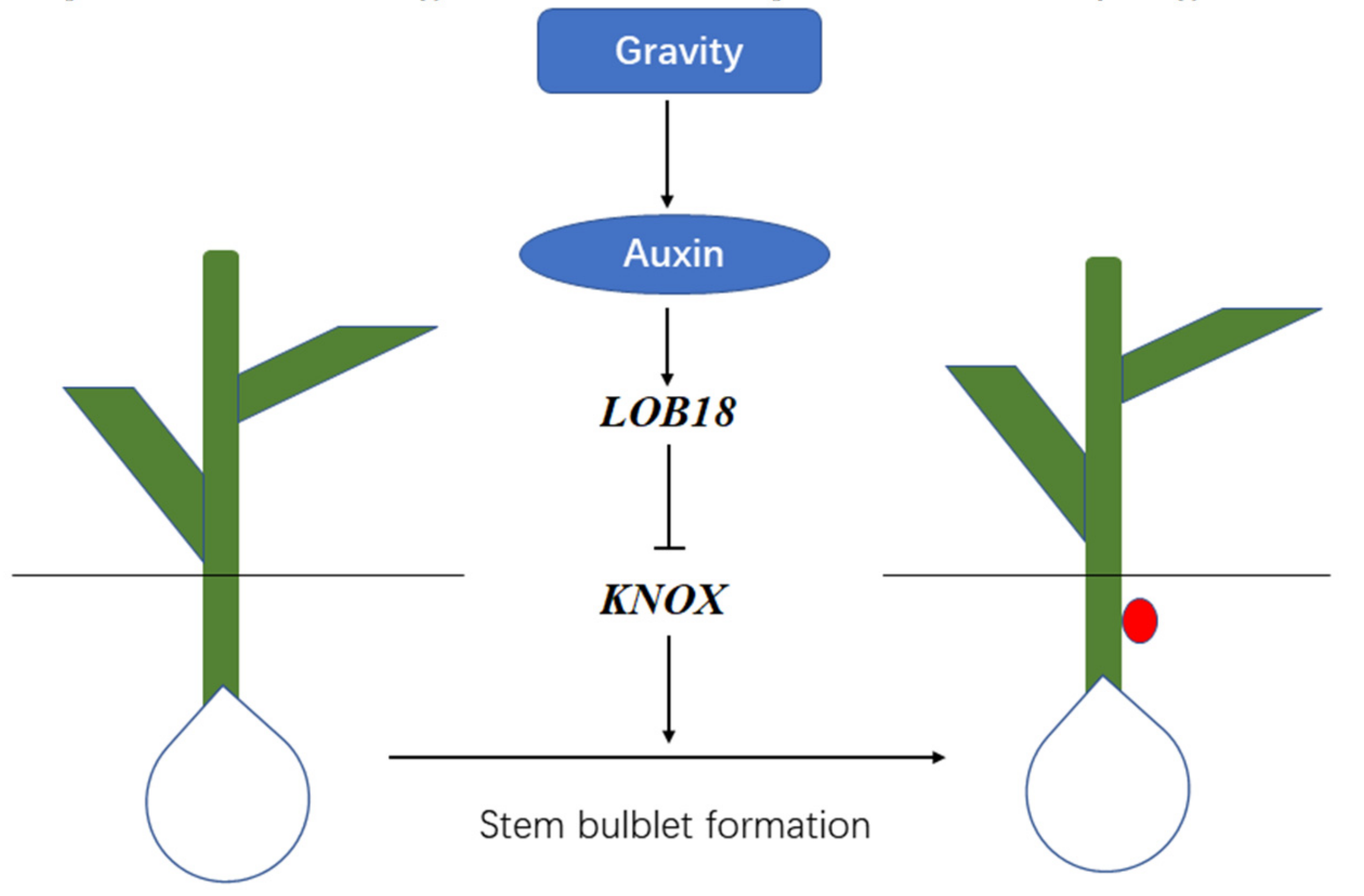
3.3. LoLOB18 Is Positive Regulator during Underground Bulblet Formation
4. Materials and Methods
4.1. Plant Material and Samples Collection
4.2. RNA Extraction and cDNA Synthesis
4.3. Library Construction and Transcriptome Sequencing
4.4. RNA-Sequencing Data Analysis
4.5. RNA Extraction and qRT-PCR
4.6. Isolation of LoLOB18 Gene
4.7. Virus-Induced Gene Silencing (VIGS)
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bakhshaie, M.; Khosravi, S.; Azadi, P.; Bagheri, H.; van Tuyl, J.M. Biotechnological Advances in Lilium. Plant Cell Rep. 2016, 35, 1799–1826. [Google Scholar] [CrossRef] [PubMed]
- Arzate-Fernández, A.-M.; Nakazaki, T.; Okumoto, Y.; Tanisaka, T. Efficient Callus Induction and Plant Regeneration from Filaments with Anther in Lily (Lilium longiflorum Thunb.). Plant Cell Rep. 1997, 16, 836–840. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ren, Z.; Gao, C.; Sun, M.; Li, S.; Min, R.; Wu, J.; Li, D.; Wang, X.; Wei, Y.; et al. Change in Sucrose Cleavage Pattern and Rapid Starch Accumulation Govern Lily Shoot-to-Bulblet Transition in Vitro. Front. Plant Sci. 2021, 11, 564713. [Google Scholar] [CrossRef] [PubMed]
- Chinestra, S.C.; Curvetto, N.R.; Marinangeli, P.A. Production of Virus-Free Plants of Lilium Spp. from Bulbs Obtained in Vitro and Ex Vitro. Sci. Hortic. 2015, 194, 304–312. [Google Scholar] [CrossRef]
- Lian, M.L.; Chakrabarty, D.; Paek, K.Y. Bulblet Formation from Bulbscale Segments of Lilium Using Bioreactor System. Biol. Plant. 2003, 46, 199–203. [Google Scholar] [CrossRef]
- Niimi, Y.; Han, D.-S.; Fujisaki, M. Production of Virus-Free Plantlets by Anther Culture Of. Sci. Hortic. 2001, 90, 325–334. [Google Scholar] [CrossRef]
- Yang, P.; Xu, L.; Xu, H.; Tang, Y.; He, G.; Cao, Y.; Feng, Y.; Yuan, S.; Ming, J. Histological and Transcriptomic Analysis during Bulbil Formation in Lilium Lancifolium. Front. Plant Sci. 2017, 8, 1508. [Google Scholar] [CrossRef] [Green Version]
- Rafiq, S.; Rather, Z.A.; Bhat, R.A.; Nazki, I.T.; AL-Harbi, M.S.; Banday, N.; Farooq, I.; Samra, B.N.; Khan, M.H.; Ahmed, A.F.; et al. Standardization of in Vitro Micropropagation Procedure of Oriental Lilium Hybrid Cv. ‘Ravenna’. Saudi J. Biol. Sci. 2021, 28, 7581–7587. [Google Scholar] [CrossRef]
- Wu, S.; Wu, J.; Jiao, X.; Zhang, Q.; Lv, Y. The Dynamics of Changes in Starch and Lipid Droplets and Sub-Cellular Localization of β-Amylase During the Growth of Lily Bulbs. J. Integr. Agric. 2012, 11, 585–592. [Google Scholar] [CrossRef]
- Raza, A.; Salehi, H.; Rahman, M.A.; Zahid, Z.; Madadkar Haghjou, M.; Najafi-Kakavand, S.; Charagh, S.; Osman, H.S.; Albaqami, M.; Zhuang, Y.; et al. Plant Hormones and Neurotransmitter Interactions Mediate Antioxidant Defenses under Induced Oxidative Stress in Plants. Front. Plant Sci. 2022, 13, 961872. [Google Scholar] [CrossRef]
- Zhang, Y.; Yong, Y.B.; Wang, Q.; Lu, Y.M. Physiological and Molecular Changes during Lily Underground Stem Axillary Bulbils Formation. Russ. J. Plant Physiol. 2018, 65, 372–383. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, Z.; Yong, Y.; Lyu, Y. Hormonal Regulatory Patterns of LaKNOXs and LaBEL1 Transcription Factors Reveal Their Potential Role in Stem Bulblet Formation in LA Hybrid Lily. Int. J. Mol. Sci. 2021, 22, 13502. [Google Scholar] [CrossRef]
- Salachna, P.; Mikiciuk, M.; Zawadzińska, A.; Piechocki, R.; Ptak, P.; Mikiciuk, G.; Pietrak, A.; Łopusiewicz, Ł. Changes in Growth and Physiological Parameters of ×Amarine Following an Exogenous Application of Gibberellic Acid and Methyl Jasmonate. Agronomy 2020, 10, 980. [Google Scholar] [CrossRef]
- Nhut, D.T. The Control of in Vitro Direct Main Stem Formation of Lilium Longiflorum Derived from Receptacle Culture, and Rapid Propagation by Using in Vitro Stem Nodes. Plant Growth Regul. 2003, 40, 179–184. [Google Scholar] [CrossRef]
- He, G.; Yang, P.; Tang, Y.; Cao, Y.; Qi, X.; Xu, L.; Ming, J. Mechanism of Exogenous Cytokinins Inducing Bulbil Formation in Lilium Lancifolium in Vitro. Plant Cell Rep. 2020, 39, 861–872. [Google Scholar] [CrossRef]
- Bandurski, R.S.; Schulze, A.; Momonoki, Y. Gravity-Induced Asymmetric Distribution of a Plant Growth Hormone. Physiologist 1984, 27, S123–S126. [Google Scholar]
- Ottenschläger, I.; Wolff, P.; Wolverton, C.; Bhalerao, R.P.; Sandberg, G.; Ishikawa, H.; Evans, M.; Palme, K. Gravity-Regulated Differential Auxin Transport from Columella to Lateral Root Cap Cells. Proc. Natl. Acad. Sci. USA 2003, 100, 2987–2991. [Google Scholar] [CrossRef] [Green Version]
- Waidmann, S.; Kleine-Vehn, J. Asymmetric Cytokinin Signaling Opposes Gravitropism in Roots. J. Integr. Plant Biol. 2020, 62, 882–886. [Google Scholar] [CrossRef]
- He, G.; Yang, P.; Cao, Y.; Tang, Y.; Wang, L.; Song, M.; Wang, J.; Xu, L.; Ming, J. Cytokinin Type-B Response Regulators Promote Bulbil Initiation in Lilium Lancifolium. Int. J. Mol. Sci. 2021, 22, 3320. [Google Scholar] [CrossRef]
- He, G.; Cao, Y.; Wang, J.; Song, M.; Bi, M.; Tang, Y.; Xu, L.; Ming, J.; Yang, P. WUSCHEL -Related Homeobox Genes Cooperate with Cytokinin to Promote Bulbil Formation in Lilium Lancifolium. Plant Physiol. 2022, 190, 387–402. [Google Scholar] [CrossRef]
- Abraham-Juárez, M.J.; Martínez-Hernández, A.; Leyva-González, M.A.; Herrera-Estrella, L.; Simpson, J. Class I KNOX Genes Are Associated with Organogenesis during Bulbil Formation in Agave Tequilana. J. Exp. Bot. 2010, 61, 4055–4067. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chai, M.; Zhu, X.; Zhang, X.; Li, H.; Wang, D.; Xing, Q.; Zhang, J.; Sun, M.; Shi, L. Cloning and Expression Analysis of LoCCD8 during IAA-Induced Bulbils Outgrowth in Lily (Oriental Hybrid ‘Sorbonne’). J. Plant Physiol. 2019, 236, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Pasare, S.A.; Ducreux, L.J.M.; Morris, W.L.; Campbell, R.; Sharma, S.K.; Roumeliotis, E.; Kohlen, W.; Krol, S.; Bramley, P.M.; Roberts, A.G.; et al. The Role of the Potato (S Olanum Tuberosum) CCD8 Gene in Stolon and Tuber Development. New Phytol. 2013, 198, 1108–1120. [Google Scholar] [CrossRef] [PubMed]
- Majer, C.; Hochholdinger, F. Defining the Boundaries: Structure and Function of LOB Domain Proteins. Trends Plant Sci. 2011, 16, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 Regulate Lateral Root Formation via Direct Activation of LBD/ASL Genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.W.; Kim, N.Y.; Lee, D.J.; Kim, J. LBD18/ASL20 Regulates Lateral Root Formation in Combination with LBD16/ASL18 Downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009, 151, 1377–1389. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.; Zhu, J.; Du, X.; Cui, X. Effects of Three Auxin-Inducible LBD Members on Lateral Root Formation in Arabidopsis Thaliana. Planta 2012, 236, 1227–1237. [Google Scholar] [CrossRef]
- Fan, M.; Xu, C.; Xu, K.; Hu, Y. LATERAL ORGAN BOUNDARIES DOMAIN Transcription Factors Direct Callus Formation in Arabidopsis Regeneration. Cell Res. 2012, 22, 1169–1180. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Luo, F.; Hochholdinger, F. LOB Domain Proteins: Beyond Lateral Organ Boundaries. Trends Plant Sci. 2016, 21, 159–167. [Google Scholar] [CrossRef]
- Benková, E.; Ivanchenko, M.G.; Friml, J.; Shishkova, S.; Dubrovsky, J.G. A Morphogenetic Trigger: Is There an Emerging Concept in Plant Developmental Biology? Trends Plant Sci. 2009, 14, 189–193. [Google Scholar] [CrossRef]
- Irfan, M.; Kumar, P.; Kumar, V.; Datta, A. Fruit Ripening Specific Expression of β-D-N-Acetylhexosaminidase (β-Hex) Gene in Tomato Is Transcriptionally Regulated by Ethylene Response Factor SlERF.E4. Plant Sci. 2022, 323, 111380. [Google Scholar] [CrossRef]
- Sharma, S.; Shree, B.; Aditika; Sharma, A.; Irfan, M.; Kumar, P. Nanoparticle-Based Toxicity in Perishable Vegetable Crops: Molecular Insights, Impact on Human Health and Mitigation Strategies for Sustainable Cultivation. Environ. Res. 2022, 212, 113168. [Google Scholar] [CrossRef]
- Kumari, C.; Sharma, M.; Kumar, V.; Sharma, R.; Kumar, V.; Sharma, P.; Kumar, P.; Irfan, M. Genome Editing Technology for Genetic Amelioration of Fruits and Vegetables for Alleviating Post-Harvest Loss. Bioengineering 2022, 9, 176. [Google Scholar] [CrossRef]
- Tayal, R.; Kumar, V.; Irfan, M. Harnessing the Power of Hydrogen Sulphide (H2S) for Improving Fruit Quality Traits. Plant Biol. 2022, 24, 594–601. [Google Scholar] [CrossRef]
- Kumar, S.; Awasthi, V.; Kanwar, J.K. Influence of Growth Regulators and Nitrogenous Compounds on in Vitro Bulblet Formation and Growth in Oriental Lily. Hortic. Sci. 2007, 34, 77–83. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Irfan, M.; Ghosh, S.; Chakraborty, N.; Chakraborty, S.; Datta, A. Fruit Ripening Mutants Reveal Cell Metabolism and Redox State during Ripening. Protoplasma 2016, 253, 581–594. [Google Scholar] [CrossRef]
- Irfan, M.; Ghosh, S.; Kumar, V.; Chakraborty, N.; Chakraborty, S.; Datta, A. Insights into Transcriptional Regulation of -D-N-Acetylhexosaminidase, an N-Glycan-Processing Enzyme Involved in Ripening-Associated Fruit Softening. J. Exp. Bot. 2014, 65, 5835–5848. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, J.; Shi, B.; Yu, T.; Qi, J.; Meyerowitz, E.M.; Jiao, Y. The Stem Cell Niche in Leaf Axils Is Established by Auxin and Cytokinin in Arabidopsis. Plant Cell 2014, 26, 2055–2067. [Google Scholar] [CrossRef] [Green Version]
- Reinhardt, D.; Pesce, E.-R.; Stieger, P.; Mandel, T.; Baltensperger, K.; Bennett, M.; Traas, J.; Friml, J.; Kuhlemeier, C. Regulation of Phyllotaxis by Polar Auxin Transport. Nature 2003, 426, 255–260. [Google Scholar] [CrossRef]
- Greb, T.; Clarenz, O.; Schäfer, E.; Müller, D.; Herrero, R.; Schmitz, G.; Theres, K. Molecular Analysis of the LATERAL SUPPRESSOR Gene in Arabidopsis Reveals a Conserved Control Mechanism for Axillary Meristem Formation. Genes Dev. 2003, 17, 1175–1187. [Google Scholar] [CrossRef] [Green Version]
- Roumeliotis, E.; Kloosterman, B.; Oortwijn, M.; Kohlen, W.; Bouwmeester, H.J.; Visser, R.G.F.; Bachem, C.W.B. The Effects of Auxin and Strigolactones on Tuber Initiation and Stolon Architecture in Potato. J. Exp. Bot. 2012, 63, 4539–4547. [Google Scholar] [CrossRef]
- Hao, J.; Li, C.; Sun, X.; Yang, Q. Phylogeny and Divergence Time Estimation of Cheilostome Bryozoans Based on Mitochodrial 16S RRNA Sequences. Chin. Sci. Bull. 2005, 50, 1205–1211. [Google Scholar] [CrossRef]
- Navarro, C.; Cruz-Oró, E.; Prat, S. Conserved Function of FLOWERING LOCUS T (FT) Homologues as Signals for Storage Organ Differentiation. Curr. Opin. Plant Biol. 2015, 23, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Zhu, Z.; Guo, Q.; Yang, X.; Liu, L.; Sun, Y.; Wang, C. Dynamic Changes in Carbohydrate Metabolism and Endogenous Hormones during Tulipa Edulis Stolon Development into a New Bulb. J. Plant Biol. 2016, 59, 121–132. [Google Scholar] [CrossRef]
- Pan, T.; Ali, M.M.; Gong, J.; She, W.; Pan, D.; Guo, Z.; Yu, Y.; Chen, F. Fruit Physiology and Sugar-Acid Profile of 24 Pomelo (Citrus Grandis (L.) Osbeck) Cultivars Grown in Subtropical Region of China. Agronomy 2021, 11, 2393. [Google Scholar] [CrossRef]
- Yu, X.; Ali, M.M.; Li, B.; Fang, T.; Chen, F. Transcriptome Data-based Identification of Candidate Genes Involved in Metabolism and Accumulation of Soluble Sugars during Fruit Development in ‘Huangguan’ Plum. J. Food Biochem. 2021, 45, e13878. [Google Scholar] [CrossRef]
- Ali, M.M.; Javed, T.; Mauro, R.P.; Shabbir, R.; Afzal, I.; Yousef, A.F. Effect of Seed Priming with Potassium Nitrate on the Performance of Tomato. Agriculture 2020, 10, 498. [Google Scholar] [CrossRef]
- He, X.; Shi, L.; Yuan, Z.; Xu, Z.; Zhang, Z.; Yi, M. Effects of Lipoxygenase on the Corm Formation and Enlargement in Gladiolus Hybridus. Sci. Hortic. 2008, 118, 60–69. [Google Scholar] [CrossRef]
- Yu, Z.; Chen, L.C.; Suzuki, H.; Ariyada, O.; Erra-Balsells, R.; Nonami, H.; Hiraoka, K. Direct Profiling of Phytochemicals in Tulip Tissues and in Vivo Monitoring of the Change of Carbohydrate Content in Tulip Bulbs by Probe Electrospray Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2009, 20, 2304–2311. [Google Scholar] [CrossRef] [Green Version]
- Tuncel, A.; Okita, T.W. Improving Starch Yield in Cereals by Over-Expression of ADPglucose Pyrophosphorylase: Expectations and Unanticipated Outcomes. Plant Sci. 2013, 211, 52–60. [Google Scholar] [CrossRef]
- Irfan, M.; Kumar, P.; Ahmad, I.; Datta, A. Unraveling the Role of Tomato Bcl-2-Associated Athanogene (BAG) Proteins during Abiotic Stress Response and Fruit Ripening. Sci. Rep. 2021, 11, 21734. [Google Scholar] [CrossRef]
- Kumar, V.; Irfan, M.; Datta, A. Manipulation of Oxalate Metabolism in Plants for Improving Food Quality and Productivity. Phytochemistry 2019, 158, 103–109. [Google Scholar] [CrossRef]
- Irfan, M.; Ghosh, S.; Meli, V.S.; Kumar, A.; Kumar, V.; Chakraborty, N.; Chakraborty, S.; Datta, A. Fruit Ripening Regulation of α-Mannosidase Expression by the MADS Box Transcription Factor RIPENING INHIBITOR and Ethylene. Front. Plant Sci. 2016, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Giroux, M.J.; Shaw, J.; Barry, G.; Cobb, B.G.; Greene, T.; Okita, T.; Hannah, L.C. A Single Mutation That Increases Maize Seed Weight. Proc. Natl. Acad. Sci. USA 1996, 93, 5824–5829. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-M.; Lee, Y.-H.; Kim, H.; Seo, S.; Kwon, S.; Cho, H.; Kim, S.-I.; Okita, T.; Kim, D. Characterization of the Potato Upreg1gene, Encoding a Mutated ADP-Glucose Pyrophosphorylase Large Subunit, in Transformed Rice. Plant Cell Tissue Organ Cult. 2010, 102, 171–179. [Google Scholar] [CrossRef]
- Meyer, F.D.; Smidansky, E.D.; Beecher, B.; Greene, T.W.; Giroux, M.J. The Maize Sh2r6hs ADP-Glucose Pyrophosphorylase (AGP) Large Subunit Confers Enhanced AGP Properties in Transgenic Wheat (Triticum Aestivum). Plant Sci. 2004, 167, 899–911. [Google Scholar] [CrossRef]
- Lin, W.-c. The Arabidopsis LATERAL ORGAN BOUNDARIES-Domain Gene ASYMMETRIC LEAVES2 Functions in the Repression of KNOX Gene Expression and in Adaxial-Abaxial Patterning. Plant Cell Online 2003, 15, 2241–2252. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Li, Z.; Zhu, Y.; Wang, H.; Ma, H.; Dong, A.; Huang, H. Arabidopsis Genes AS1, AS2, and JAG Negatively Regulate Boundary-Specifying Genes to Promote Sepal and Petal Development. Plant Physiol. 2008, 146, 323–324. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 Are Involved in the First-Step Cell Fate Transition during de Novo Root Organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.-R. Members of the LBD Family of Transcription Factors Repress Anthocyanin Synthesis and Affect Additional Nitrogen Responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.-L.; Shi, M.-Z.; Xie, D.-Y. Regulation of Anthocyanin Biosynthesis by Nitrogen in TTG1–GL3/TT8–PAP1-Programmed Red Cells of Arabidopsis Thaliana. Planta 2012, 236, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Kareem, A.; Durgaprasad, K.; Sugimoto, K.; Du, Y.; Pulianmackal, A.J.; Trivedi, Z.B.; Abhayadev, P.V.; Pinon, V.; Meyerowitz, E.M.; Scheres, B.; et al. PLETHORA Genes Control Regeneration by a Two-Step Mechanism. Curr. Biol. 2015, 25, 1017–1030. [Google Scholar] [CrossRef] [Green Version]
- Chatfield, S.P.; Capron, R.; Severino, A.; Penttila, P.-A.; Alfred, S.; Nahal, H.; Provart, N.J. Incipient Stem Cell Niche Conversion in Tissue Culture: Using a Systems Approach to Probe Early Events in WUSCHEL -Dependent Conversion of Lateral Root Primordia into Shoot Meristems. Plant J. 2013, 73, 798–813. [Google Scholar] [CrossRef]
- Kareem, A.; Radhakrishnan, D.; Wang, X.; Bagavathiappan, S.; Trivedi, Z.B.; Sugimoto, K.; Xu, J.; Mähönen, A.P.; Prasad, K. Protocol: A Method to Study the Direct Reprogramming of Lateral Root Primordia to Fertile Shoots. Plant Methods 2016, 12, 27. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Ali, M.M.; He, Y.; Ma, S.; Rizwan, H.M.; Yang, Q.; Li, B.; Lin, Z.; Chen, F. Flavonoids Accumulation in Fruit Peel and Expression Profiling of Related Genes in Purple (Passiflora Edulis f. Edulis) and Yellow (Passiflora Edulis f. Flavicarpa) Passion Fruits. Plants 2021, 10, 2240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wei, X.; Ali, M.M.; Rizwan, H.M.; Li, B.; Li, H.; Jia, K.; Yang, X.; Ma, S.; Li, S.; et al. Changes in the Content of Organic Acids and Expression Analysis of Citric Acid Accumulation-Related Genes during Fruit Development of Yellow (Passiflora Edulis f. Flavicarpa) and Purple (Passiflora Edulis f. Edulis) Passion Fruits. Int. J. Mol. Sci. 2021, 22, 5765. [Google Scholar] [CrossRef]
- Ghosh, S.; Singh, U.K.; Meli, V.S.; Kumar, V.; Kumar, A.; Irfan, M.; Chakraborty, N.; Chakraborty, S.; Datta, A. Induction of Senescence and Identification of Differentially Expressed Genes in Tomato in Response to Monoterpene. PLoS ONE 2013, 8, e76029. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.F.; Liu, Q.; Jia, G.X. Reference Gene Selection for Gene Expression Studies in Lily Using Quantitative Real-Time PCR. Genet. Mol. Res. 2016, 15, gmr.15027982. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| ID | Homologous Gene/Protein | Regulate Pattern (BIII vs. BI) | Regulate Pattern (BII vs. BI) | Log2 FC | |
|---|---|---|---|---|---|
| BIII vs. BI | BII vs. BI | ||||
| Cluster-98963.5096 | SWEET14 | UP | UP | 25.294 | 14.737 |
| Cluster-98963.36497 | SWEET14 | UP | UP | 13.14 | 11.302 |
| Cluster-95907.0 | HSR201 | DOWN | DOWN | −12.161 | −12.058 |
| Cluster-98963.11855 | SLC50A | UP | UP | 12.024 | 14.801 |
| Cluster-98963.32942 | SWEET14 | UP | UP | 12.001 | 14.567 |
| Cluster-94100.0 | CYP89A2 | DOWN | DOWN | −11.983 | −11.879 |
| Cluster-98963.3555 | E2 binding domain | UP | UP | 11.954 | 10.721 |
| Cluster-98963.31619 | GAUT12S | UP | UP | 11.589 | 11.511 |
| Cluster-98963.30325 | PER64 | UP | UP | 10.841 | 11.787 |
| Cluster-39434.0 | LOB18 | UP | UP | 10.258 | 11.872 |
| Cluster-98963.3508 | RSI1 | UP | UP | 10.732 | 12.466 |
| Cluster-98156.0 | non-specific lipid-transfer protein | UP | UP | 10.667 | 11.265 |
| Cluster-89656.0 | PBL28 | UP | UP | 10.59 | 10.543 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, S.; Yang, C.; Ali, M.M.; Lin, M.; Tian, S.; Zhang, L.; Chen, F.; Lin, Z. Transcriptome Analysis Reveals the Molecular Regularity Mechanism Underlying Stem Bulblet Formation in Oriental Lily ‘Siberia’; Functional Characterization of the LoLOB18 Gene. Int. J. Mol. Sci. 2022, 23, 15246. https://doi.org/10.3390/ijms232315246
Fang S, Yang C, Ali MM, Lin M, Tian S, Zhang L, Chen F, Lin Z. Transcriptome Analysis Reveals the Molecular Regularity Mechanism Underlying Stem Bulblet Formation in Oriental Lily ‘Siberia’; Functional Characterization of the LoLOB18 Gene. International Journal of Molecular Sciences. 2022; 23(23):15246. https://doi.org/10.3390/ijms232315246
Chicago/Turabian StyleFang, Shaozhong, Chenglong Yang, Muhammad Moaaz Ali, Mi Lin, Shengnan Tian, Lijuan Zhang, Faxing Chen, and Zhimin Lin. 2022. "Transcriptome Analysis Reveals the Molecular Regularity Mechanism Underlying Stem Bulblet Formation in Oriental Lily ‘Siberia’; Functional Characterization of the LoLOB18 Gene" International Journal of Molecular Sciences 23, no. 23: 15246. https://doi.org/10.3390/ijms232315246
APA StyleFang, S., Yang, C., Ali, M. M., Lin, M., Tian, S., Zhang, L., Chen, F., & Lin, Z. (2022). Transcriptome Analysis Reveals the Molecular Regularity Mechanism Underlying Stem Bulblet Formation in Oriental Lily ‘Siberia’; Functional Characterization of the LoLOB18 Gene. International Journal of Molecular Sciences, 23(23), 15246. https://doi.org/10.3390/ijms232315246








