Insights into the Structure of the Highly Glycosylated Ffase from Rhodotorula dairenensis Enhance Its Biotechnological Potential
Abstract
1. Introduction
2. Results and Discussion
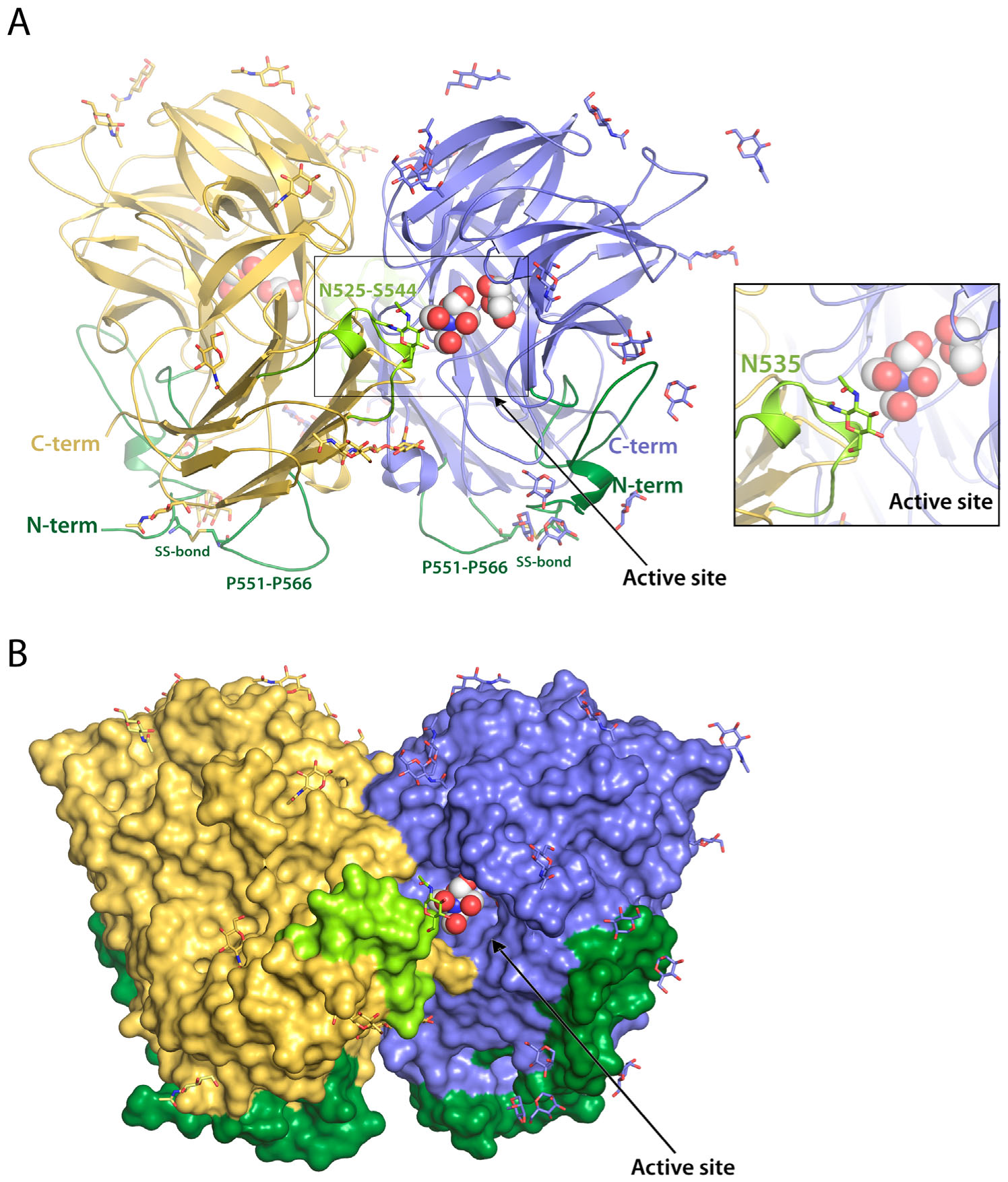
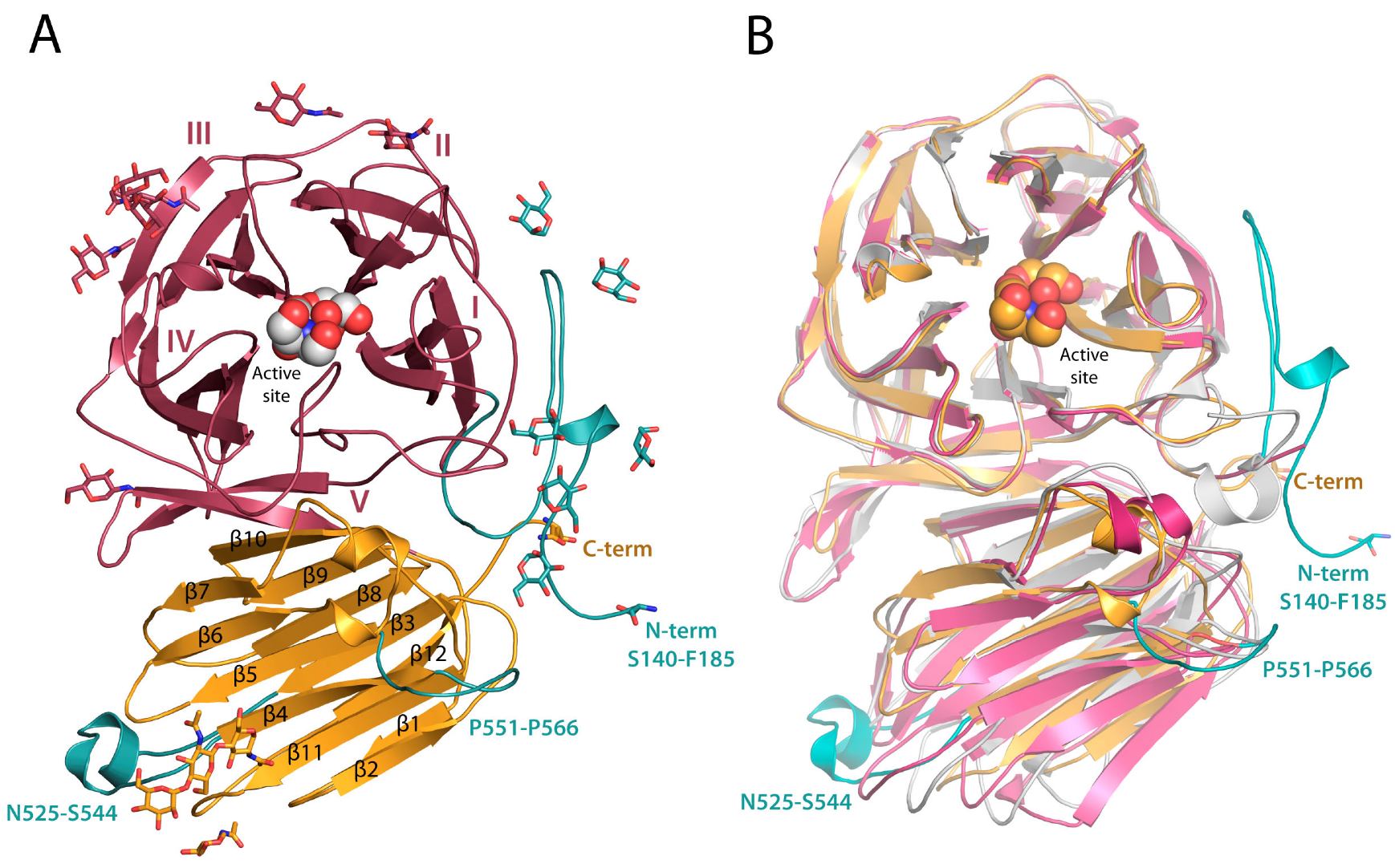
2.1. RdINV Overall Folding
2.2. RdINV N-Terminus Analysis
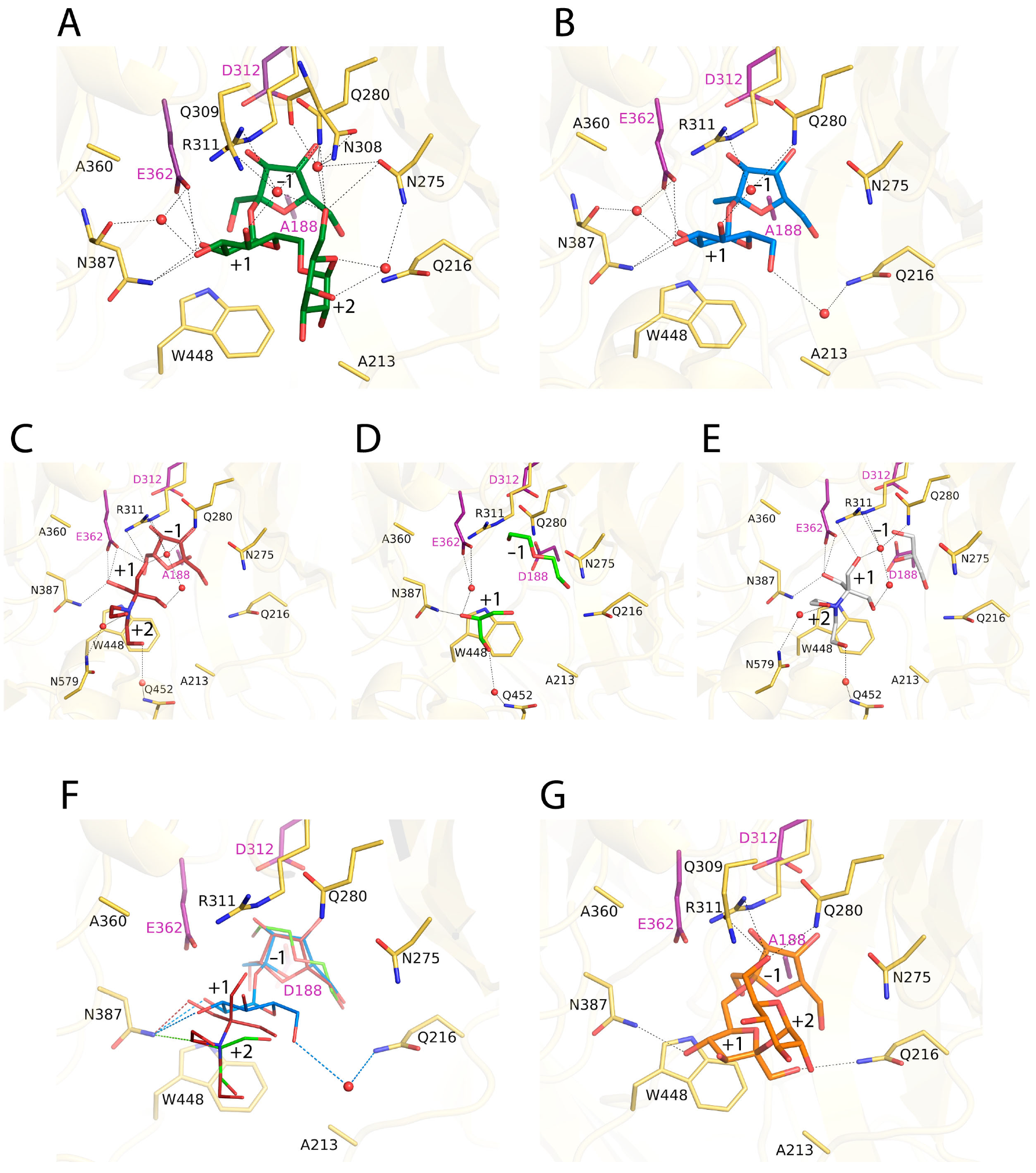
2.3. Complexes of RdINV Depict Its Active Site
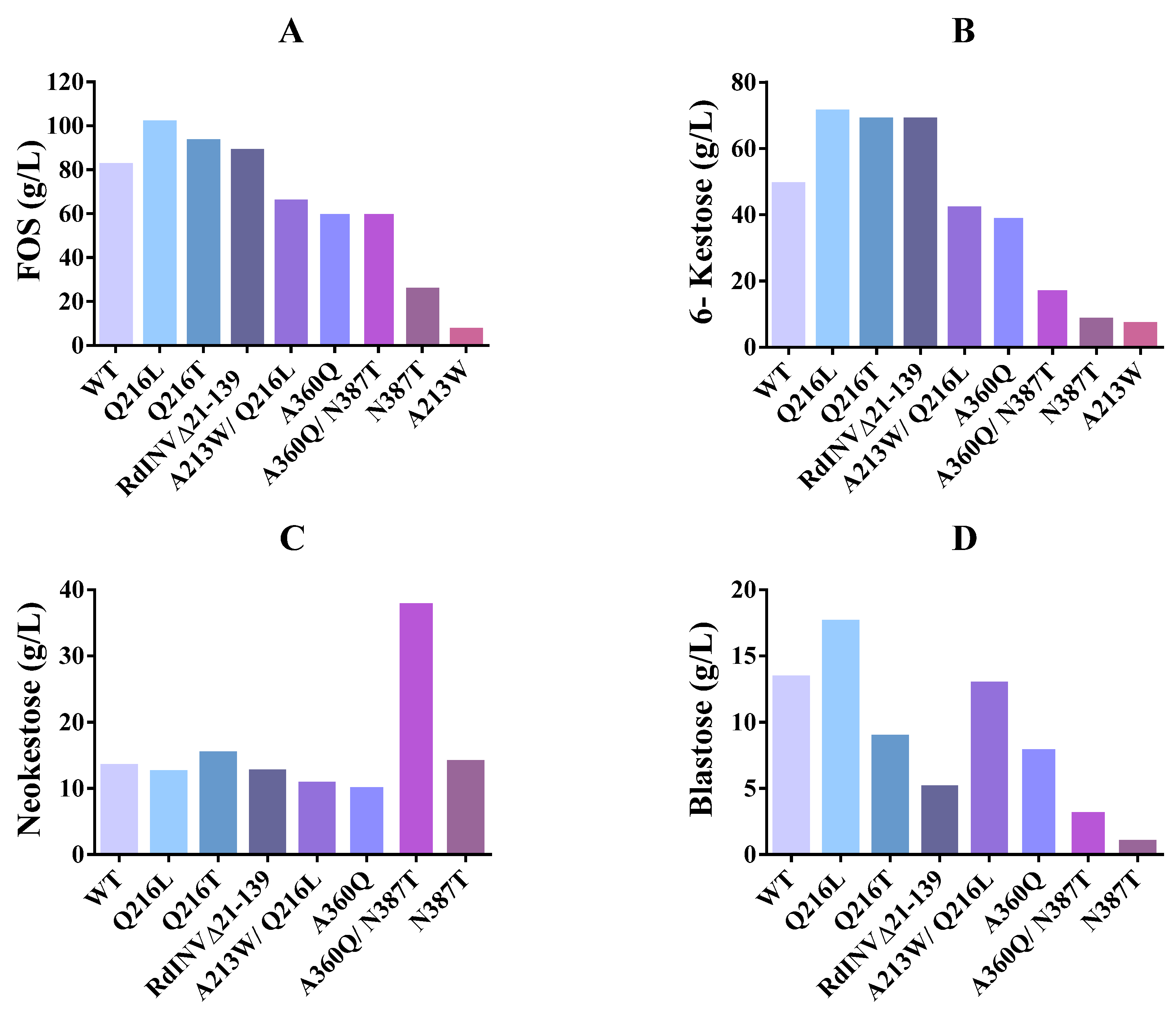
2.4. Design of the Mutations and Study of Their Effect on RdINV Hydrolytic Activity
2.5. Impact of Mutations on RdINV Transfructosylating Productivity
3. Material and Methods
3.1. Organisms, Media, Growth and Mutagenesis
3.2. RdINV Expression and Purification
3.3. Hydrolase Activity and Kinetic Analysis
3.4. Transferase Activity, Fructooligosaccharides Production and HPLC Analysis
3.5. Crystallization and X-ray Structure Determination of RdINV
3.6. Automated Docking of 6-Kestose into D188A-RdINVΔ21–139
3.7. Differential Scanning Fluorimetry
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Gimeno-Pérez, M.; Merdzo, Z.; Castillo-Rosa, E.; Hijas, C.; Fernández-Lobato, M. The β-Fructofuranosidase from Rhodotorula dairenensis: Molecular Cloning, Heterologous Expression, and Evaluation of Its Transferase Activity. Catalysts 2021, 11, 476. [Google Scholar] [CrossRef]
- Ramírez-Escudero, M.; Gimeno-Pérez, M.; González, B.; Linde, D.; Merdzo, Z.; Fernández-Lobato, M.; Sanz-Aparicio, J. Structural Analysis of β-Fructofuranosidase from Xanthophyllomyces dendrorhous Reveals Unique Features and the Crucial Role of N-Glycosylation in Oligomerization and Activity. J. Biol. Chem. 2016, 291, 6843–6857. [Google Scholar] [CrossRef] [PubMed]
- Sabater-Molina, M.; Larqué, E.; Torrella, F.; Zamora, S. Dietary fructooligosaccharides and potential benefits on health. J. Physiol. Biochem. 2009, 65, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Costa, G.; Vasconcelos, Q.; Abreu, G.; Albuquerque, A.; Vilar, J.; Aragão, G. Systematic review of the ingestion of fructooligosaccharides on the absorption of minerals and trace elements versus control groups. Clin. Nutr. ESPEN 2021, 41, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Delzenne, N.M. Oligosaccharides: State of the art. Proc. Nutr. Soc. 2003, 62, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Kelly, G. Inulin-type prebiotics: A review (Part 2). Altern. Med. Rev. 2009, 14, 36–55. [Google Scholar]
- O’Keefe, S.J.D. Diet, Microorganisms and Their Metabolites, and Colon Cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar]
- Taylor, A.M.; Holscher, H.D. A review of dietary and microbial connections to depression, anxiety, and stress. Nutr. Neurosci. 2020, 23, 237–250. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, P.; Xia, C.; Wu, Z.; Zhong, Z.; Xu, Y.; Zeng, Y.; Liu, H.; Liu, R.; Liao, M. Fructooligosaccharides supplementation mitigated chronic stress-induced intestinal barrier impairment and neuroinflammation in mice. J. Funct. Foods 2020, 72, 1–7. [Google Scholar] [CrossRef]
- Kumar, C.G.; Sripada, S.; Poornachandra, Y. Status and Future Prospects of Fructooligosaccharides as Nutraceuticals. In Role of Materials Science in Food Bioengineering; Elsevier: New York, NY, USA, 2018; Volume 19, pp. 451–503. ISBN 9780128114483. [Google Scholar]
- Kilian, S.; Kritzinger, S.; Rycroft, C.; Gibson, G.; Du Preez, J. The effects of the novel bifidogenic trisaccharide, neokestose, on the human colonic microbiota. World J. Microbiol. Biotechnol. 2002, 18, 637–644. [Google Scholar] [CrossRef]
- Taştan, Ö.; Sözgen, G.; Baysal, T.; Türköz, B.K. Production of prebiotic 6-kestose using Zymomonas mobilis levansucrase in carob molasses and its effect on 5-HMF levels during storage. Food Chem. 2019, 297, 124897. [Google Scholar] [CrossRef]
- Lee, S.-M.; Chang, J.-Y.; Wu, J.-S.; Sheu, D.-C. Antineoplastic effect of a novel chemopreventive agent, neokestose, on the Caco-2 cell line via inhibition of expression of nuclear factor-κB and cyclooxygenase-2. Mol. Med. Rep. 2015, 12, 1114–1118. [Google Scholar] [CrossRef]
- Wu, J.-S.; Chang, J.-Y.; Chen, C.W.; Lin, M.-T.; Sheu, D.-C.; Lee, S.-M. Neokestose suppresses the growth of human melanoma A2058 cells via inhibition of the nuclear factor-κB signaling pathway. Mol. Med. Rep. 2017, 16, 295–300. [Google Scholar] [CrossRef]
- Henrissat, B.; Sulzenbacher, G.; Bourne, Y. Glycosyltransferases, glycoside hydrolases: Surprise, surprise! Curr. Opin. Struct. Biol. 2008, 18, 527–533. [Google Scholar] [CrossRef]
- Toledo, L.E.T.; García, D.M.; Cruz, E.P.; Intriago, L.M.R.; Pérez, J.N.; Chanfrau, J.M.P. Fructosyltransferases and Invertases: Useful Enzymes in the Food and Feed Industries; Elsevier Inc.: New York, NY, USA, 2018; ISBN 9780128132807. [Google Scholar]
- Manoochehri, H.; Hosseini, N.F.; Saidijam, M.; Taheri, M.; Rezaee, H.; Nouri, F. A review on invertase: Its potentials and applications. Biocatal. Agric. Biotechnol. 2020, 25, 101599. [Google Scholar] [CrossRef]
- Trollope, K.M.; van Wyk, N.; Kotjomela, M.A.; Volschenk, H. Sequence and structure-based prediction of fructosyltransferase activity for functional subclassification of fungal GH32 enzymes. FEBS J. 2015, 282, 4782–4796. [Google Scholar] [CrossRef]
- Pons, T.; Naumoff, D.G.; Martínez-Fleites, C.; Hernández, L. Three acidic residues are at the active site of a β-propeller architecture in glycoside hydrolase families 32, 43, 62, and 68. Proteins Struct. Funct. Bioinform. 2004, 54, 424–432. [Google Scholar] [CrossRef]
- Bujacz, A.; Jedrzejczak-Krzepkowska, M.; Bielecki, S.; Redzynia, I.; Bujacz, G. Crystal structures of the apo form of β-fructofuranosidase from Bifidobacterium longum and its complex with fructose. FEBS J. 2011, 278, 1728–1744. [Google Scholar] [CrossRef]
- Park, J.; Kim, M.-I.; Park, Y.-D.; Shin, I.; Cha, J.; Kim, C.H.; Rhee, S. Structural and Functional Basis for Substrate Specificity and Catalysis of Levan Fructotransferase. J. Biol. Chem. 2012, 287, 31233–31241. [Google Scholar] [CrossRef]
- Kubota, A.; Kawai, R.; Li, D.; Kozono, T.; Sasaki, N.; Nishikawa, A.; Fujii, T.; Tochio, T.; Tonozuka, T. Enzymatic and structural characterization of β-fructofuranosidase from the honeybee gut bacterium Frischella perrara. Appl. Microbiol. Biotechnol. 2022, 106, 2455–2470. [Google Scholar] [CrossRef]
- Chuankhayan, P.; Hsieh, C.-Y.; Huang, Y.-C.; Hsieh, Y.-Y.; Guan, H.-H.; Hsieh, Y.-C.; Tien, Y.-C.; Chen, C.-D.; Chiang, C.-M.; Chen, C.-J. Crystal Structures of Aspergillus japonicus Fructosyltransferase Complex with Donor/Acceptor Substrates Reveal Complete Subsites in the Active Site for Catalysis. J. Biol. Chem. 2010, 285, 23251–23264. [Google Scholar] [CrossRef]
- Pouyez, J.; Mayard, A.; Vandamme, A.-M.; Roussel, G.; Perpète, E.A.; Wouters, J.; Housen, I.; Michaux, C. First crystal structure of an endo-inulinase, INU2, from Aspergillus ficuum: Discovery of an extra-pocket in the catalytic domain responsible for its endo-activity. Biochimie 2012, 94, 2423–2430. [Google Scholar] [CrossRef]
- Nagaya, M.; Kimura, M.; Gozu, Y.; Sato, S.; Hirano, K.; Tochio, T.; Nishikawa, A.; Tonozuka, T. Crystal structure of a β-fructofuranosidase with high transfructosylation activity from Aspergillus kawachii. Biosci. Biotechnol. Biochem. 2017, 81, 1786–1795. [Google Scholar] [CrossRef]
- Tarkowski, Ł.P.; Tsirkone, V.G.; Osipov, E.M.; Beelen, S.; Lammens, W.; Vergauwen, R.; Van den Ende, W.; Strelkov, S.V. Crystal structure of Arabidopsis thaliana neutral invertase 2. Acta Crystallogr. 2020, F76, 152–157. [Google Scholar] [CrossRef]
- Verhaest, M.; Lammens, W.; Le Roy, K.; De Coninck, B.; De Ranter, C.J.; Van Laere, A.; Ende, W.V.D.; Rabijns, A. X-ray diffraction structure of a cell-wall invertase from Arabidopsis thaliana. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 1555–1563. [Google Scholar] [CrossRef]
- Lammens, W.; Le Roy, K.; Van Laere, A.; Rabijns, A.; Ende, W.V.D. Crystal Structures of Arabidopsis thaliana Cell-Wall Invertase Mutants in Complex with Sucrose. J. Mol. Biol. 2008, 377, 378–385. [Google Scholar] [CrossRef]
- Miyazaki, T.; Oba, N.; Park, E.Y. Structural insight into the substrate specificity of Bombyx mori β-fructofuranosidase belonging to the glycoside hydrolase family 32. Insect Biochem. Mol. Biol. 2020, 127, 103494. [Google Scholar] [CrossRef]
- Álvaro-Benito, M.; Polo, A.; González, B.; Fernández-Lobato, M.; Sanz-Aparicio, J. Structural and Kinetic Analysis of Schwanniomyces occidentalis Invertase Reveals a New Oligomerization Pattern and the Role of Its Supplementary Domain in Substrate Binding. J. Biol. Chem. 2010, 285, 13930–13941. [Google Scholar] [CrossRef]
- Sainz-Polo, M.; Escudero, M.R.; Lafraya, A.; González, B.; Marín-Navarro, J.; Polaina, J.; Sanz-Aparicio, J. Three-dimensional Structure of Saccharomyces Invertase. J. Biol. Chem. 2013, 288, 9755–9766. [Google Scholar] [CrossRef]
- Gutiérrez-Alonso, P.; Fernández-Arrojo, L.; Plou, F.J.; Fernández-Lobato, M. Biochemical characterization of a β-fructofuranosidase from Rhodotorula dairenensis with transfructosylating activity. FEMS Yeast Res. 2009, 9, 768–773. [Google Scholar] [CrossRef][Green Version]
- Fernández-Álvarez, A.; Marín-Menguiano, M.; Lanver, D.; Jiménez-Martín, A.; Elías-Villalobos, A.; Pérez-Pulido, A.J.; Kahmann, R.; Ibeas, J.I. Identification of O-mannosylated Virulence Factors in Ustilago maydis. PLoS Pathog. 2012, 8, e1002563. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.; Sodhi, J.; McGuffin, L.; Buxton, B.; Jones, D. Prediction and Functional Analysis of Native Disorder in Proteins from the Three Kingdoms of Life. J. Mol. Biol. 2004, 337, 635–645. [Google Scholar] [CrossRef]
- Wootton, J.C. Non-globular domains in protein sequences: Automated segmentation using complexity measures. Comput. Chem. 1994, 18, 269–285. [Google Scholar] [CrossRef]
- González, M.; Brito, N.; González, C. High abundance of Serine/Threonine-rich regions predicted to be hyper-O-glycosylated in the secretory proteins coded by eight fungal genomes. BMC Microbiol. 2012, 12, 213. [Google Scholar] [CrossRef]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.-B.G.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2019, 31, 455–461. [Google Scholar] [CrossRef]
- Álvaro-Benito, M.; Polo, A.; González, B.; Fernández-Lobato, M.; Sanz-Aparicio, J.; Kilian, S.; Kritzinger, S.; Rycroft, C.; Gibson, G.; du Preez, J.; et al. Structural and Kinetic Insights Reveal That the Amino Acid Pair Gln-228/Asn-254 Modulates the Transfructosylating Specificity of Schwanniomyces occidentalis β-Fructofuranosidase, an Enzyme That Produces Prebiotics. J. Biol. Chem. 2012, 287, 527–533. [Google Scholar] [CrossRef]
- Liu, H.; Naismith, J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008, 8, 91. [Google Scholar] [CrossRef]
- Erijman, A.; Dantes, A.; Bernheim, R.; Shifman, J.M.; Peleg, Y. Transfer-PCR (TPCR): A highway for DNA cloning and protein engineering. J. Struct. Biol. 2011, 175, 171–177. [Google Scholar] [CrossRef]
- Gimeno-Pérez, M.; Linde, D.; Fernandez-Arrojo, L.; Plou, F.J.; Lobato, M.F. Heterologous overproduction of β-fructofuranosidase from yeast Xanthophyllomyces dendrorhous, an enzyme producing prebiotic sugars. Appl. Microbiol. Biotechnol. 2015, 99, 3459–3467. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1204–1214. [Google Scholar] [CrossRef] [PubMed]
- Lebedev, A.A.; Vagin, A.A.; Murshudov, G.N. Model preparation in MOLREP and examples of model improvement using X-ray data. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 64, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Terwilliger, T.C. Finding non-crystallographic symmetry in density maps of macromolecular structures. J. Struct. Funct. Genom. 2013, 14, 91–95. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Struct. Biol. Crystallogr. 2004, D60, 2126–2132. [Google Scholar] [CrossRef]
- DeLano, L.W. Pymol: An Open-Source Molecular Graphics Tool. In {CCP4} Newsletter on Protein Crystallography; BBSRC Collaborative Computational Project: Swindon, UK, 2002; Volume 40, pp. 1–8. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]

| RdINV Mutant | Km (mM) | kcat (1/s) | kcat/Km (1/s mM) |
|---|---|---|---|
| WT | 5 ± 0.1 | 4751 ± 25 | 1011 ± 10 |
| Q216L | 24 ± 1 | 2324 ± 13 | 98 ± 3 |
| Q216T | 13 ± 1 | 11,496 ± 182 | 864 ± 39 |
| A360Q | 15 ± 0.2 | 3814 ± 16 | 251 ± 21 |
| N387T | 17 ± 0.8 | 583 ± 5 | 35 ± 1 |
| A360Q/N387T | 14 ± 0.6 | 486 ± 4 | 34 ± 1 |
| A213W | 47 ± 3 | 2716 ± 45 | 58 ± 3 |
| A213W/Q216L | 88 ± 9 | 10,523 ± 329 | 120 ± 4 |
| RdINVΔ21–139 | 9 ± 1 | 16,384 ± 216 | 1781 ± 16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-Ortega, E.; Narmontaite, E.; González-Pérez, B.; Plou, F.J.; Fernández-Lobato, M.; Sanz-Aparicio, J. Insights into the Structure of the Highly Glycosylated Ffase from Rhodotorula dairenensis Enhance Its Biotechnological Potential. Int. J. Mol. Sci. 2022, 23, 14981. https://doi.org/10.3390/ijms232314981
Jiménez-Ortega E, Narmontaite E, González-Pérez B, Plou FJ, Fernández-Lobato M, Sanz-Aparicio J. Insights into the Structure of the Highly Glycosylated Ffase from Rhodotorula dairenensis Enhance Its Biotechnological Potential. International Journal of Molecular Sciences. 2022; 23(23):14981. https://doi.org/10.3390/ijms232314981
Chicago/Turabian StyleJiménez-Ortega, Elena, Egle Narmontaite, Beatriz González-Pérez, Francisco J. Plou, María Fernández-Lobato, and Julia Sanz-Aparicio. 2022. "Insights into the Structure of the Highly Glycosylated Ffase from Rhodotorula dairenensis Enhance Its Biotechnological Potential" International Journal of Molecular Sciences 23, no. 23: 14981. https://doi.org/10.3390/ijms232314981
APA StyleJiménez-Ortega, E., Narmontaite, E., González-Pérez, B., Plou, F. J., Fernández-Lobato, M., & Sanz-Aparicio, J. (2022). Insights into the Structure of the Highly Glycosylated Ffase from Rhodotorula dairenensis Enhance Its Biotechnological Potential. International Journal of Molecular Sciences, 23(23), 14981. https://doi.org/10.3390/ijms232314981







