Multi-Faceted Roles of DNAJB Protein in Cancer Metastasis and Clinical Implications
Abstract
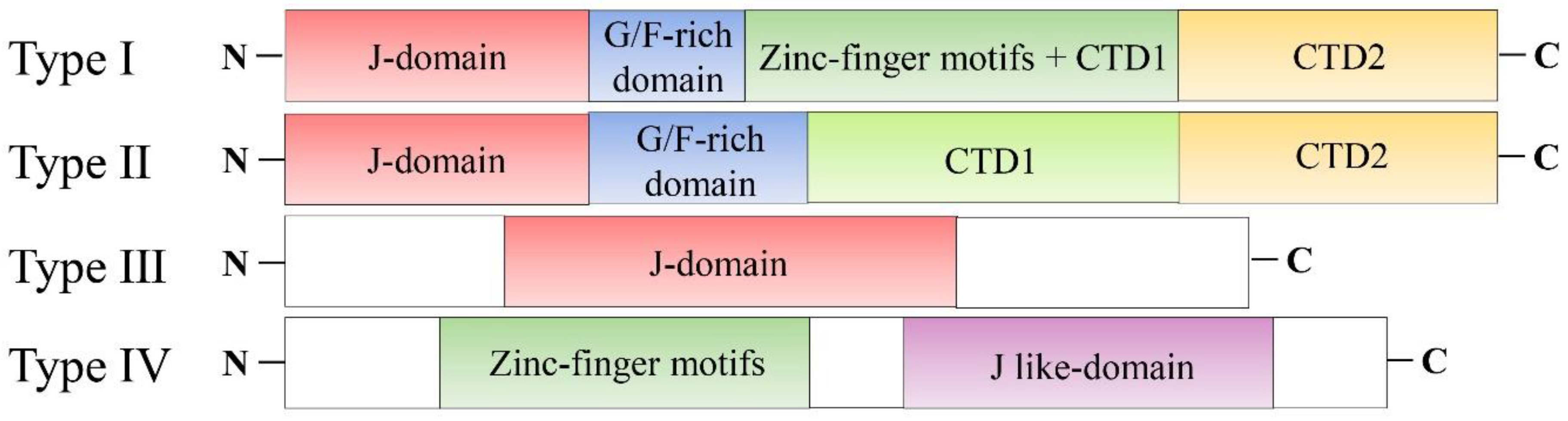
1. Introduction
2. Regulation of DNAJB Proteins at RNA Level
2.1. Transcriptional Regulation of DNAJB
2.2. Post-Transcriptional Regulation of DNAJB
3. Post-Translational Regulation of DNAJB Proteins
3.1. Phosphorylation of DNAJB Protein
3.2. Glycosylation of DNAJB Protein
3.3. Acetylation of DNAJB Protein
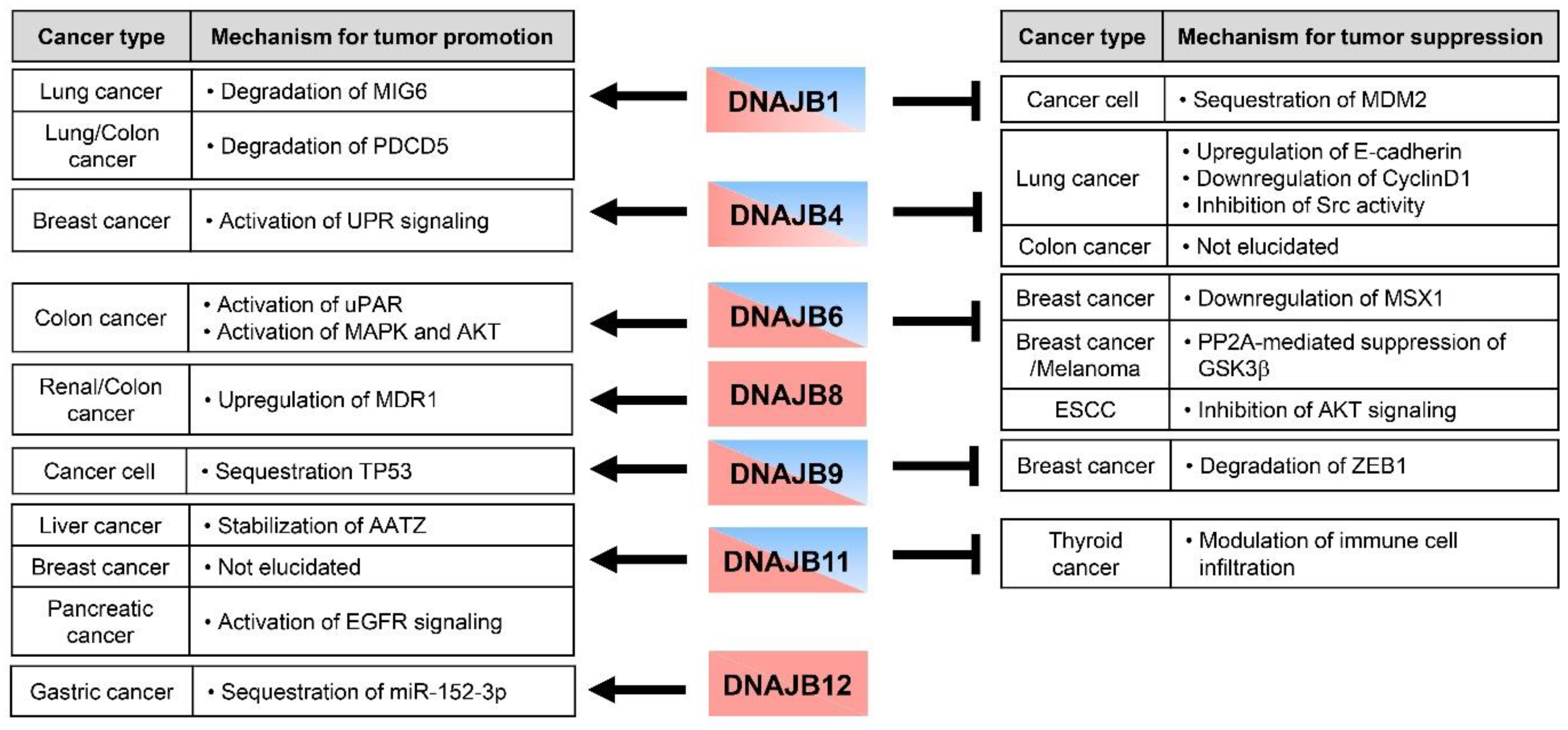
4. Multi-Faceted Roles of DNAJB Protein in Cancer Metastasis
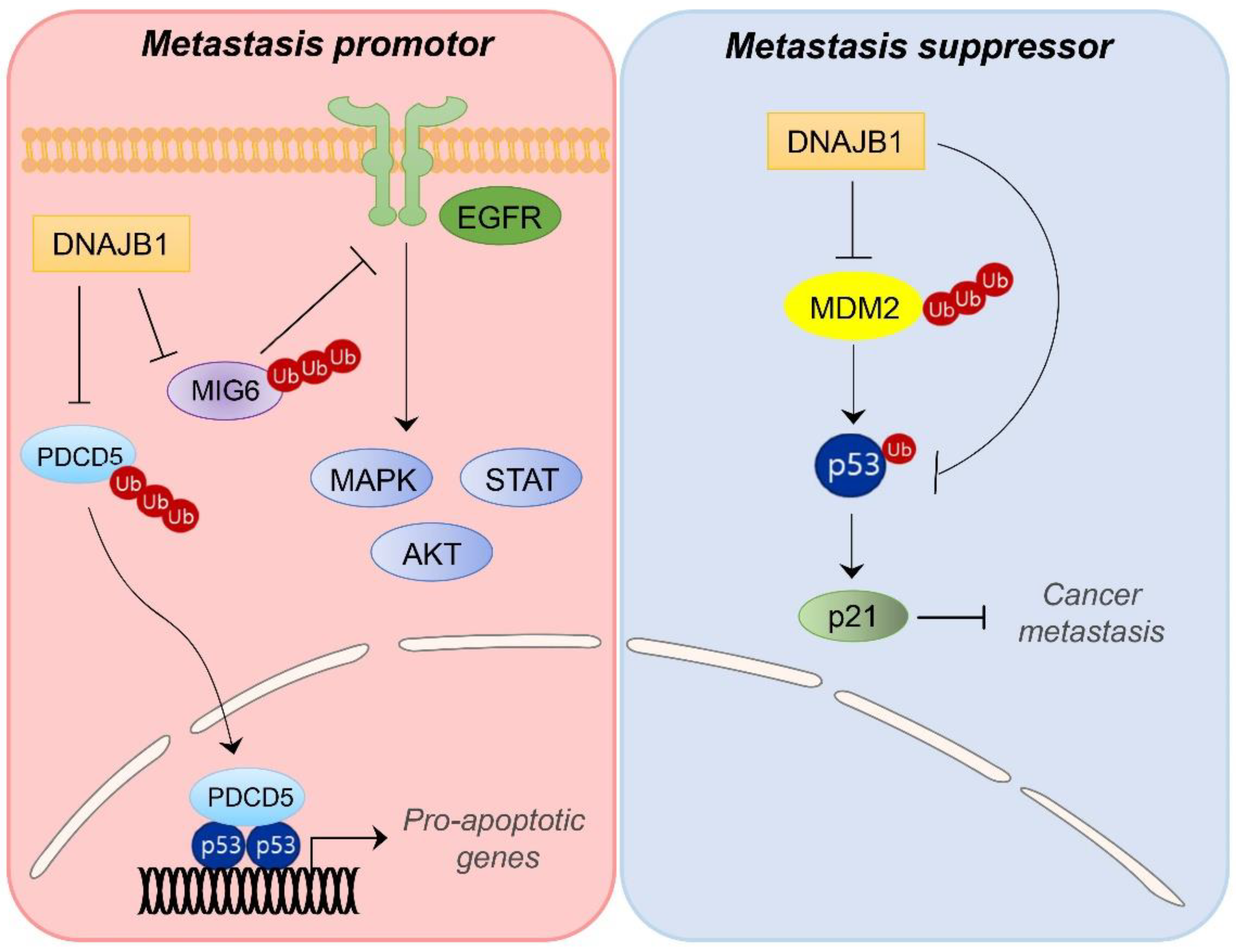
4.1. DNAJB1/HDJ1
4.2. DNAJB4/HLJ1
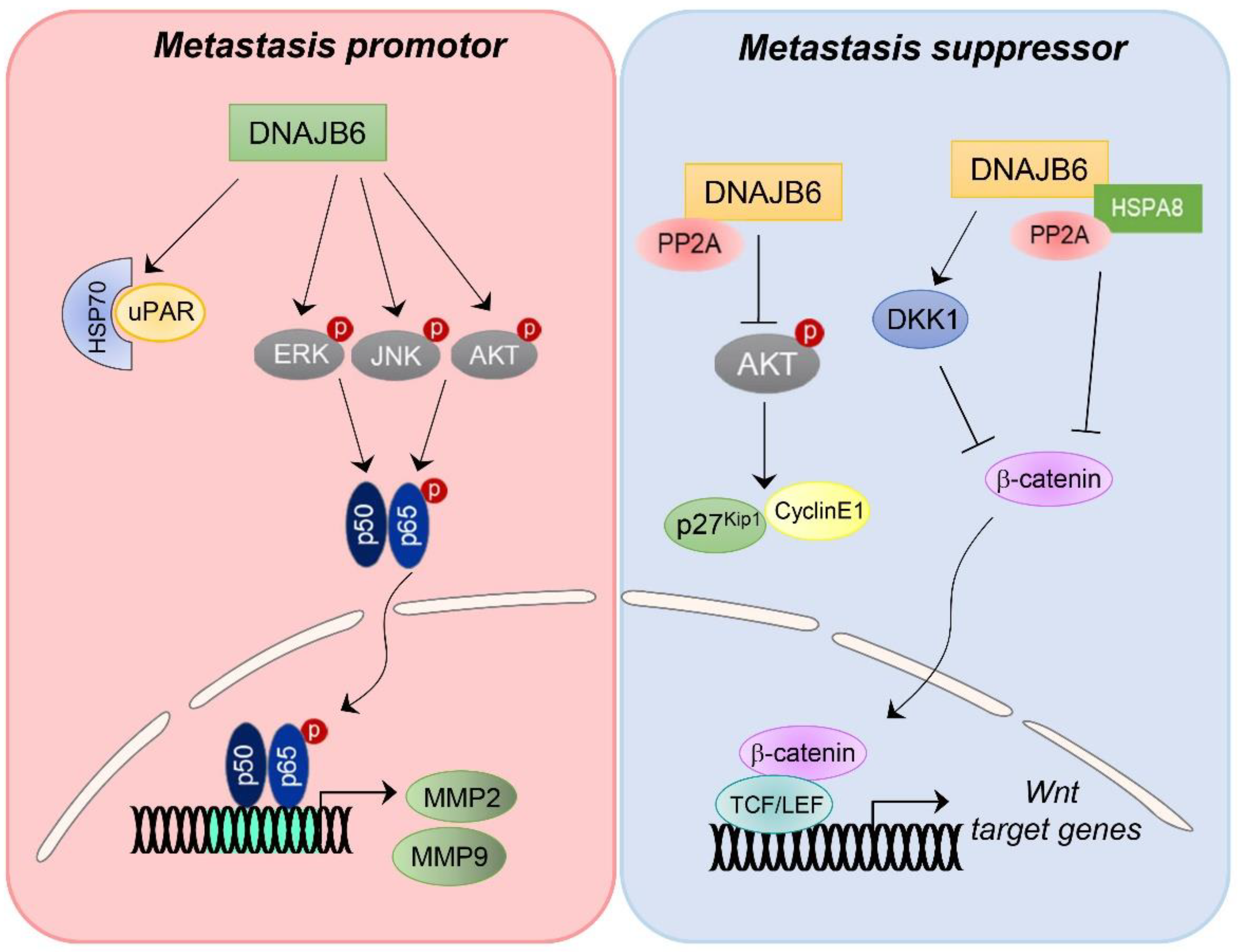
4.3. DNAJB6/MRJ
4.4. DNAJB8
4.5. DNAJB9/MDG1
4.6. DNAJB11/ERdj3
4.7. DNAJB12
5. Current Approaches and Challenges for Targeting DNAJB Protein in Cancer Therapy
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AATZ | Z mutant of alpha-1-antitrypsin |
| AML | Acute myeloid leukemia |
| CCA | Cholangiocarcinoma |
| CIC | Cancer-initiating cell |
| CRC | Colorectal cancer |
| CSC | Cancer stem cell |
| CTD | Client-binding domain |
| EGFR | Epidermal growth factor receptor |
| EMT | Epithelial–mesenchymal transition |
| ER | Endoplasmic reticulum |
| ESCC | Esophageal squamous cell carcinoma |
| FBXO45 | F-box/SPRY domain-containing protein 1 |
| HBV | Hepatitis B virus |
| HCC | Hepatocellular carcinoma |
| HSP | Heat shock protein |
| LGMDD1 | Limb-girdle muscular dystrophy type D1 |
| MIG6 | Mitogen-inducible gene 6 |
| miRNA | MicroRNA |
| MK5 | Mitogen-activated protein kinase-activated protein kinase 5 |
| NSCLC | Non-small-cell lung carcinoma |
| PROTAC | Proteolysis-targeting chimera |
| PTM | Post-translational modification |
| sEV | Small extracellular vesicle |
| SNHG5 | Small nucleolar RNA host gene 5 |
| TCGA | The Cancer Genome Atlas |
| UPR | Unfolded protein response |
References
- Rylander, M.N.; Feng, Y.; Bass, J.; Diller, K.R. Thermally induced injury and heat-shock protein expression in cells and tissues. Ann. N. Y. Acad. Sci. 2005, 1066, 222–242. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Ma, F.; Wei, J.; Li, H.; Ma, H.; Sun, P. Physiological Functions of Heat Shock Proteins. Curr. Protein Pept. Sci. 2020, 21, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Dubrez, L.; Causse, S.; Borges Bonan, N.; Dumetier, B.; Garrido, C. Heat-shock proteins: Chaperoning DNA repair. Oncogene 2020, 39, 516–529. [Google Scholar] [CrossRef]
- Saibil, H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef]
- Sonna, L.A.; Fujita, J.; Gaffin, S.L.; Lilly, C.M. Invited review: Effects of heat and cold stress on mammalian gene expression. J. Appl. Physiol. 2002, 92, 1725–1742. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Yang, J.; Qi, Z.; Wu, H.; Wang, B.; Zou, F.; Mei, H.; Liu, J.; Wang, W.; Liu, Q. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm 2022, 3, e161. [Google Scholar] [CrossRef] [PubMed]
- Bohen, S.P.; Kralli, A.; Yamamoto, K.R. Hold ‘em and fold ‘em: Chaperones and signal transduction. Science 1995, 268, 1303–1304. [Google Scholar] [CrossRef]
- Jee, H. Size dependent classification of heat shock proteins: A mini-review. J. Exerc. Rehabil. 2016, 12, 255–259. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- Schmitt, E.; Parcellier, A.; Gurbuxani, S.; Cande, C.; Hammann, A.; Morales, M.C.; Hunt, C.R.; Dix, D.J.; Kroemer, R.T.; Giordanetto, F.; et al. Chemosensitization by a non-apoptogenic heat shock protein 70-binding apoptosis-inducing factor mutant. Cancer Res. 2003, 63, 8233–8240. [Google Scholar]
- Burrows, F.; Zhang, H.; Kamal, A. Hsp90 activation and cell cycle regulation. Cell Cycle 2004, 3, 1530–1536. [Google Scholar] [CrossRef] [PubMed]
- Hoter, A.; El-Sabban, M.E.; Naim, H.Y. The HSP90 Family: Structure, Regulation, Function, and Implications in Health and Disease. Int. J. Mol. Sci. 2018, 19, 2560. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Faust, O.; Abayev-Avraham, M.; Wentink, A.S.; Maurer, M.; Nillegoda, N.B.; London, N.; Bukau, B.; Rosenzweig, R. HSP40 proteins use class-specific regulation to drive HSP70 functional diversity. Nature 2020, 587, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Jolly, C.; Morimoto, R.I. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J. Natl. Cancer Inst. 2000, 92, 1564–1572. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.Y.; Lee, S.; Ren, H.Y.; Cyr, D.M. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol. Biol. Cell 2004, 15, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Hageman, J.; Kampinga, H.H. Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones 2009, 14, 1–21. [Google Scholar] [CrossRef]
- Rajan, V.B.; D’Silva, P. Arabidopsis thaliana J-class heat shock proteins: Cellular stress sensors. Funct. Integr. Genom. 2009, 9, 433–446. [Google Scholar] [CrossRef]
- Velasco, L.; Dublang, L.; Moro, F.; Muga, A. The Complex Phosphorylation Patterns that Regulate the Activity of Hsp70 and Its Cochaperones. Int. J. Mol. Sci. 2019, 20, 4122. [Google Scholar] [CrossRef]
- Wang, C.C.; Tsai, M.F.; Hong, T.M.; Chang, G.C.; Chen, C.Y.; Yang, W.M.; Chen, J.J.; Yang, P.C. The transcriptional factor YY1 upregulates the novel invasion suppressor HLJ1 expression and inhibits cancer cell invasion. Oncogene 2005, 24, 4081–4093. [Google Scholar] [CrossRef]
- Zhang, L.; Cai, X.; Chen, K.; Wang, Z.; Wang, L.; Ren, M.; Huang, A.; Tang, H. Hepatitis B virus protein up-regulated HLJ1 expression via the transcription factor YY1 in human hepatocarcinoma cells. Virus Res. 2011, 157, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Lara-Pezzi, E.; Gómez-Gaviro, M.V.; Gálvez, B.G.; Mira, E.; Iñiguez, M.A.; Fresno, M.; Martínez, A.C.; Arroyo, A.G.; López-Cabrera, M. The hepatitis B virus X protein promotes tumor cell invasion by inducing membrane-type matrix metalloproteinase-1 and cyclooxygenase-2 expression. J. Clin. Investig. 2002, 110, 1831–1838. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.P.; Liang, H.F.; Chen, X.P.; Zhang, W.G.; Yang, S.L.; Xu, T.; Ren, L. The role of NF-kappaB in Hepatitis b virus X protein-mediated upregulation of VEGF and MMPs. Cancer Investig. 2010, 28, 443–451. [Google Scholar] [CrossRef]
- Moon, E.J.; Jeong, C.H.; Jeong, J.W.; Kim, K.R.; Yu, D.Y.; Murakami, S.; Kim, C.W.; Kim, K.W. Hepatitis B virus X protein induces angiogenesis by stabilizing hypoxia-inducible factor-1alpha. FASEB J. 2004, 18, 382–384. [Google Scholar] [CrossRef] [PubMed]
- Szremska, A.P.; Kenner, L.; Weisz, E.; Ott, R.G.; Passegue, E.; Artwohl, M.; Freissmuth, M.; Stoxreiter, R.; Theussl, H.C.; Parzer, S.B.; et al. JunB inhibits proliferation and transformation in B-lymphoid cells. Blood 2003, 102, 4159–4165. [Google Scholar] [CrossRef] [PubMed]
- Ott, R.G.; Simma, O.; Kollmann, K.; Weisz, E.; Zebedin, E.M.; Schorpp-Kistner, M.; Heller, G.; Zöchbauer, S.; Wagner, E.F.; Freissmuth, M.; et al. JunB is a gatekeeper for B-lymphoid leukemia. Oncogene 2007, 26, 4863–4871. [Google Scholar] [CrossRef][Green Version]
- Eferl, R.; Wagner, E.F. AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer 2003, 3, 859–868. [Google Scholar] [CrossRef]
- Wang, C.C.; Tsai, M.F.; Dai, T.H.; Hong, T.M.; Chan, W.K.; Chen, J.J.; Yang, P.C. Synergistic activation of the tumor suppressor, HLJ1, by the transcription factors YY1 and activator protein 1. Cancer Res. 2007, 67, 4816–4826. [Google Scholar] [CrossRef]
- Chen, H.W.; Lee, J.Y.; Huang, J.Y.; Wang, C.C.; Chen, W.J.; Su, S.F.; Huang, C.W.; Ho, C.C.; Chen, J.J.; Tsai, M.F.; et al. Curcumin inhibits lung cancer cell invasion and metastasis through the tumor suppressor HLJ1. Cancer Res. 2008, 68, 7428–7438. [Google Scholar] [CrossRef]
- Khan, S.Y.; Vasanth, S.; Kabir, F.; Gottsch, J.D.; Khan, A.O.; Chaerkady, R.; Lee, M.C.; Leitch, C.C.; Ma, Z.; Laux, J.; et al. FOXE3 contributes to Peters anomaly through transcriptional regulation of an autophagy-associated protein termed DNAJB1. Nat. Commun. 2016, 7, 10953. [Google Scholar] [CrossRef]
- Nishizawa, S.; Hirohashi, Y.; Torigoe, T.; Takahashi, A.; Tamura, Y.; Mori, T.; Kanaseki, T.; Kamiguchi, K.; Asanuma, H.; Morita, R.; et al. HSP DNAJB8 controls tumor-initiating ability in renal cancer stem-like cells. Cancer Res. 2012, 72, 2844–2854. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Kong, S.; Hu, X.; Li, X.; Xu, B.; Yue, Q.; Fu, K.; Ye, L.; Bai, S. Dnajb8, a target gene of SOX30, is dispensable for male fertility in mice. PeerJ 2020, 8, e10582. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Rostas, J.W.; Dyess, D.L.; Shevde, L.A.; Samant, R.S. Micro-RNA-632 downregulates DNAJB6 in breast cancer. Lab. Investig. 2012, 92, 1310–1317. [Google Scholar] [CrossRef] [PubMed]
- Costa Mdo, C.; Paulson, H.L. Toward understanding Machado-Joseph disease. Prog. Neurobiol. 2012, 97, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Evert, B.O.; Nalavade, R.; Jungverdorben, J.; Matthes, F.; Weber, S.; Rajput, A.; Bonn, S.; Brüstle, O.; Peitz, M.; Krauß, S. Upregulation of miR-370 and miR-543 is associated with reduced expression of heat shock protein 40 in spinocerebellar ataxia type 3. PLoS ONE 2018, 13, e0201794. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Wang, D.; Zeng, T.; Lin, Z.; Yan, L.; Wang, F.; Tang, L.; Wang, L.; Tang, D.; Chen, P.; Yang, M. Long non-coding RNA SNHG5 regulates chemotherapy resistance through the miR-32/DNAJB9 axis in acute myeloid leukemia. Biomed. Pharmacother. 2020, 123, 109802. [Google Scholar] [CrossRef]
- Ma, P.; Li, L.; Liu, F.; Zhao, Q. HNF1A-Induced lncRNA HCG18 Facilitates Gastric Cancer Progression by Upregulating DNAJB12 via miR-152-3p. Onco. Targets Ther. 2020, 13, 7641–7652. [Google Scholar] [CrossRef]
- Xu, H.; Wang, Y.; Lin, S.; Deng, W.; Peng, D.; Cui, Q.; Xue, Y. PTMD: A Database of Human Disease-associated Post-translational Modifications. Genom. Proteom. Bioinform. 2018, 16, 244–251. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, Y.; Xu, X. A novel method for predicting post-translational modifications on serine and threonine sites by using site-modification network profiles. Mol. Biosyst. 2015, 11, 3092–3100. [Google Scholar] [CrossRef]
- Strumillo, M.; Beltrao, P. Towards the computational design of protein post-translational regulation. Bioorg. Med. Chem. 2015, 23, 2877–2882. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.J.; Dammer, E.B.; Wang, G.; Seyfried, N.T.; Levey, A.I. Proteomics of protein post-translational modifications implicated in neurodegeneration. Transl. Neurodegener. 2014, 3, 23. [Google Scholar] [CrossRef] [PubMed]
- Truman, A.W.; Kristjansdottir, K.; Wolfgeher, D.; Hasin, N.; Polier, S.; Zhang, H.; Perrett, S.; Prodromou, C.; Jones, G.W.; Kron, S.J. CDK-dependent Hsp70 Phosphorylation controls G1 cyclin abundance and cell-cycle progression. Cell 2012, 151, 1308–1318. [Google Scholar] [CrossRef]
- Woodford, M.R.; Truman, A.W.; Dunn, D.M.; Jensen, S.M.; Cotran, R.; Bullard, R.; Abouelleil, M.; Beebe, K.; Wolfgeher, D.; Wierzbicki, S.; et al. Mps1 Mediated Phosphorylation of Hsp90 Confers Renal Cell Carcinoma Sensitivity and Selectivity to Hsp90 Inhibitors. Cell Rep. 2016, 14, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Poon, I.K.; Jans, D.A. Regulation of nuclear transport: Central role in development and transformation? Traffic 2005, 6, 173–186. [Google Scholar] [CrossRef]
- Götz, C.; Müller, A.; Montenarh, M.; Zimmermann, R.; Dudek, J. The ER-membrane-resident Hsp40 ERj1 is a novel substrate for protein kinase CK2. Biochem. Biophys. Res. Commun. 2009, 388, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Kostenko, S.; Jensen, K.L.; Moens, U. Phosphorylation of heat shock protein 40 (Hsp40/DnaJB1) by mitogen-activated protein kinase-activated protein kinase 5 (MK5/PRAK). Int. J. Biochem. Cell Biol. 2014, 47, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Dudek, J.; Greiner, M.; Muller, A.; Hendershot, L.M.; Kopsch, K.; Nastainczyk, W.; Zimmermann, R. ERj1p has a basic role in protein biogenesis at the endoplasmic reticulum. Nat. Struct. Mol. Biol. 2005, 12, 1008–1014. [Google Scholar] [CrossRef]
- Zupicich, J.; Brenner, S.E.; Skarnes, W.C. Computational prediction of membrane-tethered transcription factors. Genome Biol. 2001, 2, RESEARCH0050. [Google Scholar] [CrossRef] [PubMed]
- Stowell, S.R.; Ju, T.; Cummings, R.D. Protein glycosylation in cancer. Annu. Rev. Pathol. 2015, 10, 473–510. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Yang, C.; Chen, S.; Huang, Y.; Liu, C.; Liu, J.; Yin, W. Comparison of the glycopattern alterations of mitochondrial proteins in cerebral cortex between rat Alzheimer’s disease and the cerebral ischemia model. Sci. Rep. 2017, 7, 39948. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, K.; Takamatsu, S.; Minowa, M.T.; Yoshida, A.; Takeuchi, M.; Marth, J.D. Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell 2005, 123, 1307–1321. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Haslam, R.H.; Haslam, D.B. HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J. Biol. Chem. 2000, 275, 24984–24992. [Google Scholar] [CrossRef] [PubMed]
- Kopito, R.R. ER quality control: The cytoplasmic connection. Cell 1997, 88, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Soto, C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 2003, 4, 49–60. [Google Scholar] [CrossRef]
- Mukherjee, A.; Morales-Scheihing, D.; Butler, P.C.; Soto, C. Type 2 diabetes as a protein misfolding disease. Trends Mol. Med. 2015, 21, 439–449. [Google Scholar] [CrossRef]
- Peng, C.; Trojanowski, J.Q.; Lee, V.M. Protein transmission in neurodegenerative disease. Nat. Rev. Neurol. 2020, 16, 199–212. [Google Scholar] [CrossRef]
- Zoghbi, H.Y.; Orr, H.T. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 2000, 23, 217–247. [Google Scholar] [CrossRef]
- Roth, D.M.; Balch, W.E. Modeling general proteostasis: Proteome balance in health and disease. Curr. Opin. Cell Biol. 2011, 23, 126–134. [Google Scholar] [CrossRef]
- Hageman, J.; Rujano, M.A.; van Waarde, M.A.; Kakkar, V.; Dirks, R.P.; Govorukhina, N.; Oosterveld-Hut, H.M.; Lubsen, N.H.; Kampinga, H.H. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol. Cell 2010, 37, 355–369. [Google Scholar] [CrossRef]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target Ther. 2020, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, T.N.; Huysentruyt, L.C. On the origin of cancer metastasis. Crit. Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.W.; Kim, H.J.; Lim, J.H.; Lee, S.H. Heat Shock Proteins: Agents of Cancer Development and Therapeutic Targets in Anti-Cancer Therapy. Cells 2019, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Uchiumi, A.; Katagata, Y. Hsp40 regulates the amount of keratin proteins via ubiquitin-proteasome pathway in cultured human cells. Int. J. Mol. Med. 2012, 29, 165–168. [Google Scholar]
- Lenna, S.; Farina, A.G.; Martyanov, V.; Christmann, R.B.; Wood, T.A.; Farber, H.W.; Scorza, R.; Whitfield, M.L.; Lafyatis, R.; Trojanowska, M. Increased expression of endoplasmic reticulum stress and unfolded protein response genes in peripheral blood mononuclear cells from patients with limited cutaneous systemic sclerosis and pulmonary arterial hypertension. Arthritis Rheum. 2013, 65, 1357–1366. [Google Scholar] [CrossRef]
- Batra, J.; Tripathi, S.; Kumar, A.; Katz, J.M.; Cox, N.J.; Lal, R.B.; Sambhara, S.; Lal, S.K. Human Heat shock protein 40 (Hsp40/DnaJB1) promotes influenza A virus replication by assisting nuclear import of viral ribonucleoproteins. Sci. Rep. 2016, 6, 19063. [Google Scholar] [CrossRef]
- Tracz-Gaszewska, Z.; Klimczak, M.; Biecek, P.; Herok, M.; Kosinski, M.; Olszewski, M.B.; Czerwinska, P.; Wiech, M.; Wiznerowicz, M.; Zylicz, A.; et al. Molecular chaperones in the acquisition of cancer cell chemoresistance with mutated TP53 and MDM2 up-regulation. Oncotarget 2017, 8, 82123–82143. [Google Scholar] [CrossRef]
- Park, S.Y.; Choi, H.K.; Seo, J.S.; Yoo, J.Y.; Jeong, J.W.; Choi, Y.; Choi, K.C.; Yoon, H.G. DNAJB1 negatively regulates MIG6 to promote epidermal growth factor receptor signaling. Biochim. Biophys. Acta 2015, 1853, 2722–2730. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lalazar, G.; Houlihan, S.L.; Tschaharganeh, D.F.; Baslan, T.; Chen, C.C.; Requena, D.; Tian, S.; Bosbach, B.; Wilkinson, J.E.; et al. DNAJB1-PRKACA fusion kinase interacts with β-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 2017, 114, 13076–13084. [Google Scholar] [CrossRef]
- Cui, X.; Choi, H.K.; Choi, Y.S.; Park, S.Y.; Sung, G.J.; Lee, Y.H.; Lee, J.; Jun, W.J.; Kim, K.; Choi, K.C.; et al. DNAJB1 destabilizes PDCD5 to suppress p53-mediated apoptosis. Cancer Lett. 2015, 357, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Luo, M.; Chen, J.; Zhou, Y.; Li, X.; Zhan, Y.; Shen, D.; Chen, B. Identification of TPD52 and DNAJB1 as two novel bile biomarkers for cholangiocarcinoma by iTRAQbased quantitative proteomics analysis. Oncol. Rep. 2019, 42, 2622–2634. [Google Scholar] [PubMed]
- Qi, M.; Zhang, J.; Zeng, W.; Chen, X. DNAJB1 stabilizes MDM2 and contributes to cancer cell proliferation in a p53-dependent manner. Biochim. Biophys. Acta 2014, 1839, 62–69. [Google Scholar] [CrossRef]
- Inoue, M.; Noguchi, S.; Inoue, Y.U.; Iida, A.; Ogawa, M.; Bengoechea, R.; Pittman, S.K.; Hayashi, S.; Watanabe, K.; Hosoi, Y.; et al. Distinctive chaperonopathy in skeletal muscle associated with the dominant variant in DNAJB4. bioRxiv 2022, bioRxiv:2022.07.26.501446. [Google Scholar]
- Lei, J.X.; Cassone, C.G.; Luebbert, C.; Liu, Q.Y. A novel neuron-enriched protein SDIM1 is down regulated in Alzheimer’s brains and attenuates cell death induced by DNAJB4 over-expression in neuro-progenitor cells. Mol. Neurodegener. 2011, 6, 9. [Google Scholar] [CrossRef]
- Tsai, M.F.; Wang, C.C.; Chang, G.C.; Chen, C.Y.; Chen, H.Y.; Cheng, C.L.; Yang, Y.P.; Wu, C.Y.; Shih, F.Y.; Liu, C.C.; et al. A new tumor suppressor DnaJ-like heat shock protein, HLJ1, and survival of patients with non-small-cell lung carcinoma. J. Natl. Cancer Inst. 2006, 98, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Chang, W.H.; Su, K.Y.; Ku, W.H.; Chang, G.C.; Hong, Q.S.; Hsiao, Y.J.; Chen, H.C.; Chen, H.Y.; Wu, R.; et al. HLJ1 is an endogenous Src inhibitor suppressing cancer progression through dual mechanisms. Oncogene 2016, 35, 5674–5685. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, J.; Zhang, C.; Fu, W.; Xiao, X.; Ruan, S.; Zhang, Y.; Luo, X.; Tang, M. HLJ1 is a novel biomarker for colorectal carcinoma progression and overall patient survival. Int. J. Clin. Exp. Pathol. 2014, 7, 969–977. [Google Scholar]
- Uretmen Kagiali, Z.C.; Sanal, E.; Karayel, Ö.; Polat, A.N.; Saatci, Ö.; Ersan, P.G.; Trappe, K.; Renard, B.Y.; Önder, T.T.; Tuncbag, N.; et al. Systems-level Analysis Reveals Multiple Modulators of Epithelial-mesenchymal Transition and Identifies DNAJB4 and CD81 as Novel Metastasis Inducers in Breast Cancer. Mol. Cell Proteom. 2019, 18, 1756–1771. [Google Scholar] [CrossRef]
- Feng, Y.X.; Sokol, E.S.; Del Vecchio, C.A.; Sanduja, S.; Claessen, J.H.; Proia, T.A.; Jin, D.X.; Reinhardt, F.; Ploegh, H.L.; Wang, Q.; et al. Epithelial-to-mesenchymal transition activates PERK-eIF2α and sensitizes cells to endoplasmic reticulum stress. Cancer Discov. 2014, 4, 702–715. [Google Scholar] [CrossRef]
- Sarparanta, J.; Jonson, P.H.; Golzio, C.; Sandell, S.; Luque, H.; Screen, M.; McDonald, K.; Stajich, J.M.; Mahjneh, I.; Vihola, A.; et al. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat. Genet. 2012, 44, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Winter, L.; Goldmann, W.H. Biomechanical characterization of myofibrillar myopathies. Cell Biol. Int. 2015, 39, 361–363. [Google Scholar] [CrossRef] [PubMed]
- Harms, M.B.; Sommerville, R.B.; Allred, P.; Bell, S.; Ma, D.; Cooper, P.; Lopate, G.; Pestronk, A.; Weihl, C.C.; Baloh, R.H. Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann. Neurol. 2012, 71, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Belshan, M.; Ratner, L. Hsp40 facilitates nuclear import of the human immunodeficiency virus type 2 Vpx-mediated preintegration complex. J. Virol. 2008, 82, 1229–1237. [Google Scholar] [CrossRef]
- Mitra, A.; Menezes, M.E.; Shevde, L.A.; Samant, R.S. DNAJB6 induces degradation of beta-catenin and causes partial reversal of mesenchymal phenotype. J. Biol. Chem. 2010, 285, 24686–24694. [Google Scholar] [CrossRef]
- Mitra, A.; Fillmore, R.A.; Metge, B.J.; Rajesh, M.; Xi, Y.; King, J.; Ju, J.; Pannell, L.; Shevde, L.A.; Samant, R.S. Large isoform of MRJ (DNAJB6) reduces malignant activity of breast cancer. Breast Cancer Res. 2008, 10, R22. [Google Scholar] [CrossRef]
- Mitra, A.; Menezes, M.E.; Pannell, L.K.; Mulekar, M.S.; Honkanen, R.E.; Shevde, L.A.; Samant, R.S. DNAJB6 chaperones PP2A mediated dephosphorylation of GSK3β to downregulate β-catenin transcription target, osteopontin. Oncogene 2012, 31, 4472–4483. [Google Scholar] [CrossRef]
- Yu, V.Z.; Wong, V.C.; Dai, W.; Ko, J.M.; Lam, A.K.; Chan, K.W.; Samant, R.S.; Lung, H.L.; Shuen, W.H.; Law, S.; et al. Nuclear Localization of DNAJB6 Is Associated with Survival of Patients with Esophageal Cancer and Reduces AKT Signaling and Proliferation of Cancer Cells. Gastroenterology 2015, 149, 1825–1836.e5. [Google Scholar] [CrossRef]
- Lin, Y.; Peng, N.; Zhuang, H.; Zhang, D.; Wang, Y.; Hua, Z.C. Heat shock proteins HSP70 and MRJ cooperatively regulate cell adhesion and migration through urokinase receptor. BMC Cancer 2014, 14, 639. [Google Scholar] [CrossRef]
- Morita, R.; Nishizawa, S.; Torigoe, T.; Takahashi, A.; Tamura, Y.; Tsukahara, T.; Kanaseki, T.; Sokolovskaya, A.; Kochin, V.; Kondo, T.; et al. Heat shock protein DNAJB8 is a novel target for immunotherapy of colon cancer-initiating cells. Cancer Sci. 2014, 105, 389–395. [Google Scholar] [CrossRef]
- Kusumoto, H.; Hirohashi, Y.; Nishizawa, S.; Yamashita, M.; Yasuda, K.; Murai, A.; Takaya, A.; Mori, T.; Kubo, T.; Nakatsugawa, M.; et al. Cellular stress induces cancer stem-like cells through expression of DNAJB8 by activation of heat shock factor 1. Cancer Sci. 2018, 109, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Y.; Mao, R.; Zhang, Y.; Wen, J.; Liu, Q.; Liu, Y.; Zhang, T. DNAJB8 in small extracellular vesicles promotes Oxaliplatin resistance through TP53/MDR1 pathway in colon cancer. Cell Death Dis. 2022, 13, 151. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Meunier, L.; Hendershot, L.M. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 2002, 277, 15947–15956. [Google Scholar] [CrossRef] [PubMed]
- Berger, B.J.; Müller, T.S.; Buschmann, I.R.; Peters, K.; Kirsch, M.; Christ, B.; Pröls, F. High levels of the molecular chaperone Mdg1/ERdj4 reflect the activation state of endothelial cells. Exp. Cell Res. 2003, 290, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Kurisu, J.; Honma, A.; Miyajima, H.; Kondo, S.; Okumura, M.; Imaizumi, K. MDG1/ERdj4, an ER-resident DnaJ family member, suppresses cell death induced by ER stress. Genes Cells 2003, 8, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Pröls, F.; Mayer, M.P.; Renner, O.; Czarnecki, P.G.; Ast, M.; Gässler, C.; Wilting, J.; Kurz, H.; Christ, B. Upregulation of the cochaperone Mdg1 in endothelial cells is induced by stress and during in vitro angiogenesis. Exp. Cell Res. 2001, 269, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Kim, J.M.; Kim, K.H.; Heo, J.I.; Kwak, S.J.; Han, J.A. Genotoxic stress/p53-induced DNAJB9 inhibits the pro-apoptotic function of p53. Cell Death Differ. 2015, 22, 86–95. [Google Scholar] [CrossRef]
- Lee, H.J.; Jung, Y.J.; Lee, S.; Kim, J.I.; Han, J.A. DNAJB9 Inhibits p53-Dependent Oncogene-Induced Senescence and Induces Cell Transformation. Mol. Cells 2020, 43, 397–407. [Google Scholar]
- Huang, Y.; Arora, K.; Mun, K.S.; Yang, F.; Moon, C.; Yarlagadda, S.; Jegga, A.; Weaver, T.; Naren, A.P. Targeting DNAJB9, a novel ER luminal co-chaperone, to rescue ΔF508-CFTR. Sci. Rep. 2019, 9, 9808. [Google Scholar] [CrossRef]
- Kim, H.Y.; Kim, Y.M.; Hong, S. DNAJB9 suppresses the metastasis of triple-negative breast cancer by promoting FBXO45-mediated degradation of ZEB1. Cell Death Dis. 2021, 12, 461. [Google Scholar] [CrossRef]
- Genereux, J.C.; Qu, S.; Zhou, M.; Ryno, L.M.; Wang, S.; Shoulders, M.D.; Kaufman, R.J.; Lasmézas, C.I.; Kelly, J.W.; Wiseman, R.L. Unfolded protein response-induced ERdj3 secretion links ER stress to extracellular proteostasis. EMBO J. 2015, 34, 4–19. [Google Scholar] [CrossRef] [PubMed]
- Khodayari, N.; Marek, G.; Lu, Y.; Krotova, K.; Wang, R.L.; Brantly, M. Erdj3 Has an Essential Role for Z Variant Alpha-1-Antitrypsin Degradation. J. Cell Biochem. 2017, 118, 3090–3101. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Cao, D.; Gong, J. The endoplasmic reticulum co-chaperone ERdj3/DNAJB11 promotes hepatocellular carcinoma progression through suppressing AATZ degradation. Future Oncol. 2018, 14, 3001–3013. [Google Scholar] [CrossRef] [PubMed]
- Tolga, A. DNAJB11 (ERDJ3) Expression is a Novel Risk Factor for Breast Cancer Survival. J. Int. Transl. Med. 2020, 8, 13–18. [Google Scholar]
- Liu, P.; Zu, F.; Chen, H.; Yin, X.; Tan, X. Exosomal DNAJB11 promotes the development of pancreatic cancer by modulating the EGFR/MAPK pathway. Cell Mol. Biol. Lett. 2022, 27, 87. [Google Scholar] [CrossRef]
- Sun, R.; Yang, L.; Wang, Y.; Zhang, Y.; Ke, J.; Zhao, D. DNAJB11 predicts a poor prognosis and is associated with immune infiltration in thyroid carcinoma: A bioinformatics analysis. J. Int. Med. Res. 2021, 49, 3000605211053722. [Google Scholar] [CrossRef]
- Sopha, P.; Ren, H.Y.; Grove, D.E.; Cyr, D.M. Endoplasmic reticulum stress-induced degradation of DNAJB12 stimulates BOK accumulation and primes cancer cells for apoptosis. J. Biol. Chem. 2017, 292, 11792–11803. [Google Scholar] [CrossRef]
- Yokota, S.; Kitahara, M.; Nagata, K. Benzylidene lactam compound, KNK437, a novel inhibitor of acquisition of thermotolerance and heat shock protein induction in human colon carcinoma cells. Cancer Res. 2000, 60, 2942–2948. [Google Scholar]
- Wu, J.; Liu, T.; Rios, Z.; Mei, Q.; Lin, X.; Cao, S. Heat Shock Proteins and Cancer. Trends Pharmacol. Sci. 2017, 38, 226–256. [Google Scholar] [CrossRef]
- Yang, S.; Ren, X.; Liang, Y.; Yan, Y.; Zhou, Y.; Hu, J.; Wang, Z.; Song, F.; Wang, F.; Liao, W.; et al. KNK437 restricts the growth and metastasis of colorectal cancer via targeting DNAJA1/CDC45 axis. Oncogene 2020, 39, 249–261. [Google Scholar] [CrossRef]
- de La Motte Rouge, T.; Galluzzi, L.; Olaussen, K.A.; Zermati, Y.; Tasdemir, E.; Robert, T.; Ripoche, H.; Lazar, V.; Dessen, P.; Harper, F.; et al. A novel epidermal growth factor receptor inhibitor promotes apoptosis in non-small cell lung cancer cells resistant to erlotinib. Cancer Res. 2007, 67, 6253–6262. [Google Scholar] [CrossRef] [PubMed]
- Cassel, J.A.; Ilyin, S.; McDonnell, M.E.; Reitz, A.B. Novel inhibitors of heat shock protein Hsp70-mediated luciferase refolding that bind to DnaJ. Bioorg. Med. Chem. 2012, 20, 3609–3614. [Google Scholar] [CrossRef] [PubMed]
- Izbicka, E.; Campos, D.; Carrizales, G.; Patnaik, A. Biomarkers of anticancer activity of R115777 (Tipifarnib, Zarnestra) in human breast cancer models in vitro. Anticancer Res. 2005, 25, 3215–3223. [Google Scholar] [PubMed]
- Wang, C.C.; Liao, Y.P.; Mischel, P.S.; Iwamoto, K.S.; Cacalano, N.A.; McBride, W.H. HDJ-2 as a target for radiosensitization of glioblastoma multiforme cells by the farnesyltransferase inhibitor R115777 and the role of the p53/p21 pathway. Cancer Res. 2006, 66, 6756–6762. [Google Scholar] [CrossRef]
- Yamashita, M.; Hirohashi, Y.; Torigoe, T.; Kusumoto, H.; Murai, A.; Imagawa, T.; Sato, N. Dnajb8, a Member of the Heat Shock Protein 40 Family Has a Role in the Tumor Initiation and Resistance to Docetaxel but Is Dispensable for Stress Response. PLoS ONE 2016, 11, e0146501. [Google Scholar] [CrossRef]
- Zhang, T.T.; Jiang, Y.Y.; Shang, L.; Shi, Z.Z.; Liang, J.W.; Wang, Z.; Zhang, Y.; Hao, J.J.; Jia, X.M.; Xu, X.; et al. Overexpression of DNAJB6 promotes colorectal cancer cell invasion through an IQGAP1/ERK-dependent signaling pathway. Mol. Carcinog. 2015, 54, 1205–1213. [Google Scholar] [CrossRef]
- Bengoechea, R.; Findlay, A.R.; Bhadra, A.K.; Shao, H.; Stein, K.C.; Pittman, S.K.; Daw, J.A.; Gestwicki, J.E.; True, H.L.; Weihl, C.C. Inhibition of DNAJ-HSP70 interaction improves strength in muscular dystrophy. J. Clin. Investig. 2020, 130, 4470–4485. [Google Scholar] [CrossRef]
- Chang, L.; Miyata, Y.; Ung, P.M.; Bertelsen, E.B.; McQuade, T.J.; Carlson, H.A.; Zuiderweg, E.R.; Gestwicki, J.E. Chemical screens against a reconstituted multiprotein complex: Myricetin blocks DnaJ regulation of DnaK through an allosteric mechanism. Chem. Biol. 2011, 18, 210–221. [Google Scholar] [CrossRef]
- Lai, Y.H.; Yu, S.L.; Chen, H.Y.; Wang, C.C.; Chen, H.W.; Chen, J.J. The HLJ1-targeting drug screening identified Chinese herb andrographolide that can suppress tumour growth and invasion in non-small-cell lung cancer. Carcinogenesis 2013, 34, 1069–1080. [Google Scholar] [CrossRef]
- Dale, B.; Cheng, M.; Park, K.S.; Kaniskan, H.; Xiong, Y.; Jin, J. Advancing targeted protein degradation for cancer therapy. Nat. Rev. Cancer 2021, 21, 638–654. [Google Scholar] [CrossRef]
- Li, K.; Crews, C.M. PROTACs: Past, present and future. Chem. Soc. Rev. 2022, 51, 5214–5236. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; He, M.; Wang, L.; He, Y.; Rao, Y. Chemistries of bifunctional PROTAC degraders. Chem. Soc. Rev. 2022, 51, 7066–7114. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Tu, G.; Hu, Y.; Jiang, Q.; Liu, J.; Lin, S.; Yu, Z.; Li, G.; Wu, X.; Tang, Y.; et al. Discovery of BP3 as an efficacious proteolysis targeting chimera (PROTAC) degrader of HSP90 for treating breast cancer. Eur. J. Med. Chem. 2022, 228, 114013. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-Y.; Hong, S. Multi-Faceted Roles of DNAJB Protein in Cancer Metastasis and Clinical Implications. Int. J. Mol. Sci. 2022, 23, 14970. https://doi.org/10.3390/ijms232314970
Kim H-Y, Hong S. Multi-Faceted Roles of DNAJB Protein in Cancer Metastasis and Clinical Implications. International Journal of Molecular Sciences. 2022; 23(23):14970. https://doi.org/10.3390/ijms232314970
Chicago/Turabian StyleKim, Hye-Youn, and Suntaek Hong. 2022. "Multi-Faceted Roles of DNAJB Protein in Cancer Metastasis and Clinical Implications" International Journal of Molecular Sciences 23, no. 23: 14970. https://doi.org/10.3390/ijms232314970
APA StyleKim, H.-Y., & Hong, S. (2022). Multi-Faceted Roles of DNAJB Protein in Cancer Metastasis and Clinical Implications. International Journal of Molecular Sciences, 23(23), 14970. https://doi.org/10.3390/ijms232314970






