Xylanase Inhibitors: Defense Players in Plant Immunity with Implications in Agro-Industrial Processing
Abstract
:1. Introduction
2. XIs Genomic Organization
3. Inhibition Activity of XIs
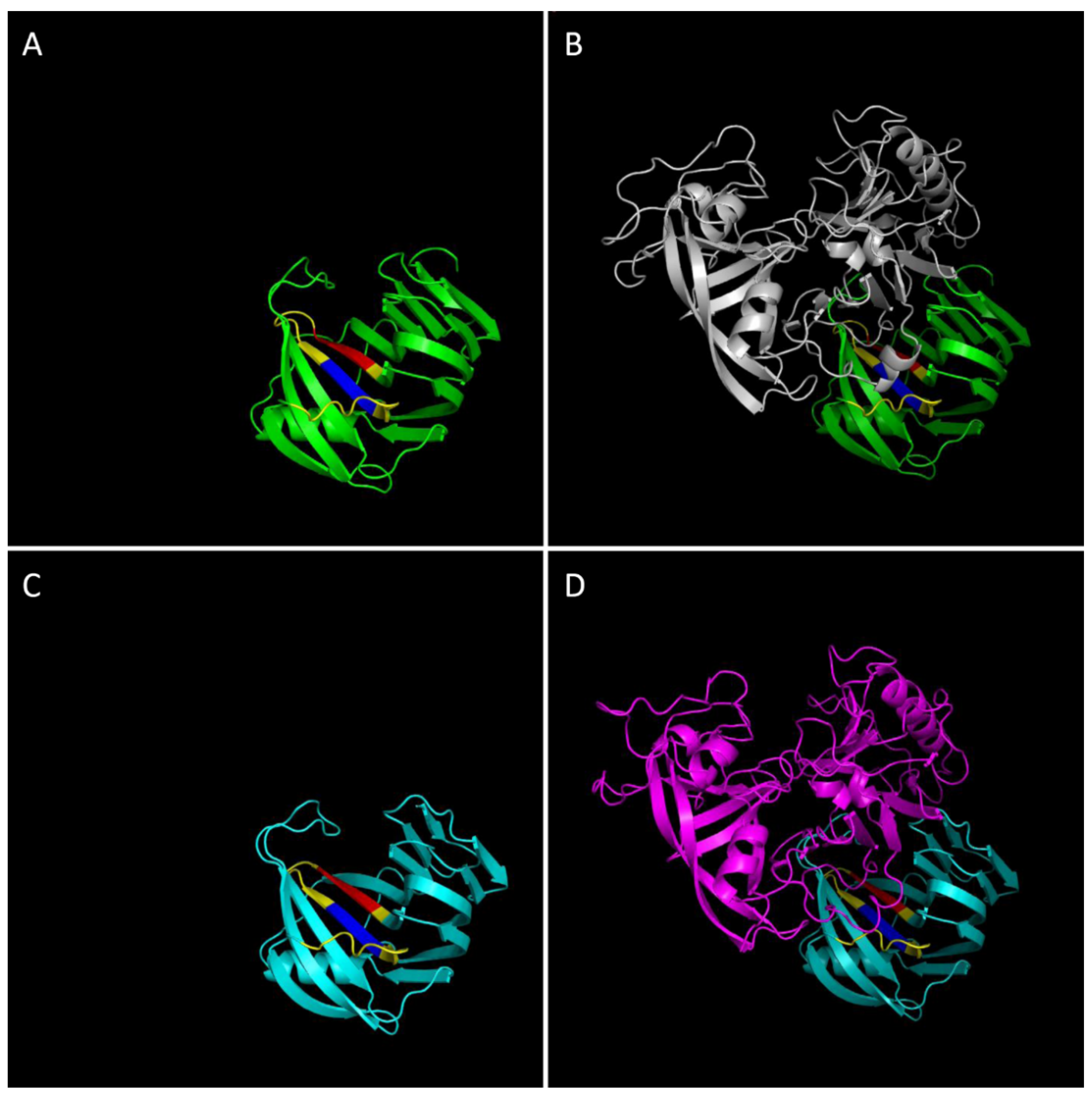
4. Structural Properties of Xylanase-XI Interaction
5. XI Regulation and Localization
6. Role of XIs in Plant Defense and Crop Engineering for Plant Protection
7. Strategies to Reduce the Effect of XIs in Food Processing and Quality
| Strategy | References |
|---|---|
| Heat treatment of matrices containing XIs | [105] |
| Selection of plant row materials with low xylanase inhibitory activity | [106] |
| Application of xylanases that escape the inhibition of xylanase inhibitors | [64] |
| Engineering xylanases that escape the inhibition of xylanase inhibitors and/or with improved catalytic activity and thermostability | [60,92,107,112,113,114] |
| Adjustment of xylanase dosage | [115,116] |
| Targeted disruption of specific XIs in plant species used in agro-industrial processes | [42] |
8. Involvement of XIs in Food Allergy
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferrari, S.; Savatin, D.V.; Sicilia, F.; Gramegna, G.; Cervone, F.; de Lorenzo, G. Oligogalacturonides: Plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 2013, 4, 49. [Google Scholar] [CrossRef] [Green Version]
- Houston, K.; Tucker, M.R.; Chowdhury, J.; Shirley, N.; Little, A. The plant cell wall: A complex and dynamic structure as revealed by the responses of genes under stress conditions. Front. Plant Sci. 2016, 7, 984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malinovsky, F.G.; Fangel, J.U.; Willats, W.G.T. The role of the cell wall in plant immunity. Front. Plant Sci. 2014, 5, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giovannoni, M.; Gramegna, G.; Benedetti, M.; Mattei, B. Industrial use of cell wall degrading enzymes: The fine line between production strategy and economic feasibility. Front. Bioeng. Biotechnol. 2020, 8, 356. [Google Scholar] [CrossRef]
- Choquer, M.; Fournier, E.; Kunz, C.; Levis, C.; Pradier, J.M.; Simon, A.; Viaud, M. Botrytis cinerea virulence factors: New insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 2007, 277, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant cell wall–degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef]
- Chang, H.X.; Yendrek, C.R.; Caetano-Anolles, G.; Hartman, G.L. Genomic characterization of plant cell wall degrading enzymes and in silico analysis of xylanses and polygalacturonases of Fusarium virguliforme. BMC Microbiol. 2016, 16, 147. [Google Scholar] [CrossRef] [Green Version]
- Rai, K.M.; Balasubramanian, V.K.; Welker, C.M.; Pang, M.; Hii, M.M.; Mendu, V. Genome wide comprehensive analysis and web resource development on cell wall degrading enzymes from phyto-parasitic nematodes. BMC Plant Biol. 2015, 15, 187. [Google Scholar] [CrossRef] [Green Version]
- Blackman, L.M.; Cullerne, D.P.; Hardham, A.R. Bioinformatic characterisation of genes encoding cell wall degrading enzymes in the Phytophthora parasitica genome. BMC Genom. 2014, 15, 785. [Google Scholar] [CrossRef]
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005, 29, 3–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gírio, F.M.; Fonseca, C.; Carvalheiro, F.; Duarte, L.C.; Marques, S.; Bogel-Łukasik, R. Hemicelluloses for fuel ethanol: A review. Bioresour. Technol. 2010, 101, 4775–4800. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J. Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 2008, 11, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Perrot, T.; Pauly, M.; Ramírez, V. Emerging roles of beta-glucanases in plant development and adaptative responses. Plants 2022, 11, 1119. [Google Scholar] [CrossRef]
- Kabel, M.A.; Frommhagen, M.; Sun, P.; Schols, H.A. Modification of plant carbohydrates using fungal enzymes. In Encyclopedia of Mycology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 370–384. [Google Scholar] [CrossRef]
- Padayachee, A.; Day, L.; Howell, K.; Gidley, M.J. Complexity and health functionality of plant cell wall fibers from fruits and vegetables. Crit. Rev. Food Sci. Nutr. 2017, 57, 59–81. [Google Scholar] [CrossRef]
- Kumar, A.; Naraian, R. Differential expression of the microbial β-1,4-xylanase, and β-1,4-endoglucanase genes. In New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Genes Biochemistry and Applications; Singh, H.B., Gupta, V.K., Jogaiah, S., Eds.; Elsevier: Chennai, India, 2019; pp. 95–111. ISBN 9780444635037. [Google Scholar]
- Noda, J.; Brito, N.; González, C. The Botrytis cinerea xylanase Xyn11A contributes to virulence with its necrotizing activity, not with its catalytic activity. BMC Plant Biol. 2010, 10, 38. [Google Scholar] [CrossRef] [Green Version]
- Rotblat, B.; Enshell-Seijffers, D.; Gershoni, J.M.; Schuster, S.; Avni, A. Identification of an essential component of the elicitation active site of the EIX protein elicitor. Plant J. 2002, 32, 1049–1055. [Google Scholar] [CrossRef] [Green Version]
- Xin, Z.; Wang, Q.; Yu, Z.; Hu, L.; Li, J.; Xiang, C.; Wang, B.; Lou, Y. Overexpression of a xylanase inhibitor gene, OSHI-XIP, enhances resistance in rice to herbivores. Plant Mol. Biol. Rep. 2014, 32, 465–475. [Google Scholar] [CrossRef]
- Moscetti, I.; Tundo, S.; Janni, M.; Sella, L.; Gazzetti, K.; Tauzin, A.; Giardina, T.; Masci, S.; Favaron, F.; D’Ovidio, R. Constitutive expression of the xylanase inhibitor TAXI-III delays Fusarium head blight symptoms in durum wheat transgenic plants. Mol. Plant Microbe Interact. 2013, 26, 1464–1472. [Google Scholar] [CrossRef]
- Hou, C.; Lv, T.; Zhan, Y.; Peng, Y.; Huang, Y.; Jiang, D.; Weng, X. Overexpression of the RIXI xylanase inhibitor improves disease resistance to the fungal pathogen, Magnaporthe oryzae, in rice. Plant Cell Tissue Organ. Cult. 2015, 120, 167–177. [Google Scholar] [CrossRef]
- Tundo, S.; Paccanaro, M.C.; Elmaghraby, I.; Moscetti, I.; D’ovidio, R.; Favaron, F.; Sella, L. The xylanase inhibitor TAXI-I increases plant resistance to Botrytis cinerea by inhibiting the BcXyn11a xylanase necrotizing activity. Plants 2020, 9, 601. [Google Scholar] [CrossRef] [PubMed]
- Tundo, S.; Kalunke, R.; Janni, M.; Volpi, C.; Lionetti, V.; Bellincampi, D.; Favaron, F.; D’Ovidio, R. Pyramiding PvPGIP2 and TAXI-III But Not PvPGIP2 and PMEI enhances resistance against Fusarium graminearum. Mol. Plant Microbe Interact. 2016, 29, 629–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tundo, S.; Janni, M.; Moscetti, I.; Mandalá, G.; Savatin, D.; Blechl, A.; Favaron, F.; D’Ovidio, R. PvPGIP2 accumulation in specific floral tissues but not in the endosperm limits Fusarium graminearum infection in wheat. Mol. Plant Microbe Interact. 2016, 29, 815–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volpi, C.; Janni, M.; Lionetti, V.; Bellincampi, D.; Favaron, F.; D’Ovidio, R. The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol. Plant Microbe Interact. 2011, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandalà, G.; Ceoloni, C.; Busato, I.; Favaron, F.; Tundo, S. Transgene pyramiding in wheat: Combination of deoxynivalenol detoxification with inhibition of cell wall degrading enzymes to contrast Fusarium Head Blight and Crown Rot. Plant Sci. 2021, 313, 111059. [Google Scholar] [CrossRef] [PubMed]
- Debyser, W.; Derdelinckx, G.; Delcour, J.A. Arabinoxylan solubilization and inhibition of the barley malt xylanolytic system by wheat during mashing with wheat wholemeal adjunct: Evidence for a new class of enzyme inhibitors in wheat. J. Am. Soc. Brew. Chem. 1997, 55, 153–156. [Google Scholar] [CrossRef]
- Debyser, W.; Peumanst, W.J.; van Damme, E.J.M.; Delcour, J.A. Triticum aestivum Xylanase Inhibitor (TAXI), a new class of enzyme inhibitor affecting breadmaking performance. J. Cereal Sci. 1999, 30, 39–43. [Google Scholar] [CrossRef]
- McLauchlan, W.R.; Garcia-Conesa, M.T.; Williamson, G.; Roza, M.; Ravestein, P.; Maat, J. A novel class of protein from wheat which inhibits xylanases. Biochem. J. 1999, 338, 441. [Google Scholar] [CrossRef]
- Fierens, E.; Rombouts, S.; Gebruers, K.; Goesaert, H.; Brijs, K.; Beaugrand, J.; Volckaert, G.; van Campenhout, S.; Proost, P.; Courtin, C.M.; et al. TLXI, a novel type of xylanase inhibitor from wheat (Triticum aestivum) belonging to the thaumatin family. Biochem. J. 2007, 403, 583–591. [Google Scholar] [CrossRef]
- Sun, W.; Zhou, Y.; Movahedi, A.; Wei, H.; Zhuge, Q. Thaumatin-like protein (Pe-TLP) acts as a positive factor in transgenic poplars enhanced resistance to spots disease. Physiol. Mol. Plant Pathol. 2020, 112, 101512. [Google Scholar] [CrossRef]
- Dornez, E.; Croes, E.; Gebruers, K.; de Coninck, B.; Cammue, B.P.A.; Delcour, J.A.; Courtin, C.M. Accumulated evidence substantiates a role for three classes of wheat Xylanase Inhibitors in plant defense. Crit. Rev. Plant Sci. 2010, 29, 244–264. [Google Scholar] [CrossRef]
- Ong, Q.; Nguyen, P.; Phuong Thao, N.; Le, L. Bioinformatics approach in plant genomic research. Curr. Genom. 2016, 17, 368–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalunke, R.M.; Cenci, A.; Volpi, C.; O’Sullivan, D.M.; Sella, L.; Favaron, F.; Cervone, F.; de Lorenzo, G.; D’Ovidio, R. The pgip family in soybean and three other legume species: Evidence for a birth-and-death model of evolution. BMC Plant Biol. 2014, 14, 189. [Google Scholar] [CrossRef] [Green Version]
- Juge, N.; Svensson, B. Proteinaceous inhibitors of carbohydrate-active enzymes in cereals: Implication in agriculture, cereal processing and nutrition. J. Sci. Food Agric. 2006, 86, 1573–1586. [Google Scholar] [CrossRef]
- York, W.S.; Qin, Q.; Rose, J.K.C. Proteinaceous inhibitors of endo-β-glucanases. Biochim. Biophys. Acta Proteins Proteom. 2004, 1696, 223–233. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Han, N.; Wang, S.; Chen, C.; Lu, J.; Riaz, M.W.; Si, H.; Sun, G.; Ma, C. Genome-wide identification of Triticum aestivum Xylanase Inhibitor gene family and inhibitory effects of XI-2 subfamily proteins on Fusarium graminearum GH11 Xylanase. Front. Plant Sci. 2021, 12, 1508. [Google Scholar] [CrossRef]
- Wolfe, D.; Dudek, S.; Ritchie, M.D.; Pendergrass, S.A. Visualizing genomic information across chromosomes with PhenoGram. BioData Min. 2013, 6, 18. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhang, Y.; Wei, Z.; Chen, H.; Jia, X. Molecular cloning and characterizations of Xylanase Inhibitor Protein from wheat (Triticum aestivum). J. Food Sci. 2017, 82, 1582–1587. [Google Scholar] [CrossRef]
- Sun, Z.; Zhang, M.; An, Y.; Han, X.; Guo, B.; Lv, G.; Zhao, Y.; Guo, Y.; Li, S. CRISPR/Cas9-mediated disruption of Xylanase inhibitor protein (XIP) gene improved the dough quality of common wheat. Front. Plant Sci. 2022, 13, 783. [Google Scholar] [CrossRef]
- Sonah, H.; Chavan, S.; Katara, J.; Chaudhary, J.; Kadam, S.; Patil, G.; Deshmukh, R. Genome-wide identification and characterization of Xylanase Inhibitor Protein (XIP) genes in cereals. Indian J. Genet. Plant Breed. 2016, 76, 159–166. [Google Scholar] [CrossRef] [Green Version]
- Flatman, R.; McLauchlan, W.R.; Juge, N.; Furniss, C.; Berrin, J.G.; Hughes, R.K.; Manzanares, P.; Ladbury, J.E.; O’Brien, R.; Williamson, G. Interactions defining the specificity between fungal xylanases and the xylanase-inhibiting protein XIP-I from wheat. Biochem. J. 2002, 365, 773–781. [Google Scholar] [CrossRef] [Green Version]
- Channamallikarjuna, V.; Sonah, H.; Prasad, M.; Rao, G.J.N.; Chand, S.; Upreti, H.C.; Singh, N.K.; Sharma, T.R. Identification of major quantitative trait loci qSBR11-1 for sheath blight resistance in rice. Mol. Breed. 2010, 25, 155–166. [Google Scholar] [CrossRef]
- Kalunke, R.M.; Tundo, S.; Benedetti, M.; Cervone, F.; de Lorenzo, G.; D’Ovidio, R. An update on polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein that protects crop plants against pathogens. Front. Plant Sci. 2015, 6, 146. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, L.K.; Mylroie, J.E.; Oliveira, D.A.; Smith, J.S.; Ozkan, S.; Windham, G.L.; Williams, W.P.; Warburton, M.L. Characterization of the maize chitinase genes and their effect on Aspergillus flavus and aflatoxin accumulation resistance. PLoS ONE 2015, 10, e0126185. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Lv, Y.; Hou, Z.; Li, X.; Ding, L. Expansion and evolution of thaumatin-like protein (TLP) gene family in six plants. Plant Growth Regul. 2015, 79, 299–307. [Google Scholar] [CrossRef]
- Gebruers, K.; Brijs, K.; Courtin, C.M.; Fierens, K.; Goesaert, H.; Rabijns, A.; Raedschelders, G.; Robben, J.; Sansen, S.; Sørensen, J.F.; et al. Properties of TAXI-type endoxylanase inhibitors. Biochim. Biophys. Acta Proteins Proteom. 2004, 1696, 213–221. [Google Scholar] [CrossRef]
- Gebruers, K.; Dornez, E.; Bedõ, Z.; Rakszegi, M.; Courtin, C.M.; Delcour, J.A. Variability in xylanase and xylanase inhibition activities in different cereals in the HEALTHGRAIN diversity screen and contribution of environment and genotype to this variability in common wheat. J. Agric. Food Chem. 2010, 58, 9362–9371. [Google Scholar] [CrossRef]
- Elliott, G.O.; McLauchlan, W.R.; Williamson, G.; Kroon, P.A. A Wheat Xylanase Inhibitor Protein (XIP-I) Accumulates in the Grain and has Homologues in Other Cereals. J Cereal Sci 2003, 37, 187–194. [Google Scholar] [CrossRef]
- Mendis, M.; Ohm, J.B.; Delcour, J.A.; Gebruers, K.; Meinhardt, S.; Simsek, S. Variability in Arabinoxylan, Xylanase Activity, and Xylanase Inhibitor Levels in Hard Spring Wheat. Cereal Chem. 2013, 90, 240–248. [Google Scholar] [CrossRef]
- Goesaert, H.; Elliott, G.; Kroon, P.A.; Gebruers, K.; Courtin, C.M.; Robben, J.; Delcour, J.A.; Juge, N. Occurrence of proteinaceous endoxylanase inhibitors in cereals. Biochim. Biophys. Acta Proteins Proteom. 2004, 1696, 193–202. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, M.; Li, J.; Wei, H.; Zhang, K. Experimental and in silico studies of competitive inhibition of family GH10 Aspergillus fumigatus xylanase A by Oryza sativa xylanase inhibitor protein. Int. J. Biol. Macromol. 2021, 193, 1391–1399. [Google Scholar] [CrossRef]
- Liu, M.; Wu, X.; Huo, W.; Li, J.; Weng, X.; Liu, J.; Fang, Z. Differential inhibition of GH family 11 endo-xylanase by rice xylanase inhibitor and verification by a modified yeast two-hybrid system. Int. J. Biol. Macromol. 2019, 132, 514–523. [Google Scholar] [CrossRef]
- Fierens, K.; Gils, A.; Sansen, S.; Brijs, K.; Courtin, C.M.; Declerck, P.J.; de Ranter, C.J.; Gebruers, K.; Rabijns, A.; Robben, J.; et al. His374 of wheat endoxylanase inhibitor TAXI-I stabilizes complex formation with glycoside hydrolase family 11 endoxylanases. FEBS J. 2005, 272, 5872–5882. [Google Scholar] [CrossRef]
- Dang, Y.; Liu, M.; Wu, X. Recombinant rice xylanase-inhibiting protein inhibits GH11 endo-xylanases through competitive inhibition. Protein Expr. Purif. 2019, 156, 17–24. [Google Scholar] [CrossRef]
- Zhan, Y.; Sun, R.; Sun, X.; Xu, Y.; Hou, C.; Huang, Y.; Jiang, D.; Weng, X. Expression regulation of a xylanase inhibitor gene riceXIP in rice (Oryza sativa L.). Braz. J. Bot. 2017, 40, 983–991. [Google Scholar] [CrossRef]
- Hou, C.X.; Zhan, Y.H.; Jiang, D.A.; Weng, X.Y. Functional characterization of a new pathogen induced xylanase inhibitor (RIXI) from rice. Eur. J. Plant Pathol. 2014, 138, 405–414. [Google Scholar] [CrossRef]
- Huo, W.K.; Liu, M.Q.; Weng, X.Y.; Qi, Y.P. Recombinant rice xylanase inhibitor (RIXI) expressed in Escherichia coli and its inhibitory activity on family GH11endo-xylanases. Int. J. Biol. Macromol. 2018, 117, 1343–1351. [Google Scholar] [CrossRef]
- Raedschelders, G.; Fierens, K.; Sansen, S.; Rombouts, S.; Gebruers, K.; Robben, J.; Rabijns, A.; Courtin, C.M.; Delcour, J.A.; van Campenhout, S.; et al. Molecular identification of wheat endoxylanase inhibitor TAXI-II and the determinants of its inhibition specificity. Biochem. Biophys. Res. Commun. 2005, 335, 512–522. [Google Scholar] [CrossRef]
- Liu, M.; Li, J.; Rehman, A.U.; Luo, S.; Wang, Y.; Wei, H.; Zhang, K. Sensitivity of family GH11 bacillus amyloliquefaciens xylanase A (BaxA) and the T33I mutant to Oryza sativa xylanase inhibitor protein (OsXIP): An experimental and computational study. Enzym. Microb. Technol. 2022, 156, 109998. [Google Scholar] [CrossRef]
- Brutus, A.; Reca, I.B.; Herga, S.; Mattei, B.; Puigserver, A.; Chaix, J.C.; Juge, N.; Bellincampi, D.; Giardina, T. A family 11 xylanase from the pathogen Botrytis cinerea is inhibited by plant endoxylanase inhibitors XIP-I and TAXI-I. Biochem. Biophys. Res. Commun. 2005, 337, 160–166. [Google Scholar] [CrossRef]
- Gusakov, A.V.; Ustinov, B.B. Assaying sensitivity of fungal xylanases to proteinaceous inhibitors from a rye extract: Two GH10 family xylanases resistant to XIP-like inhibitors. Ind. Biotechnol. 2009, 5, 104–109. [Google Scholar] [CrossRef]
- Dornez, E.; Verjans, P.; Arnaut, F.; Delcour, J.A.; Courtin, C.M. Use of psychrophilic xylanases provides insight into the xylanase functionality in bread making. J. Agric. Food Chem. 2011, 59, 9553–9562. [Google Scholar] [CrossRef]
- Pollet, A.; Beliën, T.; Fierens, K.; Delcour, J.A.; Courtin, C.M. Fusarium graminearum xylanases show different functional stabilities, substrate specificities and inhibition sensitivities. Enzym. Microb. Technol. 2009, 44, 189–195. [Google Scholar] [CrossRef]
- Tundo, S.; Moscetti, I.; Faoro, F.; Lafond, M.; Giardina, T.; Favaron, F.; Sella, L.; D’Ovidio, R. fusarium graminearum produces different xylanases causing host cell death that is prevented by the xylanase inhibitors XIP-I and TAXI-III in wheat. Plant Sci. 2015, 240, 161–169. [Google Scholar] [CrossRef]
- Beliën, T.; van Campenhout, S.; van Acker, M.; Volckaert, G. Cloning and characterization of two endoxylanases from the cereal phytopathogen Fusarium graminearum and their inhibition profile against endoxylanase inhibitors from wheat. Biochem. Biophys. Res. Commun. 2005, 327, 407–414. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, M.; Li, J.; Wang, Q.; Ouyang, X.; Wei, H.; Zhang, K. Exploring competitive inhibition of a family 10 xylanase derived from Hu sheep rumen microbiota by Oryza sativa xylanase inhibitor protein: In vitro and in silico perspectives. Enzym. Microb. Technol. 2022, 160, 110082. [Google Scholar] [CrossRef]
- Padilla-Hurtado, B.; Flárez-Ramos, C.; Aguilera-Gálvez, C.; Medina-Olaya, J.; Ramírez-Sanjuan, A.; Rubio-Gámez, J.; Acũa-Zornosa, R. Cloning and expression of an endo-1,4–Xylanase from the coffee berry borer, Hypothenemus hampei. BMC Res. Notes 2012, 5, 23. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Liu, X.; Xie, X.; Yang, S.; Lin, H.; Chen, H. Characteristics of a XIP-resistant xylanase from Neocallimastix sp. GMLF1 and its advantage in barley malt saccharification. Int. J. Food Sci. Technol. 2020, 55, 2152–2160. [Google Scholar] [CrossRef]
- Payan, F.; Leone, P.; Porciero, S.; Furniss, C.; Tahir, T.; Williamson, G.; Durand, A.; Manzanares, P.; Gilbert, H.J.; Juge, N.; et al. The Dual Nature of the Wheat Xylanase Protein Inhibitor XIP-I: Structural basis for the inhibition of family 10 and family 11 xylanases. J. Biol. Chem. 2004, 279, 36029–36037. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Singh, N.; Mishra, B.; Dube, D.; Sinha, M.; Singh, S.B.; Dey, S.; Kaur, P.; Sharma, S.; Singh, T.P. Modulation of inhibitory activity of xylanase–α-amylase inhibitor protein (XAIP): Binding studies and crystal structure determination of XAIP- II from Scadoxus multiflorus at 1.2 Å resolution. BMC Struct. Biol. 2010, 10, 41. [Google Scholar] [CrossRef] [Green Version]
- Furniss, C.S.M.; Williamson, G.; Kroon, P.A. The substrate specificity and susceptibility to wheat inhibitor proteins of Penicillium funiculosum xylanases from a commercial enzyme preparation. J. Sci. Food Agric. 2005, 85, 574–582. [Google Scholar] [CrossRef]
- Pollet, A.; Sansen, S.; Raedschelders, G.; Gebruers, K.; Rabijns, A.; Delcour, J.A.; Courtin, C.M. Identification of structural determinants for inhibition strength and specificity of wheat xylanase inhibitors TAXI-IA and TAXI-IIA. FEBS J. 2009, 276, 3916–3927. [Google Scholar] [CrossRef]
- Brutus, A.; Villard, C.; Durand, A.; Tahir, T.; Furniss, C.; Puigserver, A.; Juge, N.; Giardina, T. The inhibition specificity of recombinant Penicillium funiculosum xylanase B towards wheat proteinaceous inhibitors. Biochim. Biophys. Acta 2004, 1701, 121–128. [Google Scholar] [CrossRef]
- Berrin, J.G.; Ajandouz, E.H.; Georis, J.; Arnaut, F.; Juge, N. Substrate and product hydrolysis specificity in family 11 glycoside hydrolases: An analysis of Penicillium funiculosum and Penicillium griseofulvum xylanases. Appl. Microbiol. Biotechnol. 2007, 74, 1001–1010. [Google Scholar] [CrossRef]
- Driss, D.; Berrin, J.G.; Juge, N.; Bhiri, F.; Ghorbel, R.; Chaabouni, S.E. Functional characterization of Penicillium occitanis Pol6 and Penicillium funiculosum GH11 xylanases. Protein Expr. Purif. 2013, 90, 195–201. [Google Scholar] [CrossRef]
- Sun, R.J.; Xu, Y.; Hou, C.X.; Zhan, Y.H.; Liu, M.Q.; Weng, X.Y. Expression and characteristics of rice xylanase inhibitor OsXIP, a member of a new class of antifungal proteins. Biol. Plant 2018, 62, 569–578. [Google Scholar] [CrossRef]
- Fierens, E.; Gebruers, K.; Voet, A.R.D.; de Maeyer, M.; Courtin, C.M.; Delcour, J.A. Biochemical and structural characterization of TLXI, the Triticum aestivum L. thaumatin-like xylanase inhibitor. J. Enzym. Inhib. Med. Chem. 2009, 24, 646–654. [Google Scholar] [CrossRef]
- Durand, A.; Hughes, R.; Roussel, A.; Flatman, R.; Henrissat, B.; Juge, N. Emergence of a subfamily of xylanase inhibitors within glycoside hydrolase family 18. FEBS J. 2005, 272, 1745–1755. [Google Scholar] [CrossRef]
- Goesaert, H.; Gebruers, K.; Courtin, C.M.; Delcour, J.A. Purification and characterization of a XIP-type endoxylanase inhibitor from Rice (Oryza sativa). J. Enzym. Inhib. Med. Chem. 2005, 20, 95–101. [Google Scholar] [CrossRef]
- Tokunaga, T.; Esaka, M. Induction of a novel XIP-type Xylanase Inhibitor by external ascorbic acid treatment and differential expression of XIP-Family genes in rice. Plant Cell. Physiol. 2007, 48, 700–714. [Google Scholar] [CrossRef]
- Vasconcelos, E.A.R.; Santana, C.G.; Godoy, C.V.; Seixas, C.D.S.; Silva, M.S.; Moreira, L.R.S.; Oliveira-Neto, O.B.; Price, D.; Fitches, E.; Filho, E.X.F.; et al. A new chitinase-like xylanase inhibitor protein (XIP) from coffee (Coffea arabica) affects Soybean Asian rust (Phakopsora pachyrhizi) spore germination. BMC Biotechnol. 2011, 11, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Wang, Y.; Kim, S.T.; Kim, S.G.; Kang, K.Y. Characterization of a newly identified rice chitinase-like protein (OsCLP) homologous to xylanase inhibitor. BMC Biotechnol. 2013, 13, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansen, S.; de Ranter, C.J.; Gebruers, K.; Brijs, K.; Courtin, C.M.; Delcour, J.A.; Rabijns, A. Structural basis for inhibition of Aspergillus niger xylanase by Triticum aestivum Xylanase Inhibitor-I. J. Biol. Chem. 2004, 279, 36022–36028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansen, S.; de Ranter, C.J.; Gebruers, K.; Brijs, K.; Courtin, C.M.; Delcour, J.A.; Rabijns, A. Crystallization and preliminary X-ray diffraction study of two complexes of a TAXI-type xylanase inhibitor with glycoside hydrolase family 11 xylanases from Aspergillus niger and Bacillus subtilis. Acta Crystallogr. 2004, 60, 555–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tundo, S.; Paccanaro, M.C.; Bigini, V.; Savatin, D.V.; Faoro, F.; Favaron, F.; Sella, L. The Fusarium graminearum FGSG_03624 xylanase enhances plant immunity and increases resistance against bacterial and fungal pathogens. Int. J. Mol. Sci. 2021, 22, 10811. [Google Scholar] [CrossRef]
- Moscetti, I.; Faoro, F.; Moro, S.; Sabbadin, D.; Sella, L.; Favaron, F.; D’Ovidio, R. The xylanase inhibitor TAXI-III counteracts the necrotic activity of a Fusarium graminearum xylanase in vitro and in durum wheat transgenic plants. Mol. Plant Pathol. 2015, 16, 583–592. [Google Scholar] [CrossRef]
- Frías, M.; González, M.; González, C.; Brito, N. A 25-residue peptide from Botrytis cinerea xylanase bcxyn11a elicits plant defenses. Front. Plant Sci. 2019, 10, 474. [Google Scholar] [CrossRef]
- Ron, M.; Avni, A. The Receptor for the Fungal Elicitor Ethylene-Inducing Xylanase Is a Member of a Resistance-Like Gene Family in Tomato. Plant Cell 2004, 16, 1604–1615. [Google Scholar] [CrossRef]
- Denisenko, Y.A.; Gusakov, A.V.; Rozhkova, A.M.; Zorov, I.N.; Bashirova, A.V.; Matys, V.Y.; Nemashkalov, V.A.; Sinitsyn, A.P. Protein engineering of GH10 family xylanases for gaining a resistance to cereal proteinaceous inhibitors. Biocatal. Agric. Biotechnol. 2019, 17, 690–695. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, N.; Sinha, M.; Dube, D.; Singh, S.B.; Bhushan, A.; Kaur, P.; Srinivasan, A.; Sharma, S.; Singh, T.P. Crystal structure determination and inhibition studies of a novel xylanase and α-amylase inhibitor protein (XAIP) from Scadoxus multiflorus. FEBS J. 2010, 277, 2868–2882. [Google Scholar] [CrossRef]
- Sancho, A.I.; Faulds, C.B.; Svensson, B.; Bartolomé, B.; Williamson, G.; Juge, N. Cross-inhibitory activity of cereal protein inhibitors against α-amylases and xylanases. Biochim. Biophys. Acta Proteins Proteom. 2003, 1650, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Vandermarliere, E.; Lammens, W.; Schoepe, J.; Rombouts, S.; Fierens, E.; Gebruers, K.; Volckaert, G.; Rabijns, A.; Delcour, J.A.; Strelkov, S.; et al. Crystal structure of the noncompetitive xylanase inhibitor TLXI, member of the small thaumatin-like protein family. Proteins Struct. Funct. Bioinform. 2010, 78, 2391–2394. [Google Scholar] [CrossRef] [PubMed]
- Rombouts, S.; Fierens, E.; Vandermarliere, E.; Voet, A.; Gebruers, K.; Beaugrand, J.; Courtin, C.M.; Delcour, J.A.; de Maeyer, M.; Rabijns, A.; et al. His22 of TLXI plays a critical role in the inhibition of glycoside hydrolase family 11 xylanases. J. Enzym. Inhib. Med. Chem. 2009, 24, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.Q.; Sun, X.Y.; Peng, Y.Y.; Huang, Y.Y.; Liu, M.Q.; Weng, X.Y. WRKY transcription factor functions as a transcriptional regulator of Xylanase Inhibitor RIXI, involved in rice disease resistance to Magnaporthe oryzae. J. Plant Biol. 2020, 63, 177–188. [Google Scholar] [CrossRef]
- Zhan, Y.; Sun, X.; Rong, G.; Hou, C.; Huang, Y.; Jiang, D.; Weng, X. Identification of two transcription factors activating the expression of OsXIP in rice defence response. BMC Biotechnol. 2017, 17, 26. [Google Scholar] [CrossRef] [Green Version]
- Croes, E.; Gebruers, K.; Luyten, N.; Delcour, J.A.; Courtin, C.M. The three classes of wheat xylanase-inhibiting proteins accumulate in an analogous way during wheat ear development and germination. J. Plant Physiol. 2009, 166, 1253–1262. [Google Scholar] [CrossRef]
- Sella, L.; Gazzetti, K.; Faoro, F.; Odorizzi, S.; D’Ovidio, R.; Schäfer, W.; Favaron, F. A Fusarium graminearum xylanase expressed during wheat infection is a necrotizing factor but is not essential for virulence. Plant Physiol. Biochem. 2013, 64, 1–10. [Google Scholar] [CrossRef]
- Mélida, H.; Bacete, L.; Ruprecht, C.; Rebaque, D.; del Hierro, I.; López, G.; Brunner, F.; Pfrengle, F.; Molina, A. Arabinoxylan-oligosaccharides act as Damage Associated Molecular Patterns in plants regulating disease resistance. Front. Plant Sci. 2020, 11, 1210. [Google Scholar] [CrossRef]
- Federici, L.; di Matteo, A.; Fernandez-Recio, J.; Tsernoglou, D.; Cervone, F. Polygalacturonase inhibiting proteins: Players in plant innate immunity? Trends Plant Sci. 2006, 11, 65–70. [Google Scholar] [CrossRef]
- Lin, P.; Wong, J.H.; Ng, T.B.; Ho, V.S.M.; Xia, L. A sorghum xylanase inhibitor-like protein with highly potent antifungal, antitumor and HIV-1 reverse transcriptase inhibitory activities. Food Chem. 2013, 141, 2916–2922. [Google Scholar] [CrossRef]
- Fuzi, S.F.Z.M.; Mahadi, N.M.; Jahim, J.M.; Murad, A.M.A.; Bakar, F.D.A.; Jusoh, M.; Rahman, R.A.; Illias, R.M. Development and validation of a medium for recombinant endo-β-1,4- xylanase production by Kluyveromyces lactis using a statistical experimental design. Ann. Microbiol. 2012, 62, 283–292. [Google Scholar] [CrossRef]
- Smeets, N.; Nuyens, F.; Niewold, T.; van Campenhout, L. Temperature resistance of xylanase inhibitors and the presence of grain-associated xylanases affect the activity of exogenous xylanases added to pelleted wheat-based feeds. Cereal Chem. 2014, 91, 572–577. [Google Scholar] [CrossRef]
- Krogh Madsen, C.; Pettersson, D.; Hjortshøj, R.; Katholm, A.; Brinch-Pedersen, H. Superior growth rates in broilers fed wheat with low in vitro feed-xylanase inhibition. J. Agric. Food Chem. 2018, 66, 4044–4050. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Mo, Y.; Shah, S.; Xie, Y.; Mehmood, A.; Jiang, H.; Guo, Y. Characterization of Two Wheat-Derived Glycoside Hydrolase Family-10 Xylanases Resistant to Xylanase Inhibitors. J. Food Qual. 2022, 2022, 9590243. [Google Scholar] [CrossRef]
- Vázquez, M.J.; Alonso, J.L.; Domínguez, H.; Parajó, J.C. Xylooligosaccharides: Manufacture and applications. Trends Food Sci. Technol. 2000, 11, 387–393. [Google Scholar] [CrossRef]
- Berger, K.; Burleigh, S.; Lindahl, M.; Bhattacharya, A.; Patil, P.; Stålbrand, H.; Nordberg Karlsson, E.; Hållenius, F.; Nyman, M.; Adlercreutz, P. Xylooligosaccharides increase Bifidobacteria and Lachnospiraceae in mice on a high-fat diet, with a concomitant increase in short-chain fatty acids, especially butyric acid. J. Agric. Food Chem. 2021, 69, 3617–3625. [Google Scholar] [CrossRef]
- Finegold, S.M.; Li, Z.; Summanen, P.H.; Downes, J.; Thames, G.; Corbett, K.; Dowd, S.; Krak, M.; Heber, D. Xylooligosaccharide increases Bifidobacteria but not Lactobacilli in human gut microbiota. Food Funct. 2014, 5, 436–445. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X.; Ho, C.L. Recent development of probiotic Bifidobacteria for treating human diseases. Front. Bioeng. Biotechnol. 2021, 9, 770248. [Google Scholar] [CrossRef]
- Xu, X.; Liu, M.Q.; Huo, W.K.; Dai, X.J. Obtaining a mutant of Bacillus amyloliquefaciens xylanase A with improved catalytic activity by directed evolution. Enzym. Microb. Technol. 2016, 86, 59–66. [Google Scholar] [CrossRef]
- Sun, J.Y.; Liu, M.Q.; Xu, Y.L.; Xu, Z.R.; Pan, L.; Gao, H. Improvement of the thermostability and catalytic activity of a mesophilic family 11 xylanase by N-terminus replacement. Protein Expr. Purif. 2005, 42, 122–130. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, K.; Chen, X.; Chu, X.; Sun, F.; Dong, Z. Five mutations in N-terminus confer thermostability on mesophilic xylanase. Biochem. Biophys. Res. Commun. 2010, 395, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Rouau, X.; El-Hayek, M.L.; Moreau, D. Effect of an enzyme preparation containing pentosanases on the bread-making quality of flours in relation to changes in pentosan properties. J. Cereal Sci. 1994, 19, 259–272. [Google Scholar] [CrossRef]
- Leys, S.; de Bondt, Y.; Schreurs, L.; Courtin, C.M. Sensitivity of the Bacillus subtilis Xyn A xylanase and its mutants to different xylanase inhibitors determines their activity profile and functionality during bread making. J. Agric. Food Chem. 2019, 67, 11198–11209. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Li, X.; Cai, G.; Wang, D.; Li, F.; Lu, J.; Yu, X. Production of recombinant barley xylanase inhibitor in Pichia pastoris and its inhibitory effect on premature yeast flocculation. J. Inst. Brew. 2022, 128, 59–65. [Google Scholar] [CrossRef]
- Gupta, R.S.; Warren, C.M.; Smith, B.M.; Jiang, J.; Blumenstock, J.A.; Davis, M.M.; Schleimer, R.P.; Nadeau, K.C. Prevalence and severity of food allergies among US adults. JAMA Netw. Open 2019, 2, e185630. [Google Scholar] [CrossRef] [Green Version]
- Šotkovský, P.; Sklenář, J.; Halada, P.; Cinová, J.; Šetinová, I.; Kainarová, A.; Goliáš, J.; Pavlásková, K.; Honzová, S.; Tučková, L. A new approach to the isolation and characterization of wheat flour allergens. Clin. Exp. Allergy 2011, 41, 1031–1043. [Google Scholar] [CrossRef]
- Hirano, K.; Hino, S.; Oshima, K.; Nadano, D.; Urisu, A.; Takaiwa, F.; Matsuda, T. Evaluation of allergenic potential for rice seed protein components utilizing a rice proteome database and an allergen database in combination with IgE-binding of recombinant proteins. Biosci. Biotechnol. Biochem. 2016, 80, 564–573. [Google Scholar] [CrossRef]
- Tundo, S.; Lupi, R.; Lafond, M.; Giardina, T.; Larré, C.; Denery-Papini, S.; Morisset, M.; Kalunke, R.; Sestili, F.; Masci, S. Wheat ati cm3, cm16 and 0.28 allergens produced in Pichia pastoris display a different eliciting potential in food allergy to wheat‡. Plants 2018, 7, 101. [Google Scholar] [CrossRef]
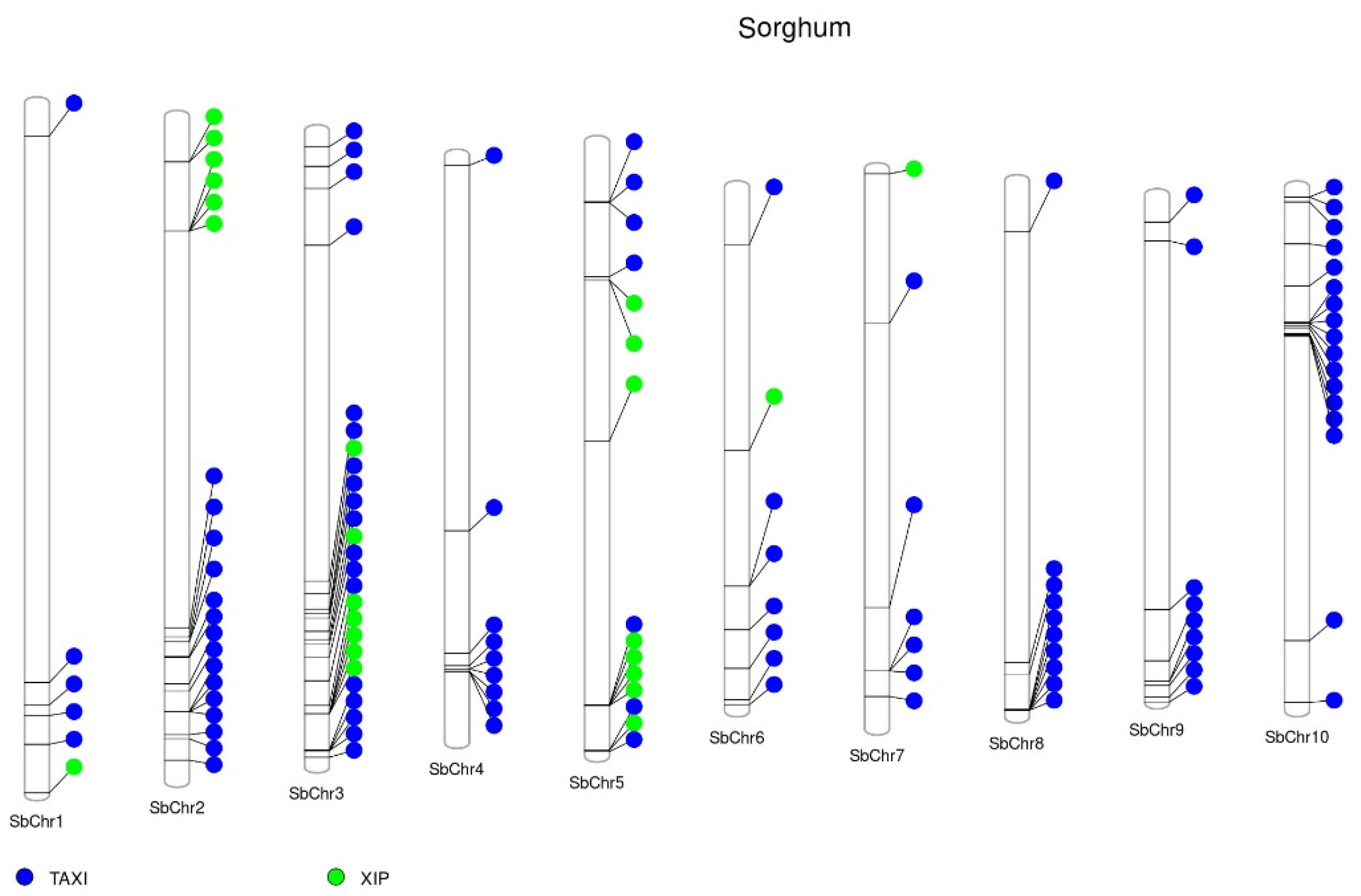



| Xylanase | GH | Origin | TAXI-I | TAXI-II | TAXI-III | TAXI-IV | XIP-I | TLXI | RIXI | riceXIP | OsXIP | OsHI-XIP | XAIP | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aspergillus aculeatus (AaXyl2) | 10 | F | no | no | no | no | [31,44,49,53] | |||||||
| Aspergillus fumigatus xylanase A (AfXylA10) | 10 | F | 177.94 | [54] | ||||||||||
| Aspergillus nidulans (AnidXlnC) | 10 | F | no | no | 9 | [44,49,53] | ||||||||
| Aspergillus niger (XynIII) | 11 | F | yes | [41] | ||||||||||
| Aspergillus niger (AnExlA/AnxA/ANX) | 11 | F | 20.1 | no | 317 | 135 | yes | yes | [31,44,49,53,55,56,57,58,59,60,61] | |||||
| Aspergillus niger hybrid mutant of AnxA xylanase and TfxA_CD (FSI-124) | 11 | F/B | yes | [60] | ||||||||||
| Aspergillus niger (AnM4Xyl) | 11 | F | 18.8 | [21] | ||||||||||
| Aspergillus niger (AnXynA) | 10 | F | no | no | yes | no | yes | [31,49,53,59] | ||||||
| Aspergillus niger (Xyn10) | 10 | F | yes | [41] | ||||||||||
| Aspergillus niger xylanase hybrid of AnxA xylanase and TfxA_CD (Atx) | 11 | F/B | yes | [55,60] | ||||||||||
| Aspergillus oryzae (AoXyn) | 10 | F | no | no | 17 | no | [31,44,53] | |||||||
| Bacillus agaradhaerens (BaXyl) | 11 | B | no | [44] | ||||||||||
| Bacillus amyloliquefaciens (reBaxA) | 11 | B | yes | yes | 54.09 | [55,57,60,62] | ||||||||
| Bacillus amyloliquefaciens xylanase mutant of reBaxA (reBaxA454) | 11 | B | yes | [57] | ||||||||||
| Bacillus amyloliquefaciens xylanase mutant of reBaxA (reBaxA50) | 11 | B | yes | [55,60] | ||||||||||
| Bacillus amyloliquefaciens xylanase mutant T331 of reBaxA (DS199) | 11 | B | 12.16 | [62] | ||||||||||
| Bacillus sp. | 10 | B | no | [44] | ||||||||||
| Bacillus subtilis (BsXynA) | 11 | B | 16.7 | yes | no | no | [31,44,49,53,56] | |||||||
| Bacillus subtilis (BSX) | 11 | B | yes | yes | yes | [59,61] | ||||||||
| Botrytis cinerea (BcXyn11a) | 11 | F | 6 | no | 2.1 | [63] | ||||||||
| Chrysosporium lucknowense (ClXynB) | 10 | F | no | no | no | [64] | ||||||||
| Chrysosporium lucknowense (ClXynC) | 10 | F | no | no | no | [64] | ||||||||
| Fibrobacter succinogenes (FsXynC) | 11 | B | no | [44,53] | ||||||||||
| Flavobacterium sp. MSY-2 (rXFH) | 10 | B | no | no | [65] | |||||||||
| Fusarium graminearum (FCSG_11487) | 10 | F | no | no | no | no | no | no | [39,66,67] | |||||
| Fusarium graminearum (FGSG_03624) | 11 | F | yes | yes | 2.59 | yes | no | no | [21,39,66,67,68] | |||||
| Fusarium graminearum (FGSG_10999) | 11 | F | yes | yes | 12.04 | yes | no | no | [39,66,67,68] | |||||
| Fusarium graminearum (FGSG_11304) | 10 | F | no | no | no | no | 83.7 | no | [39,66,67] | |||||
| Hu sheep rumen microbiota (XYN-LXY_CD, catalytic domain) | 10 | B | 237.37 | [69] | ||||||||||
| Hypothenemus hampei (HhXyl) | 10 | I | yes | [70] | ||||||||||
| Neocallimastix patriciarum (NpXynA) | 11 | F | no | no | [71,72] | |||||||||
| Neocallimastix sp. GMLF1 (Xyn1B) | 11 | F | no | [71] | ||||||||||
| Oryza sativa (OSX) | 10 | P | no | [59] | ||||||||||
| Paenibacillus sp | U | B | yes | [41] | ||||||||||
| Penicillium funiculosum (not specified) | 11 | F | yes | [73] | ||||||||||
| Penicillium funiculosum (PfXynA) | 7 | F | 46 | 46 | 106 | [74] | ||||||||
| Penicillium funiculosum (PfXynB) | 11 | F | 2.9 | no | 89.7 | [75,76] | ||||||||
| Penicillium funiculosum (PfXynC) | 11 | F | 16 | 17 | 3.4 | 289.6 | [31,44,49,75,77,78] | |||||||
| Penicillium funiculosum (PfXynD) | 10 | F | no | no | yes | [74] | ||||||||
| Penicillium griseofulvum (PgXynA) | 11 | F | no | [77] | ||||||||||
| Penicillium occitanis Pol6 (PoXyn3) | 11 | F | yes | [78] | ||||||||||
| Penicillium purpurogenum (PpXynA) | 10 | F | no | no | no | [31,49,53] | ||||||||
| Penicillium purpurogenum (PpXynB) | 11 | F | yes | yes | [49,53,75] | |||||||||
| Pseudoalteromonas haloplanktis TAH3A (XPH) | 8 | B | no | no | no | no | [65] | |||||||
| Pseudomonas fluorescens (PfXynA) | 10 | B | no | no | [31,44] | |||||||||
| Talaromyces emersonii xylanase (TeX-1) | 10 | F | no | no | [49] | |||||||||
| Thermobacillus xylanilyticus (TxXyl) | 11 | B | 234.7 | [31] | ||||||||||
| Thermobacillus xylanilyticus (TxXynA) | 10 | B | no | [31] | ||||||||||
| Thermomonospora fusca (TFX) | 11 | B | no | [59] | ||||||||||
| Thermomonospora fusca TF xylanase A catalytic domain (TfxA_CD) | 11 | B | yes | yes | [55,57] | |||||||||
| Thermomonospora fusca catalytic domain TfxA_CD mutant (TfxA_CD214) | 11 | B | yes | 12.2 | [55,57,79] | |||||||||
| Thermomonospora fusca catalytic domain TfxA_CD mutant (TfxA_CD309) | 11 | B | yes | yes | [55,57] | |||||||||
| Thermomonospora fusca catalytic domain TfxA_CD mutant (TfxA_CD311) | 11 | B | yes | yes | [55,57] | |||||||||
| Thermomonospora fusca catalytic domain TfxA_CD mutant (TfxA_CD467) | 11 | B | yes | [57] | ||||||||||
| Thermomonospora fusca catalytic domain TfxA_CD mutant (TfxA_CD526) | 11 | B | yes | [57] | ||||||||||
| Thermomonospora lanuginosus (TLx) | 11 | F | yes | yes | yes | yes | [20,55,57,60,79] | |||||||
| Trichoderma harzianum (ThXyn1) | 11 | F | yes | [41,71] | ||||||||||
| Trichoderma longibrachiatum (endo-1,4-β; M3) | 11 | F | yes | yes | yes | [20,60,79] | ||||||||
| Trichoderma longibrachiatum (Xyn I) | 11 | F | 4.2 | [31] | ||||||||||
| Trichoderma longibrachiatum (Xyn II) | 11 | F | no | [31] | ||||||||||
| Trichoderma reesei (TRx) | 11 | F | yes | yes | [55,57] | |||||||||
| Trichoderma reesei (Xyn1) | 11 | F | yes | yes | [75] | |||||||||
| Trichoderma reesei (Xyn2) | 11 | F | yes | yes | [75] | |||||||||
| Trichoderma viride (TvXyl) | 11 | F | yes | yes | 610 | 170.4 | [31,44,49,53,75] | |||||||
| Triticum aestivum xylanase (from flour) | 10 | P | no | no | [49,51] | |||||||||
| Uncultured bacterium (rXyn8) | 8 | B | no | no | no | no | [65] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tundo, S.; Mandalà, G.; Sella, L.; Favaron, F.; Bedre, R.; Kalunke, R.M. Xylanase Inhibitors: Defense Players in Plant Immunity with Implications in Agro-Industrial Processing. Int. J. Mol. Sci. 2022, 23, 14994. https://doi.org/10.3390/ijms232314994
Tundo S, Mandalà G, Sella L, Favaron F, Bedre R, Kalunke RM. Xylanase Inhibitors: Defense Players in Plant Immunity with Implications in Agro-Industrial Processing. International Journal of Molecular Sciences. 2022; 23(23):14994. https://doi.org/10.3390/ijms232314994
Chicago/Turabian StyleTundo, Silvio, Giulia Mandalà, Luca Sella, Francesco Favaron, Renesh Bedre, and Raviraj M. Kalunke. 2022. "Xylanase Inhibitors: Defense Players in Plant Immunity with Implications in Agro-Industrial Processing" International Journal of Molecular Sciences 23, no. 23: 14994. https://doi.org/10.3390/ijms232314994
APA StyleTundo, S., Mandalà, G., Sella, L., Favaron, F., Bedre, R., & Kalunke, R. M. (2022). Xylanase Inhibitors: Defense Players in Plant Immunity with Implications in Agro-Industrial Processing. International Journal of Molecular Sciences, 23(23), 14994. https://doi.org/10.3390/ijms232314994






