Cell Wall Matrix Polysaccharides Contribute to Salt–Alkali Tolerance in Rice
Abstract
:1. Introduction
2. Results
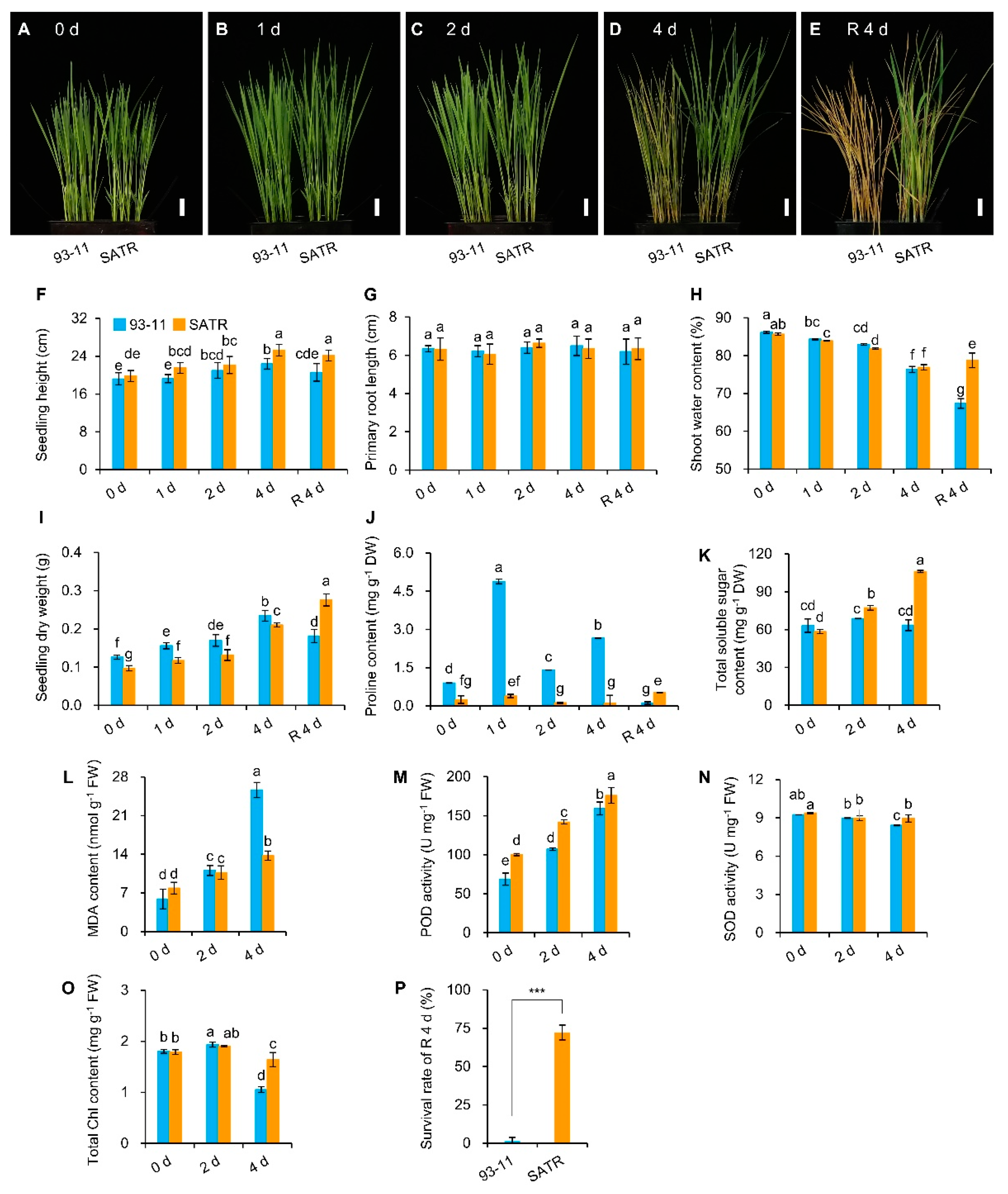
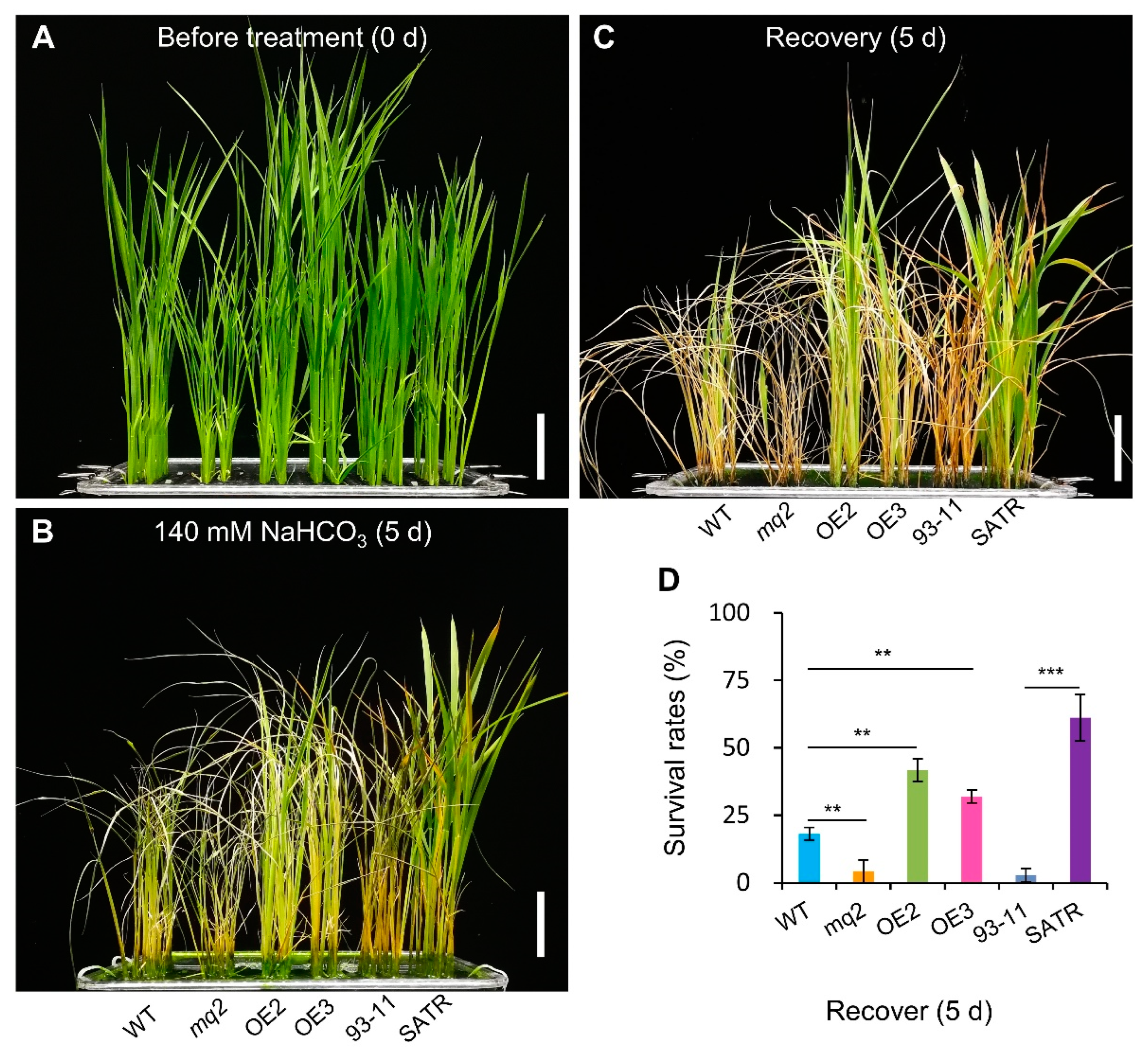
2.1. Salt–Alkali Tolerance of SATR Is Conferred by Adaptive Physio-Biochemical Responses
2.2. Enrichment of the Highlighted Cell Wall Related GO Terms and KEGG Pathways between SATR and 93-11 under SSAS
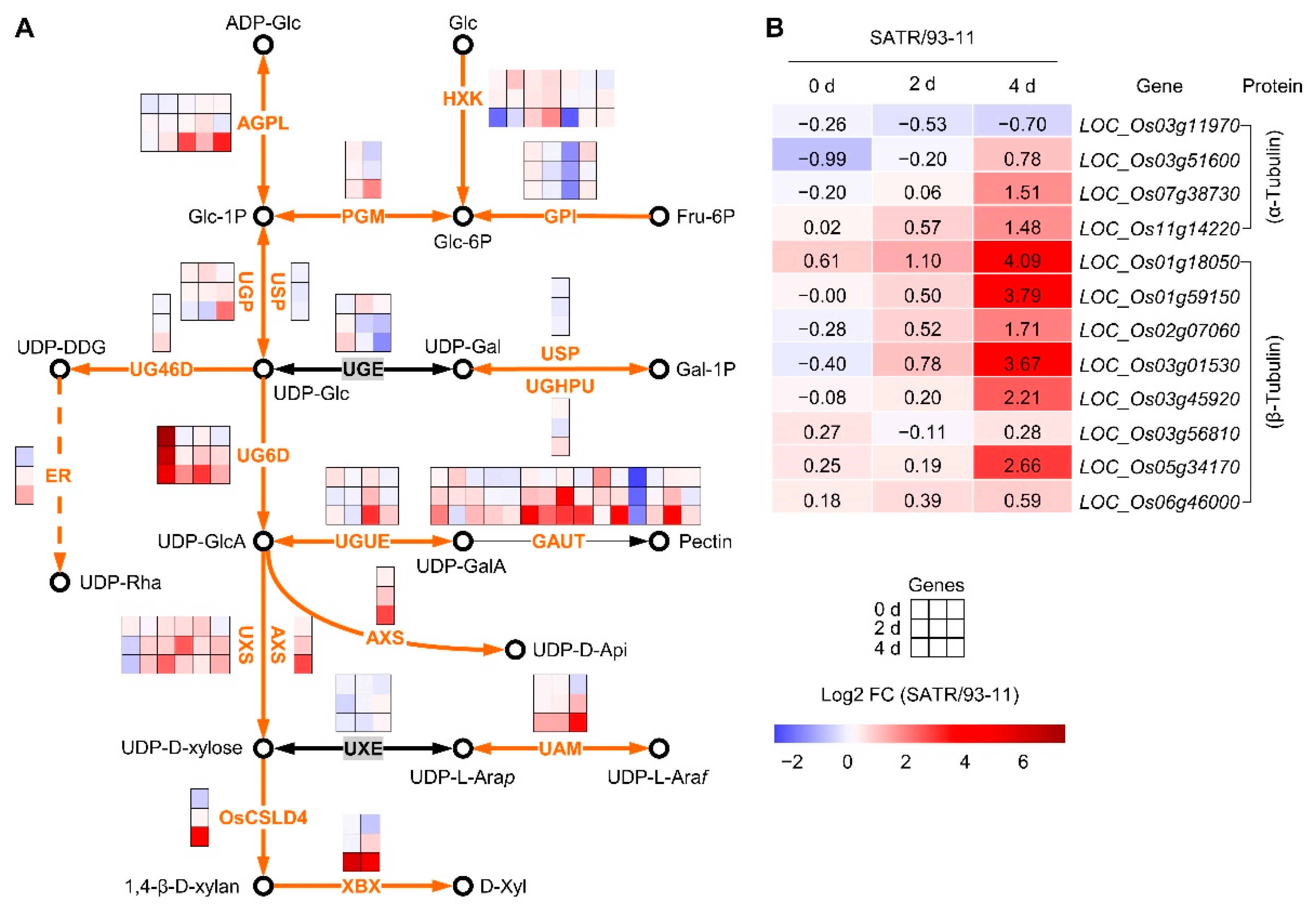
2.3. SATR Had Higher Expression Levels of Hemicellulose and Pectin Biosynthesis Pathway and Tubulin Genes under SSAS
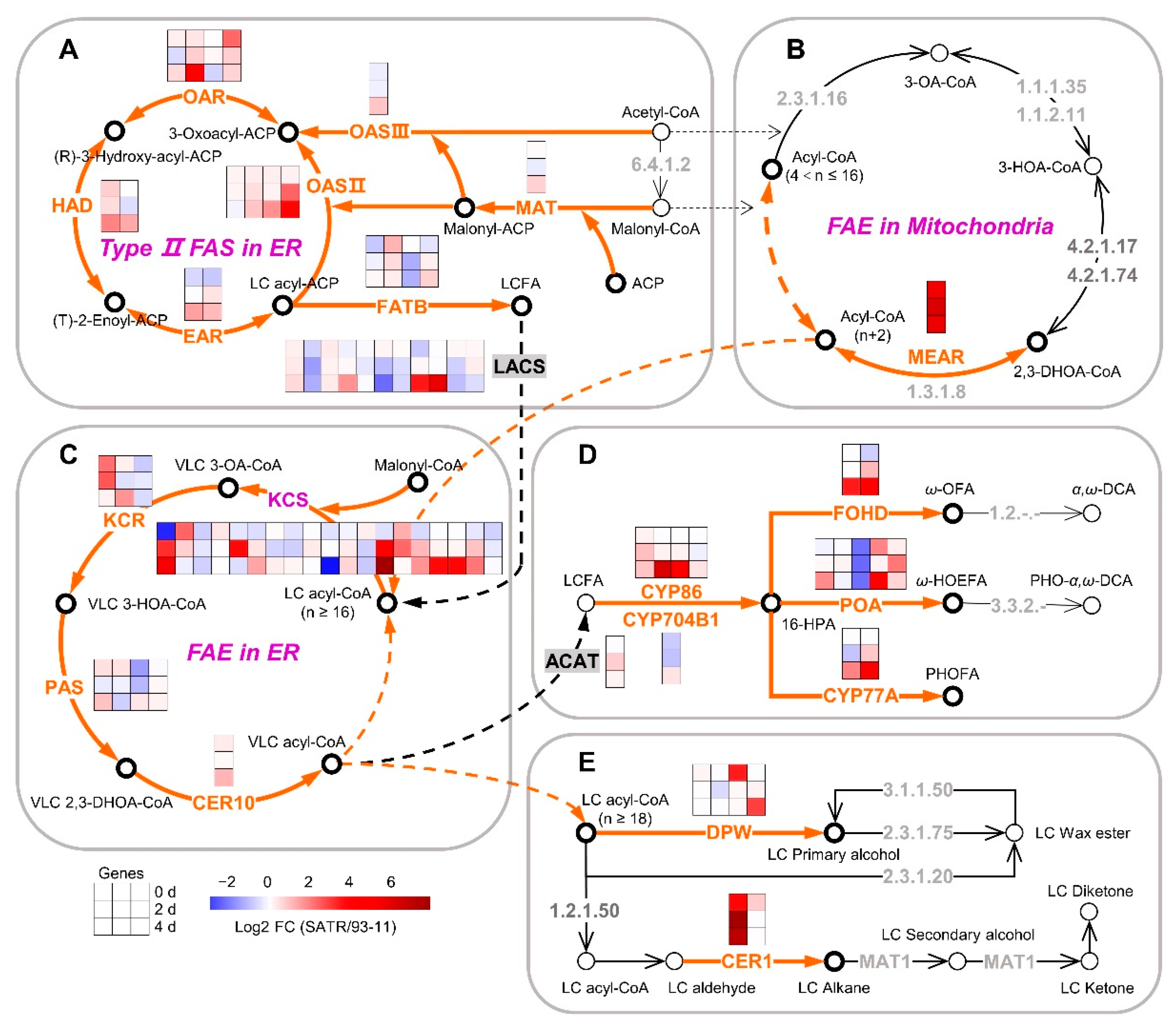
2.4. SATR Had Higher Gene Expressions in Cutin, Suberin and Wax Biosynthesis Pathway under SSAS
2.5. Mapping of the Candidate Genes Controlling Salt–Alkali Tolerance in SATR
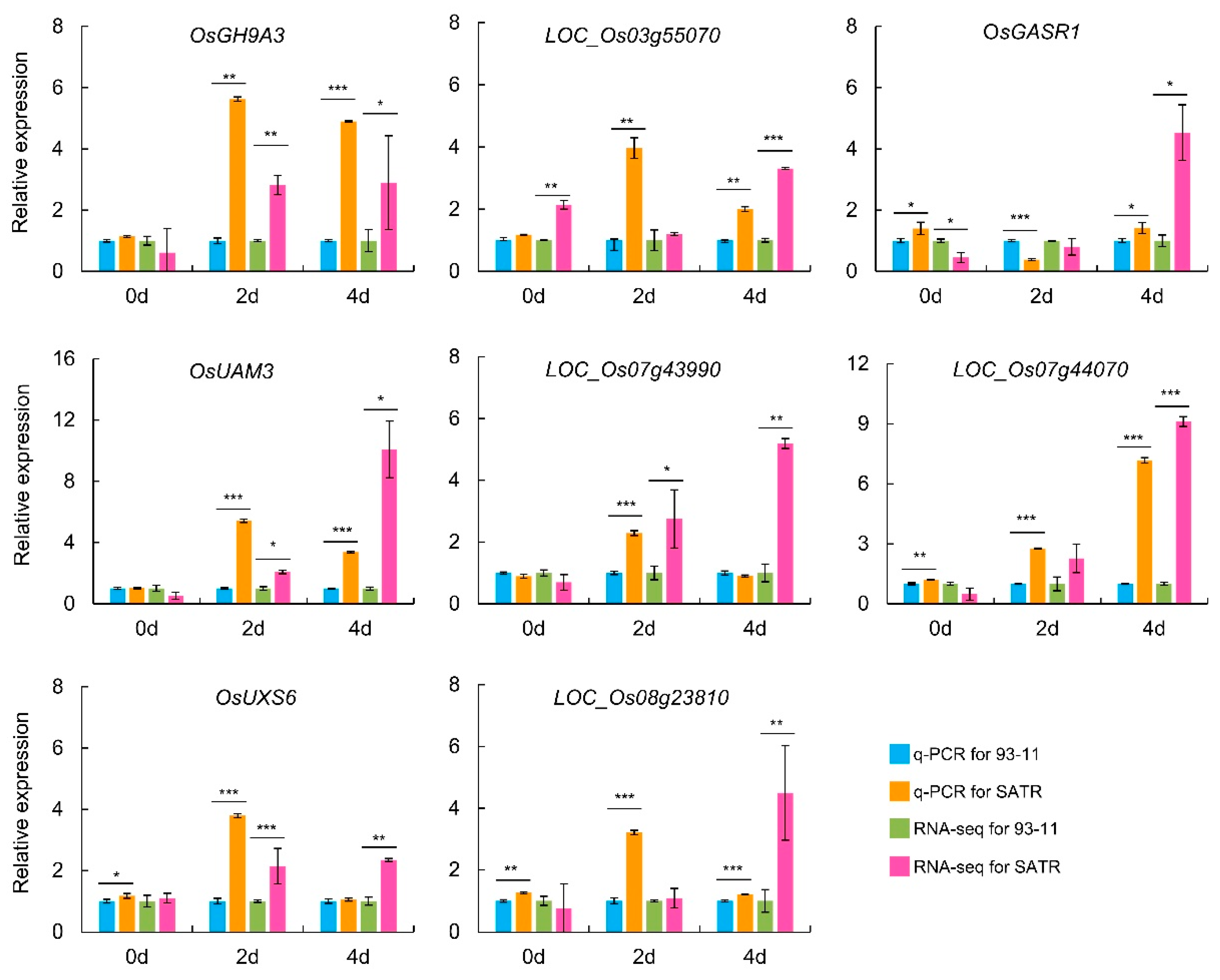
2.6. Validation of RNA-Seq Data through qRT-PCR Analysis
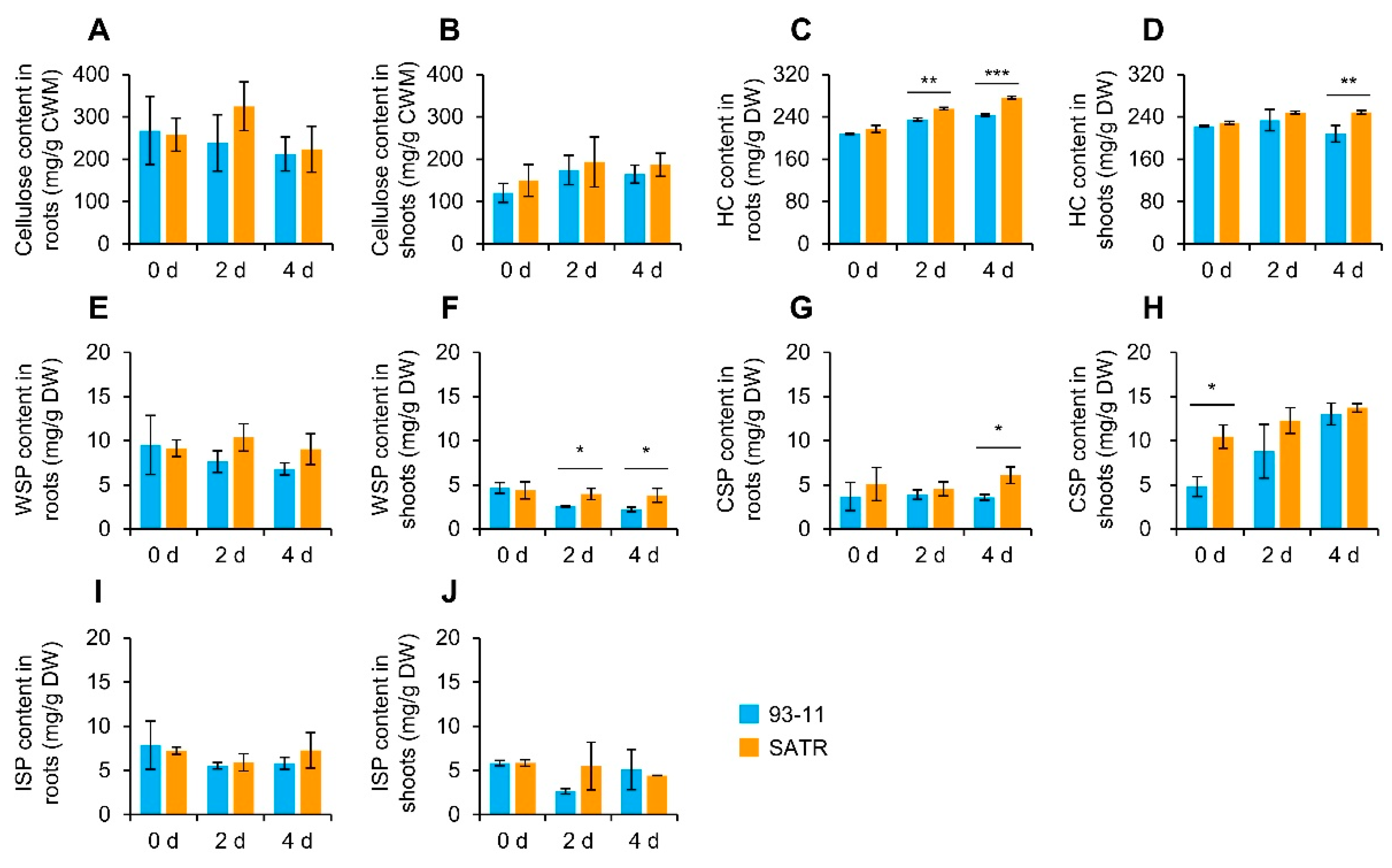
2.7. SATR Seedlings Accumulated Higher Levels of Hemicellulose and Pectin under SSAS
2.8. The Hemicellulose Biosynthesis Gene OsCSLD4 Confers Salt–Alkali Stress Tolerance in Rice
2.9. OsCSLD4 Positively Regulates Grain Yield under Salt–Alkali Stress in Rice
3. Discussion
4. Materials and Methods
4.1. Establishment of a Large-Scale Screening System for Obtaining Salt–Alkali-Tolerant Rice Germplasms
4.2. Plant Materials
4.3. Identification and Evaluation of the SSAS Tolerance between SATR and 93-11
4.4. Identification and Evaluation of the SSAS Tolerance of OsCSLD4 Function-Disrupted Mutant and Overexpressing Lines
4.5. Measurement of Phenotypic Parameters
4.6. Measurement of Physio-Biochemical Parameters
4.6.1. Measurement of Total Chlorophyll Content
4.6.2. Measurement of Malondialdehyde Content
4.6.3. Determination of POD and SOD Activity
4.6.4. Measurement of Proline and Total Soluble Sugar Content
4.7. RNA-Sequencing (RNA-Seq) and qRT-PCR
4.7.1. Analysis of RNA-Seq and Differentially Expressed Genes (DEGs)
4.7.2. RNA Isolation and qRT-PCR Assay
4.8. Measurement of Cellulose, Hemicellulose and Pectin (WSP, CSP, ISP) Contents
4.9. Mapping of the Salt–Alkali Tolerance Genes by GBTS
4.10. Analysis of Major Agronomic Traits
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, G.; Zhang, H.; Araya, K.; Jia, H.; Ohomiya, K.; Matsuda, J. Improvement of salt-affected soils, part 4: Heat transfer coefficient and thermal conductivity of salt-affected soils. Biosyst. Eng. 2007, 96, 593–603. [Google Scholar] [CrossRef]
- Kumar, P.; Sharma, P.K. Soil salinity and food security in India. Front. Sustain. Food Syst. 2020, 4, 533781. [Google Scholar] [CrossRef]
- Mandal, S.; Raju, R.; Kumar, A.; Kumar, P.; Sharma, P. Current status of research, technology response and policy needs of salt-affected soils in India—A review. J. Indian Soc. Coast. Agric. Res. 2018, 36, 40–53. [Google Scholar]
- Wang, H.; Wu, Z.; Chen, Y.; Yang, C.; Shi, D. Effects of salt and alkali stresses on growth and ion balance in rice (Oryza sativa L.). Plant Soil Environ. 2011, 57, 286–294. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Fang, B.; Yang, C.; Shi, D.; Wang, D. Effects of various salt–alkaline mixed stresses on the state of mineral elements in nutrient solutions and the growth of alkali resistant halophyte Chloris virgata. J. Plant Nutr. 2009, 32, 1137–1147. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, L.; Chen, J.; Tao, L.; An, Y.; Cai, H.; Guo, C. Overexpression of the alfalfa WRKY11 gene enhances salt tolerance in soybean. PLoS ONE 2018, 13, e0192382. [Google Scholar] [CrossRef] [Green Version]
- Bu, N.; Li, X.; Li, Y.; Ma, C.; Ma, L.; Zhang, C. Effects of Na2CO3 stress on photosynthesis and antioxidative enzymes in endophyte infected and non-infected rice. Ecotoxicol. Environ. Saf. 2012, 78, 35–40. [Google Scholar] [CrossRef]
- Fang, S.; Hou, X.; Liang, X. Response mechanisms of plants under saline-alkali stress. Front. Plant Sci. 2021, 12, 667458. [Google Scholar] [CrossRef]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.H.; Lee, S.C. Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef] [Green Version]
- Lechner, E.; Leonhardt, N.; Eisler, H.; Parmentier, Y.; Alioua, M.; Jacquet, H.; Leung, J.; Genschik, P. MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev. Cell 2011, 21, 1116–1128. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Sun, X.; Zhao, Y.; Zhao, C.; Duanmu, H.; Yu, Y.; Ji, W.; Zhu, Y. Ectopic expression of GsPPCK3 and SCMRP in Medicago sativa enhances plant alkaline stress tolerance and methionine content. PLoS ONE 2014, 9, e89578. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Yu, Y.; Duan, X.; Sun, X.; Duanmu, H.; Zhu, Y. GsSKP21, a Glycine soja S-phase kinase-associated protein, mediates the regulation of plant alkaline tolerance and ABA sensitivity. Plant Mol. Biol. 2015, 87, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liu, A.; Duan, X.; Wang, S.; Sun, X.; Duanmu, H.; Zhu, D.; Chen, C.; Cao, L.; Xiao, J.; et al. GsERF6, an ethylene-responsive factor from Glycine soja, mediates the regulation of plant bicarbonate tolerance in Arabidopsis. Planta 2016, 244, 681–698. [Google Scholar] [CrossRef]
- Cao, L.; Yu, Y.; Ding, X.; Zhu, D.; Yang, F.; Liu, B.; Sun, X.; Duan, X.; Yin, K.; Zhu, Y. The Glycine soja NAC transcription factor GsNAC019 mediates the regulation of plant alkaline tolerance and ABA sensitivity. Plant Mol. Biol. 2017, 95, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Duan, X.; Ding, X.; Chen, C.; Zhu, D.; Yin, K.; Cao, L.; Song, X.; Zhu, P.; Li, Q.; et al. A novel AP2/ERF family transcription factor from Glycine soja, GsERF71, is a DNA binding protein that positively regulates alkaline stress tolerance in Arabidopsis. Plant Mol. Biol. 2017, 94, 509–530. [Google Scholar] [CrossRef]
- Sun, N.; Song, T.; Ma, Z.; Dong, L.; Zhan, L.; Xing, Y.; Liu, J.; Song, J.; Wang, S.; Cai, H. Overexpression of MsSiR enhances alkali tolerance in alfalfa (Medicago sativa L.) by increasing the glutathione content. Plant Physiol. Biochem. 2020, 154, 538–546. [Google Scholar] [CrossRef]
- Wang, X.; Ajab, Z.; Liu, C.; Hu, S.; Liu, J.; Guan, Q. Overexpression of transcription factor SlWRKY28 improved the tolerance of Populus davidiana × P. bolleana to alkaline salt stress. BMC Genet. 2020, 21, 103. [Google Scholar] [CrossRef]
- Qu, D.; Show, P.L.; Miao, X. Transcription factor ChbZIP1 from alkaliphilic microalgae Chlorella sp. BLD enhancing alkaline tolerance in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 2387. [Google Scholar] [CrossRef]
- Ye, X.; Wang, H.; Cao, X.; Jin, X.; Cui, F.; Bu, Y.; Liu, H.; Wu, W.; Takano, T.; Liu, S. Transcriptome profiling of Puccinellia tenuiflora during seed germination under a long-term saline-alkali stress. BMC Genom. 2019, 20, 589. [Google Scholar] [CrossRef]
- Ma, S.; Lv, L.; Meng, C.; Zhou, C.; Fu, J.; Shen, X.; Zhang, C.; Li, Y. Genome-wide analysis of abscisic acid biosynthesis, catabolism, and signaling in Sorghum bicolor under saline-alkali stress. Biomolecules 2019, 9, 823. [Google Scholar] [CrossRef] [Green Version]
- Guan, Q.; Liao, X.; He, M.; Li, X.; Wang, Z.; Ma, H.; Yu, S.; Liu, S. Tolerance analysis of chloroplast OsCu/Zn-SOD overexpressing rice under NaCl and NaHCO3 stress. PLoS ONE 2017, 12, e0186052. [Google Scholar] [CrossRef] [PubMed]
- Ogden, M.; Hoefgen, R.; Roessner, U.; Persson, S.; Khan, G.A. Feeding the walls: How does nutrient availability regulate cell wall composition? Int. J. Mol. Sci. 2018, 19, 2691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harholt, J.; Suttangkakul, A.; Vibe Scheller, H. Biosynthesis of pectin. Plant Physiol. 2010, 153, 384–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broxterman, S.E.; Schols, H.A. Characterisation of pectin-xylan complexes in tomato primary plant cell walls. Carbohydr. Polym. 2018, 197, 269–276. [Google Scholar] [CrossRef]
- Shao, Y.; An, P.; Feng, X.; Muhammad, I.; Otie, V.; Li, W.; Zheng, Y.; Qiman, Y. Differential responses of roots for varying tolerance to salinity stress in wheat with special reference to elasticity. Plant Growth Regul. 2021, 94, 183–193. [Google Scholar] [CrossRef]
- Xin, A.; Herburger, K. Precursor biosynthesis regulation of lignin, suberin and cutin. Protoplasma 2021, 258, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, R.; Araújo, P.; Llerena, J.P.P.; Mazzafera, P. Suberin and hemicellulose in sugarcane cell wall architecture and crop digestibility: A biotechnological perspective. Food Energy Secur. 2019, 8, e00163. [Google Scholar] [CrossRef] [Green Version]
- Agoda-Tandjawa, G.; Durand, S.; Gaillard, C.; Garnier, C.; Doublier, J.L. Properties of cellulose/pectins composites: Implication for structural and mechanical properties of cell wall. Carbohydr. Polym. 2012, 90, 1081–1091. [Google Scholar] [CrossRef]
- Yang, B.; Hofmann, F.; Usadel, B.; Voiniciuc, C. Seed hemicelluloses tailor mucilage properties and salt tolerance. New Phytol. 2021, 229, 1946–1954. [Google Scholar] [CrossRef]
- Byrt, C.S.; Munns, R.; Burton, R.A.; Gilliham, M.; Wege, S. Root cell wall solutions for crop plants in saline soils. Plant Sci. 2018, 269, 47–55. [Google Scholar] [CrossRef]
- Kunst, L.; Samuels, L. Plant cuticles shine: Advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 2009, 12, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Riederer, M.; Schreiber, L. Protecting against water loss: Analysis of the barrier properties of plant cuticles. J. Exp. Bot. 2001, 52, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, R.; Yang, X.; Ju, Q.; Li, W.; Lu, S.; Tran, L.P.; Xu, J. The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence. Plant Cell Environ. 2020, 43, 1925–1943. [Google Scholar] [CrossRef]
- Li, C.; Lu, H.; Li, W.; Yuan, M.; Fu, Y. A ROP2-RIC1 pathway fine-tunes microtubule reorganization for salt tolerance in Arabidopsis. Plant Cell Environ. 2017, 40, 1127–1142. [Google Scholar] [CrossRef]
- Chun, H.J.; Baek, D.; Jin, B.J.; Cho, H.M.; Park, M.S.; Lee, S.H.; Lim, L.H.; Cha, Y.J.; Bae, D.W.; Kim, S.T.; et al. Microtubule dynamics plays a vital role in plant adaptation and tolerance to salt stress. Int. J. Mol. Sci. 2021, 22, 5957. [Google Scholar] [CrossRef] [PubMed]
- Gandini, A.; Pascoal Neto, C.; Silvestre, A.J.D. Suberin: A promising renewable resource for novel macromolecular materials. Prog. Polym. Sci. 2006, 31, 878–892. [Google Scholar] [CrossRef]
- Hao, Z.; Mohnen, D. A review of xylan and lignin biosynthesis: Foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 212–241. [Google Scholar] [CrossRef]
- Li, M.; Xiong, G.; Li, R.; Cui, J.; Tang, D.; Zhang, B.; Pauly, M.; Cheng, Z.; Zhou, Y. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 2009, 60, 1055–1069. [Google Scholar] [CrossRef]
- Zhao, H.; Li, Z.; Wang, Y.; Wang, J.; Xiao, M.; Liu, H.; Quan, R.; Zhang, H.; Huang, R.; Zhu, L.; et al. Cellulose synthase-like protein OsCSLD4 plays an important role in the response of rice to salt stress by mediating abscisic acid biosynthesis to regulate osmotic stress tolerance. Plant Biotechnol. J. 2021, 20, 468–484. [Google Scholar] [CrossRef]
- Schaffer, W.M.; Bronnikova, T.V. Peroxidase-ROS interactions. Nonlinear Dyn. 2012, 68, 413–430. [Google Scholar] [CrossRef]
- Hamada, T. Microtubule organization and microtubule-associated proteins in plant cells. Int. Rev. Cell Mol. Biol. 2014, 312, 1–52. [Google Scholar] [PubMed]
- Coddard, R.H.; Wick, S.M.; Silflow, C.D.; Snustad, D.P. Microtubule components of the plant cell cytoskeleton. Plant Physiol. 1994, 104, 1–6. [Google Scholar]
- Delgadillo, M.O.; Ruano, G.; Zouhar, J.; Sauer, M.; Shen, J.; Lazarova, A.; Sanmartin, M.; Lai, L.T.F.; Deng, C.; Wang, P.; et al. MTV proteins unveil ER- and microtubule-associated compartments in the plant vacuolar trafficking pathway. Proc. Natl. Acad. Sci. USA 2020, 117, 9884–9895. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Bashline, L.; Lei, L.; Li, S.; Gu, Y. Cell wall, cytoskeleton, and cell expansion in higher plants. Mol. Plant 2014, 7, 586–600. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.; Eberhard, S.; Pattathil, S.; Warder, C.; Glushka, J.; Yuan, C.; Hao, Z.; Zhu, X.; Avci, U.; Miller, J.S.; et al. An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 2013, 25, 270–287. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Zheng, H.; Wei, X.; Song, J.; Wang, B.; Sui, N. Transcriptome analysis of sweet Sorghum inbred lines differing in salt tolerance provides novel insights into salt exclusion by roots. Plant Soil 2018, 430, 423–439. [Google Scholar] [CrossRef]
- Bi, H.; Kovalchuk, N.; Langridge, P.; Tricker, P.J.; Lopato, S.; Borisjuk, N. The impact of drought on wheat leaf cuticle properties. BMC Plant Biol. 2017, 17, 85. [Google Scholar] [CrossRef] [Green Version]
- Joubès, J.; Domergue, F. Biosynthesis of the plant cuticle. In Hydrocarbons, Oils and Lipids: Diversity, Origin, Chemistry and Fate; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 139–157. [Google Scholar]
- Bado, S.; Forster, B.P.; Ghanim, A.M.A.; Jankowicz-Cieslak, J.; Berthold, G.; Luxiang, L. Protocol for screening for salt tolerance in rice. In Protocols for Pre-Field Screening of Mutants for Salt Tolerance in Rice, Wheat and Barley; Springer International Publishing: Cham, Switzerland, 2016; pp. 21–31. [Google Scholar]
- Yoshida, S.; Forno, D.A.; Cock, H.C.J. Laboratory Manual for Physiological Studies of Rice, 3rd. ed.; International Rice Research Institute: Manila, Philippine, 1976; pp. 61–66. [Google Scholar]
- Zhang, G.H.; Li, S.Y.; Wang, L.; Ye, W.J.; Zeng, D.L.; Rao, Y.C.; Peng, Y.L.; Hu, J.; Yang, Y.L.; Xu, J.; et al. LSCHL4 from Japonica Cultivar, which is allelic to NAL1, increases yield of indica super rice 93-11. Mol. Plant 2014, 7, 1350–1364. [Google Scholar] [CrossRef] [Green Version]
- Arnon, D.T. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Yin, W.; Xiao, Y.; Niu, M.; Meng, W.; Li, L.; Zhang, X.; Liu, D.; Zhang, G.; Qian, Y.; Sun, Z.; et al. ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice. Plant Cell 2020, 32, 2292–2306. [Google Scholar] [CrossRef] [PubMed]
- Kochba, J.; Spiegel-Roy, P.L.S. Differences in peroxidase activity and isoenzymes in embryogenic ane non-embryogenic ‘Shamouti’orange ovular callus lines. Plant Cell Physiol. 1977, 18, 463–467. [Google Scholar] [CrossRef]
- Luis Sequeira, L.M. Partial purification and kinetics of indoleacetic acid oxidase from tobacco roots. Plant Physiol. 1966, 41, 1200–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fridovich, C.B.I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, 3053–3065. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550–570. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yang, Q.; Zheng, H.; Xu, Y.; Sang, Z.; Guo, Z.; Peng, H.; Zhang, C.; Lan, H.; Wang, Y.; et al. Genotyping by target sequencing (GBTS) and its applications. Sci. Agric. Sin. 2020, 53, 2983–3004. [Google Scholar]
- Guo, Z.; Wang, H.; Tao, J.; Ren, Y.; Xu, C.; Wu, K.; Zou, C.; Zhang, J.; Xu, Y. Development of multiple SNP marker panels affordable to breeders through genotyping by target sequencing (GBTS) in maize. Mol. Breed. 2019, 39, 37. [Google Scholar] [CrossRef]
- Guo, Z.; Yang, Q.; Huang, F.; Zheng, H.; Sang, Z.; Xu, Y.; Zhang, C.; Wu, K.; Tao, J.; Prasanna, B.M.; et al. Development of high-resolution multiple-SNP arrays for genetic analyses and molecular breeding through genotyping by target sequencing and liquid chip. Plant Commun. 2021, 2, 100230. [Google Scholar] [CrossRef]
- Li, X.; Zheng, H.; Wu, W.; Liu, H.; Wang, J.; Jia, Y.; Li, J.; Yang, L.; Lei, L.; Zou, D.; et al. QTL mapping and candidate gene analysis for alkali tolerance in Japonica rice at the bud stage based on linkage mapping and genome-wide association study. Rice 2020, 13, 48. [Google Scholar] [CrossRef]
- Li, P.; Li, H.; Liu, Z.; Zhuang, Y.; Wei, M.; Gu, Y.; Liu, Y.; Sun, X.; Tang, Y.; Yue, L.; et al. Characterization of the ‘Oat-Like Rice’ caused by a novel allele OsMADS1Olr reveals vital importance of OsMADS1 in regulating grain shape in Oryza sativa L. Rice 2020, 13, 73. [Google Scholar] [CrossRef]
- Xie, G.; Yang, B.; Xu, Z.; Li, F.; Guo, K.; Zhang, M.; Wang, L.; Zou, W.; Wang, Y.; Peng, L. Global identification of multiple OsGH9 family members and their involvement in cellulose crystallinity modification in rice. PLoS ONE 2013, 8, e50171. [Google Scholar] [CrossRef] [Green Version]
- Todaka, D.; Nakashima, K.; Maruyama, K.; Kidokoro, S.; Osakabe, Y.; Ito, Y.; Matsukura, S.; Fujita, Y.; Yoshiwara, K.; Ohme-Takagi, M.; et al. Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc. Natl. Acad. Sci. USA 2012, 109, 15947–15952. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.; Zhang, B.; Dai, Y.; Zhang, L.; Shang-Guan, K.; Peng, Y.; Zhou, Y.; Zhu, Z. Brittle culm15 encodes a membrane-associated chitinase-like protein required for cellulose biosynthesis in rice. Plant Physiol. 2012, 159, 1440–1452. [Google Scholar] [CrossRef] [Green Version]
- Leng, Y.; Yang, Y.; Ren, D.; Huang, L.; Dai, L.; Wang, Y.; Chen, L.; Tu, Z.; Gao, Y.; Li, X.; et al. A rice PECTATE LYASE-LIKE gene is required for plant growth and leaf senescence. Plant Physiol. 2017, 174, 1151–1166. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Qin, Y.; Fang, J.; Yuan, S.; Peng, L.; Zhao, J.; Li, X. A missense mutation in the zinc finger domain of OsCESA7 deleteriously affects cellulose biosynthesis and plant growth in rice. PLoS ONE 2016, 11, e0153993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, Y.; Koshiba, T.; Tobimatsu, Y.; Suzuki, S.; Murakami, S.; Yamamura, M.; Rahman, M.M.; Takano, T.; Hattori, T.; Sakamoto, M.; et al. Regulation of CONIFERALDEHYDE 5-HYDROXYLASE expression to modulate cell wall lignin structure in rice. Planta 2017, 246, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Wenwen, Y.; Zang, G.; Kang, Z.; Zhang, Z.; Huang, J.; Wang, G. OsEXPB2, a β-expansin gene, is involved in rice root system architecture. Mol. Breed. 2015, 35, 41. [Google Scholar] [CrossRef]
- Lee, S.-C.; Han, S.-K.; Kim, S.-R. Salt- and ABA-inducible OsGASR1 is involved in salt tolerance. J. Plant Biol. 2015, 58, 96–101. [Google Scholar] [CrossRef]
- Fang, L.; Zhao, F.; Cong, Y.; Sang, X.; Du, Q.; Wang, D.; Li, Y.; Ling, Y.; Yang, Z.; He, G. Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol. J. 2012, 10, 524–532. [Google Scholar] [CrossRef]
- Kim, C.M.; Park, S.H.; Je, B.I.; Park, S.H.; Park, S.J.; Piao, H.L.; Eun, M.Y.; Dolan, L.; Han, C.D. OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiol. 2007, 143, 1220–1230. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.J.; Zhu, C. Heterologous expression of a rice syntaxin-related protein KNOLLE gene (OsKNOLLE) in yeast and its functional analysis in the role of abiotic stress. Hereditas 2011, 33, 1251–1257. [Google Scholar] [CrossRef]
- Singh, K.; Pareek, S.; Singla, P. Overexpression of rice CBS domain containing protein improves salinity, oxidative, and heavy metal tolerance in transgenic tobacco. Mol. Biotechnol. 2012, 52, 205–216. [Google Scholar] [CrossRef]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Tuteja, N.; Sahoo, R.K.; Garg, B.; Tuteja, R. OsSUV3 dual helicase functions in salinity stress tolerance by maintaining photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. IR64). Plant J. 2013, 76, 115–127. [Google Scholar] [CrossRef]
- Zhong, R.; Wang, Y.; Gai, R.; Xi, D.; Mao, C.; Ming, F. Rice SnRK protein kinase OsSAPK8 acts as a positive regulator in abiotic stress responses. Plant Sci. 2020, 292, 110373. [Google Scholar] [CrossRef] [PubMed]
- Chapagain, S.; Park, Y.C.; Kim, J.H.; Jang, C.S. Oryza sativa salt-induced RING E3 ligase 2 (OsSIRP2) acts as a positive regulator of transketolase in plant response to salinity and osmotic stress. Planta 2018, 247, 925–939. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, M.; Gu, D.; Liu, X.; Zhang, J.; Wu, K.; Zhang, X.; Teixeira da Silva, J.A.; Duan, J. Involvement of rice histone deacetylase HDA705 in seed germination and in response to ABA and abiotic stresses. Biochem. Biophys. Res. Commun. 2016, 470, 439–444. [Google Scholar] [CrossRef]
- Giri, J.; Vij, S.; Dansana, P.K.; Tyagi, A.K. Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 2011, 191, 721–732. [Google Scholar] [CrossRef]
- Wu, T.M.; Lin, W.R.; Kao, C.H.; Hong, C.Y. Gene knockout of glutathione reductase 3 results in increased sensitivity to salt stress in rice. Plant Mol. Biol. 2015, 87, 555–564. [Google Scholar] [CrossRef]
- Alfatih, A.; Wu, J.; Jan, S.U.; Zhang, Z.S.; Xia, J.Q.; Xiang, C.B. Loss of rice PARAQUAT TOLERANCE 3 confers enhanced resistance to abiotic stresses and increases grain yield in field. Plant Cell Environ. 2020, 43, 2743–2754. [Google Scholar] [CrossRef]
- Li, L.H.; Lv, M.M.; Li, X.; Ye, T.Z.; He, X.; Rong, S.H.; Dong, Y.L.; Guan, Y.; Gao, X.L.; Zhu, J.Q.; et al. The rice OsDUF810 family: OsDUF810.7 may be Involved in the tolerance to salt and drought. Mol. Biol. 2018, 52, 489–496. [Google Scholar] [CrossRef]
- Islam, M.O.; Kato, H.; Shima, S.; Tezuka, D.; Matsui, H.; Imai, R. Functional identification of a rice trehalase gene involved in salt stress tolerance. Gene 2019, 685, 42–49. [Google Scholar] [CrossRef]
- Sharma, R.; Sahoo, A.; Devendran, R.; Jain, M. Over-expression of a rice tau class glutathione s-transferase gene improves tolerance to salinity and oxidative stresses in Arabidopsis. PLoS ONE 2014, 9, e92900. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Hu, Y.; Du, A.; Yu, L.; Fu, X.; Wu, C.; Lu, L.; Liu, Y.; Wang, S.; Huang, W.; et al. Cell Wall Matrix Polysaccharides Contribute to Salt–Alkali Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 15019. https://doi.org/10.3390/ijms232315019
Liu Z, Hu Y, Du A, Yu L, Fu X, Wu C, Lu L, Liu Y, Wang S, Huang W, et al. Cell Wall Matrix Polysaccharides Contribute to Salt–Alkali Tolerance in Rice. International Journal of Molecular Sciences. 2022; 23(23):15019. https://doi.org/10.3390/ijms232315019
Chicago/Turabian StyleLiu, Zhijian, Yongzhi Hu, Anping Du, Lan Yu, Xingyue Fu, Cuili Wu, Longxiang Lu, Yangxuan Liu, Songhu Wang, Weizao Huang, and et al. 2022. "Cell Wall Matrix Polysaccharides Contribute to Salt–Alkali Tolerance in Rice" International Journal of Molecular Sciences 23, no. 23: 15019. https://doi.org/10.3390/ijms232315019
APA StyleLiu, Z., Hu, Y., Du, A., Yu, L., Fu, X., Wu, C., Lu, L., Liu, Y., Wang, S., Huang, W., Tu, S., Ma, X., & Li, H. (2022). Cell Wall Matrix Polysaccharides Contribute to Salt–Alkali Tolerance in Rice. International Journal of Molecular Sciences, 23(23), 15019. https://doi.org/10.3390/ijms232315019





