Dual-Mode Gold Nanoparticle-Based Method for Early Detection of Acanthamoeba
Abstract
:1. Introduction
2. Results and Discussion
2.1. Gold Nanoparticles Synthesis and Loading
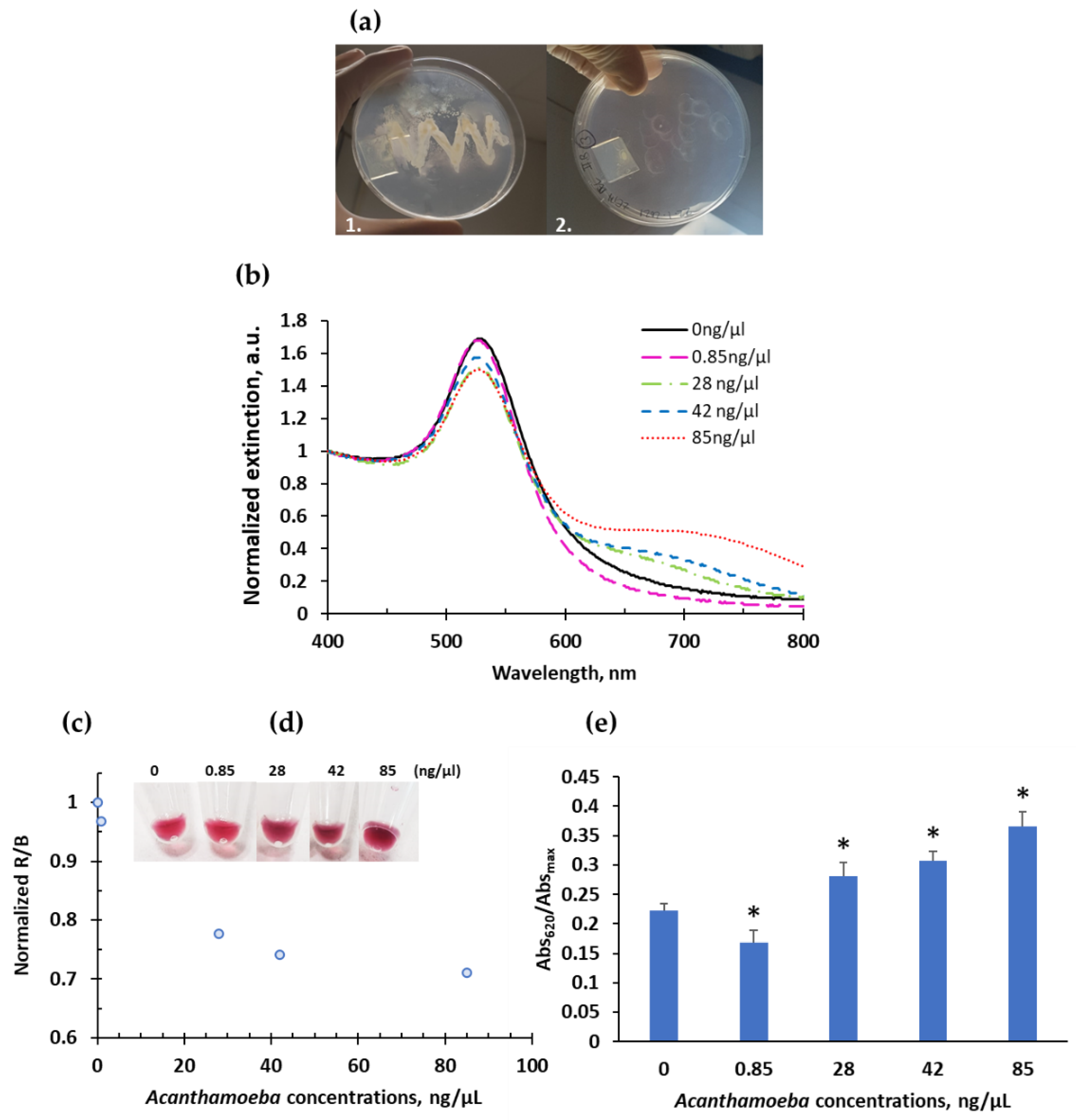
2.2. Colorimetric System and Optimizations
2.3. Real Samples Testing
3. Materials and Methods
3.1. Materials and Reagents
3.2. Selection of Specific Acanthamoeba-Sequences
3.3. Gold Nanoparticles Synthesis
3.4. AuNP Functionalized with DNA
3.4.1. pH Reduction Method
3.4.2. Salt Aging Method
3.5. Colorimetric Assay Procedure
Dual Colorimetric Strategy Based on the Naked Eye and Smartphone Digital Analysis
3.6. Microbial Culture Samples
3.7. Real Sample Analysis with the Colorimetric System
4. Conclusions
5. Patents
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Visvesvara, G.S. Infections with free-living amebae. Handb. Clin. Neurol. 2013, 114, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Visvesvara, G.S.; Moura, H.; Schuster, F.L. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol. Med. Microbiol. 2007, 50, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Lacerda, A.G.; Lira, M. Acanthamoeba keratitis: A review of biology, pathophysiology and epidemiology. Ophthalmic Physiol. Opt. 2021, 41, 116–135. [Google Scholar] [CrossRef] [PubMed]
- Szentmáry, N.; Daas, L.; Shi, L.; Laurik, K.L.; Lepper, S.; Milioti, G.; Seitz, B. Acanthamoeba keratitis—Clinical signs, differential diagnosis and treatment. J. Curr. Ophthalmol. 2018, 31, 16–23. [Google Scholar] [CrossRef]
- Jain, R.; Garg, P.; Motukupally, S.R.; Geary, M.B. Clinico-microbiological review of non-contact-lens-associated Acanthamoeba keratitis. Semin. Ophthalmol. 2015, 30, 281–288. [Google Scholar] [CrossRef]
- Brown, A.C.; Ross, J.; Jones, D.B.; Collier, S.A.; Ayers, T.L.; Hoekstra, R.M.; Backensen, B.; Roy, S.L.; Beach, M.J.; Yoder, J.S. Risk factors for Acanthamoeba keratitis—A multistate case–control study, 2008–2011. Eye Contact Lens 2018, 44, S173–S178. [Google Scholar] [CrossRef] [PubMed]
- Radford, C.F.; Bacon, A.S.; Dart, J.K.; Minassian, D.C. Risk factors for acanthamoeba keratitis in contact lens users: A case-control study. BMJ 1995, 310, 1567–1570. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, Y.W.; Boase, D.L.; Cree, I.A. Factors affecting the epidemiology of Acanthamoeba keratitis. Ophthalmic Epidemiol. 2007, 14, 53–60. [Google Scholar] [CrossRef]
- Seal, D.V. Acanthamoeba keratitis update-incidence, molecular epidemiology and new drugs for treatment. Eye 2003, 17, 893–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Putaporntip, C.; Kuamsab, N.; Nuprasert, W.; Rojrung, R.; Pattanawong, U.; Tia, T.; Yanmanee, S.; Jongwutiwes, S. Analysis of Acanthamoeba genotypes from public freshwater sources in Thailand reveals a new genotype, T23 Acanthamoeba bangkokensis sp. nov. Sci. Rep. 2021, 11, 17290. [Google Scholar] [CrossRef]
- Castro-Artavia, E.; Retana-Moreira, L.; Lorenzo-Morales, J.; Abrahams-Sandí, E. Potentially pathogenic Acanthamoeba genotype T4 isolated from dental units and emergency combination showers. Mem. Inst. Oswaldo Cruz 2017, 112, 817–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciver, S.K.; Asif, M.; Simmen, M.W.; Lorenzo-Morales, J. A systematic analysis of Acanthamoeba genotype frequency correlated with source and pathogenicity: T4 is confirmed as a pathogen-rich genotype. Eur. J. Protistol. 2013, 49, 217–221. [Google Scholar] [CrossRef]
- Yera, H.; Ok, V.; Lee Koy Kuet, F.; Dahane, N.; Ariey, F.; Hasseine, L.; Delaunay, P.; Martiano, D.; Marty, P.; Bourges, J.L. PCR and culture for diagnosis of Acanthamoeba keratitis. Br. J. Ophthalmol. 2020, 105, 1302–1306. [Google Scholar] [CrossRef] [PubMed]
- Füst, Á.; Tóth, J.; Simon, G.; Imre, L.; Nagy, Z.Z. Specificity of in vivo confocal cornea microscopy in Acanthamoeba keratitis. Eur. J. Ophthalmol. 2017, 27, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.P.; Kowalski, R.P.; Shanks, R.M.; Gordon, Y.J. Validation of real-time PCR for laboratory diagnosis of Acanthamoeba keratitis. J. Clin. Microbiol. 2008, 46, 3232–3236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, E.Y.; Joslin, C.E.; Sugar, J.; Booton, G.C.; Shoff, M.E.; Fuerst, P.A. The relative value of confocal microscopy and superficial corneal scrapings in the diagnosis of Acanthamoeba keratitis. Cornea 2008, 27, 764–772. [Google Scholar] [CrossRef] [Green Version]
- Daas, L.; Szentmáry, N.; Eppig, T.; Langenbucher, A.; Hasenfus, A.; Roth, M.; Saeger, M.; Nölle, B.; Lippmann, B.; Böhringer, D.; et al. The German Acanthamoeba keratitis register: Initial results of a multicenter study. Ophthalmologe 2015, 112, 752–763. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yan, L.; Wang, X.; Zhu, S.; Chen, C.; Gu, Z.; Zhao, Y. Progress, challenges, and future of nanomedicine. Nano Today 2020, 35, 101008. [Google Scholar] [CrossRef]
- Singh, P.; Pandit, S.; Mokkapati, V.; Garg, A.; Ravikumar, V.; Mijakovic, I. Gold Nanoparticles in Diagnostics and Therapeutics for Human Cancer. Int. J. Mol. Sci. 2018, 19, 1979. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.D.; Lai, J.-Y. Synthesis, bioactive properties, and biomedical applications of intrinsically therapeutic nanoparticles for disease treatment. Chem. Eng. J. 2022, 435, 134970. [Google Scholar] [CrossRef]
- Alex, S.; Tiwari, A. Functionalized Gold Nanoparticles: Synthesis, Properties and Applications--A Review. J. Nanosci. Nanotechnol. 2015, 15, 1869–1894. [Google Scholar] [CrossRef]
- Zanoli, L.M.; D’Agata, R.; Spoto, G. Functionalized gold nanoparticles for ultrasensitive DNA detection. Anal. Bioanal. Chem. 2012, 402, 1759–1771. [Google Scholar] [CrossRef]
- Mirkin, C.A.; Letsinger, R.L.; Mucic, R.C.; Storhoff, J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; Pang, D.W.; Zhang, Z.L.; Zheng, H.Z.; Cao, J.P.; Shen, J.T. Visual gene diagnosis of HBV and HCV based on nanoparticle probe amplification and silver staining enhancement. J. Med. Virol. 2003, 70, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Yeung, C.Y.; Liu, C.C.; Tseng, Y.T.; Tsai, K.C.; Hsieh, M.A.; Chan, W.T.; Liu, H.L.; Lee, H.C.; Hou, S.Y. Rapid identification of Salmonella using Hektoen enteric agar and 16s ribosomal DNA probe-gold nanoparticle immunochromatography assay in clinical faecal specimens. Lett. Appl. Microbiol. 2014, 58, 311–317. [Google Scholar] [CrossRef]
- Chen, S.-H.; Lin, K.-I.; Tang, C.-Y.; Peng, S.-L.; Chuang, Y.-C.; Lin, Y.-R.; Wang, J.-P.; Lin, C.-S. Optical detection of human papillomavirus type 16 and type 18 by sequence sandwich hybridization with oligonucleotide-functionalized Au nanoparticles. IEEE Trans. Nanobioscience 2009, 8, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano 2020, 14, 7617–7627. [Google Scholar] [CrossRef] [PubMed]
- Elumalai, M.; Ipatov, A.; Carvalho, J.; Guerreiro, J.; Prado, M. Dual colorimetric strategy for specific DNA detection by nicking endonuclease-assisted gold nanoparticle signal amplification. Anal. Bioanal. Chem. 2021, 414, 5239–5253. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Mungroo, M.R.; Anwar, A.; Sullivan, W.J., Jr.; Khan, N.A.; Siddiqui, R. Repositioning of Guanabenz in Conjugation with Gold and Silver Nanoparticles against Pathogenic Amoebae Acanthamoeba castellanii and Naegleria fowleri. ACS Infect. Dis. 2019, 5, 2039–2046. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Siddiqui, R.; Shah, M.R.; Khan, N.A. Gold Nanoparticle-Conjugated Cinnamic Acid Exhibits Antiacanthamoebic and Antibacterial Properties. Antimicrob. Agents Chemother. 2018, 62, e00630-18. [Google Scholar] [CrossRef] [PubMed]
- Khlebtsov, B.N.; Khlebtsov, N.G. On the measurement of gold nanoparticle sizes by the dynamic light scattering method. Colloid J. 2011, 73, 118–127. [Google Scholar] [CrossRef]
- Zhang, X.; Servos, M.R.; Liu, J. Instantaneous and quantitative functionalization of gold nanoparticles with thiolated DNA using a pH-assisted and surfactant-free route. J. Am. Chem. Soc. 2012, 134, 7266–7269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Liu, J. Methods for preparing DNA-functionalized gold nanoparticles, a key reagent of bioanalytical chemistry. Anal. Methods 2017, 9, 2633–2643. [Google Scholar] [CrossRef]
- Deka, J.; Mech, R.; Ianeselli, L.; Amenitsch, H.; Cacho-Nerin, F.; Parisse, P.; Casalis, L. Surface passivation improves the synthesis of highly stable and specific DNA-functionalized gold nanoparticles with variable DNA density. ACS Appl. Mater. Interfaces 2015, 7, 7033–7040. [Google Scholar] [CrossRef] [PubMed]
- Hurst, S.J.; Lytton-Jean, A.K.R.; Mirkin, C.A. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal. Chem. 2006, 78, 8313–8318. [Google Scholar] [CrossRef] [Green Version]
- Petryayeva, E.; Krull, U.J. Localized surface plasmon resonance: Nanostructures, bioassays and biosensing—A review. Anal. Chim. Acta 2011, 706, 8–24. [Google Scholar] [CrossRef]
- Ghosh, S.K.; Pal, T. Interparticle Coupling Effect on the Surface Plasmon Resonance of Gold Nanoparticles: From Theory to Applications. Chem. Rev. 2007, 107, 4797–4862. [Google Scholar] [CrossRef]
- Jain, P.K.; El-Sayed, M.A. Plasmonic coupling in noble metal nanostructures. Chem. Phys. Lett. 2010, 487, 153–164. [Google Scholar] [CrossRef]
- Sanromán-Iglesias, M.; Lawrie, C.H.; Schäfer, T.; Grzelczak, M.; Liz-Marzán, L.M. Sensitivity Limit of Nanoparticle Biosensors in the Discrimination of Single Nucleotide Polymorphism. ACS Sens. 2016, 1, 1110–1116. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Maestu, A.; Azinheiro, S.; Fuciños, P.; Carvalho, J.; Prado, M. Highly sensitive detection of gluten-containing cereals in food samples by real-time Loop-mediated isothermal AMPlification (qLAMP) and real-time polymerase chain reaction (qPCR). Food Chem. 2018, 246, 156–163. [Google Scholar] [CrossRef]
- Li, J.; Deng, T.; Chu, X.; Yang, R.; Jiang, J.; Shen, G.; Yu, R. Rolling circle amplification combined with gold nanoparticle aggregates for highly sensitive identification of single-nucleotide polymorphisms. Anal. Chem. 2010, 82, 2811–2816. [Google Scholar] [CrossRef] [PubMed]
- Srimongkol, G.; Ditmangklo, B.; Choopara, I.; Thaniyavarn, J.; Dean, D.; Kokpol, S.; Vilaivan, T.; Somboonna, N. Rapid colorimetric loop-mediated isothermal amplification for hypersensitive point-of-care Staphylococcus aureus enterotoxin A gene detection in milk and pork products. Sci. Rep. 2020, 10, 7768. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Shi, X.; Wu, G.; Wu, T.; Qin, R.; Wang, Y. Visual detection and differentiation of Classic Swine Fever Virus strains using nucleic acid sequence-based amplification (NASBA) and G-quadruplex DNAzyme assay. Sci. Rep. 2017, 7, 44211. [Google Scholar] [CrossRef] [Green Version]
- Liandris, E.; Gazouli, M.; Andreadou, M.; Comor, M.; Abazovic, N.; Sechi, L.A.; Ikonomopoulos, J. Direct detection of unamplified DNA from pathogenic mycobacteria using DNA-derivatized gold nanoparticles. J. Microbiol. Methods 2009, 78, 260–264. [Google Scholar] [CrossRef]
- Bakthavathsalam, P.; Rajendran, V.K.; Baquir Mohammed, J.A. A direct detection of Escherichia coli genomic DNA using gold nanoprobes. J. Nanobiotechnol. 2012, 10, 8. [Google Scholar] [CrossRef] [Green Version]
- Kalidasan, K.; Neo, J.L.; Uttamchandani, M. Direct visual detection of Salmonella genomic DNA using gold nanoparticles. Mol. BioSyst. 2013, 9, 618–621. [Google Scholar] [CrossRef]
- Kim, H.; Park, M.; Hwang, J.; Kim, J.H.; Chung, D.-R.; Lee, K.-s.; Kang, M. Development of label-free colorimetric assay for MERS-CoV using gold nanoparticles. ACS Sens. 2019, 4, 1306–1312. [Google Scholar] [CrossRef] [Green Version]
- Goh, J.W.Y.; Harrison, R.; Hau, S.; Alexander, C.L.; Tole, D.M.; Avadhanam, V.S. Comparison of In Vivo Confocal Microscopy, PCR and Culture of Corneal Scrapes in the Diagnosis of Acanthamoeba Keratitis. Cornea 2018, 37, 480–485. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Turkevich, J.; Stevenson, P.C.; Hillier, J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss. Faraday Soc. 1951, 11, 55–75. [Google Scholar] [CrossRef]
- Scarabelli, L.; Sánchez-Iglesias, A.; Pérez-Juste, J.; Liz-Marzán, L.M. A “Tips and Tricks” Practical Guide to the Synthesis of Gold Nanorods. J. Phys. Chem. Lett. 2015, 6, 4270–4279. [Google Scholar] [CrossRef] [Green Version]
- Storhoff, J.J.; Elghanian, R.; Mucic, R.C.; Mirkin, C.A.; Letsinger, R.L. One-Pot Colorimetric Differentiation of Polynucleotides with Single Base Imperfections Using Gold Nanoparticle Probes. J. Am. Chem. Soc. 1998, 120, 1959–1964. [Google Scholar] [CrossRef]
- Dairawan, M.; Shetty, P.J. The evolution of DNA extraction methods. Am. J. Biomed. Sci. Res. 2020, 8, 39–46. [Google Scholar]
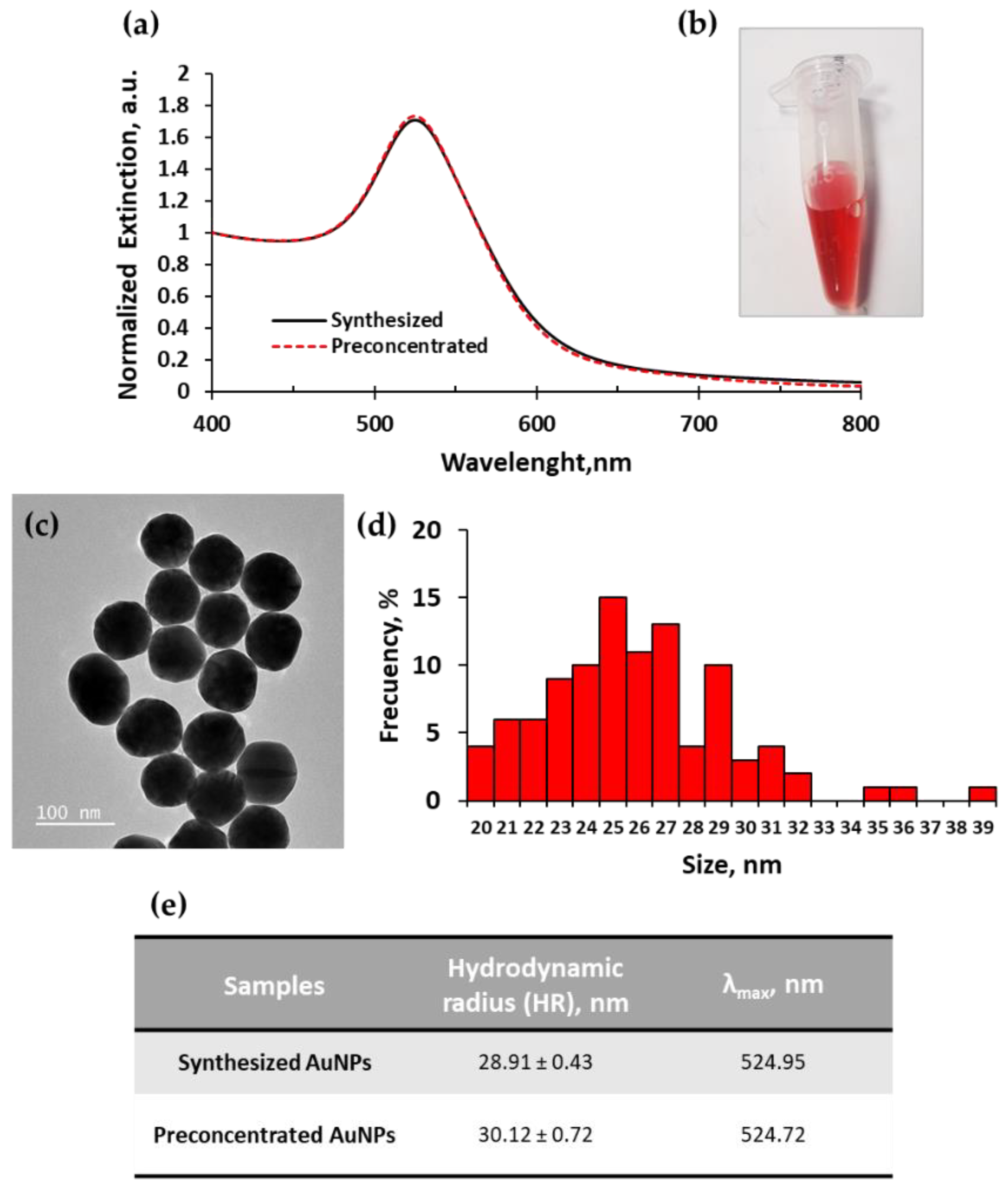
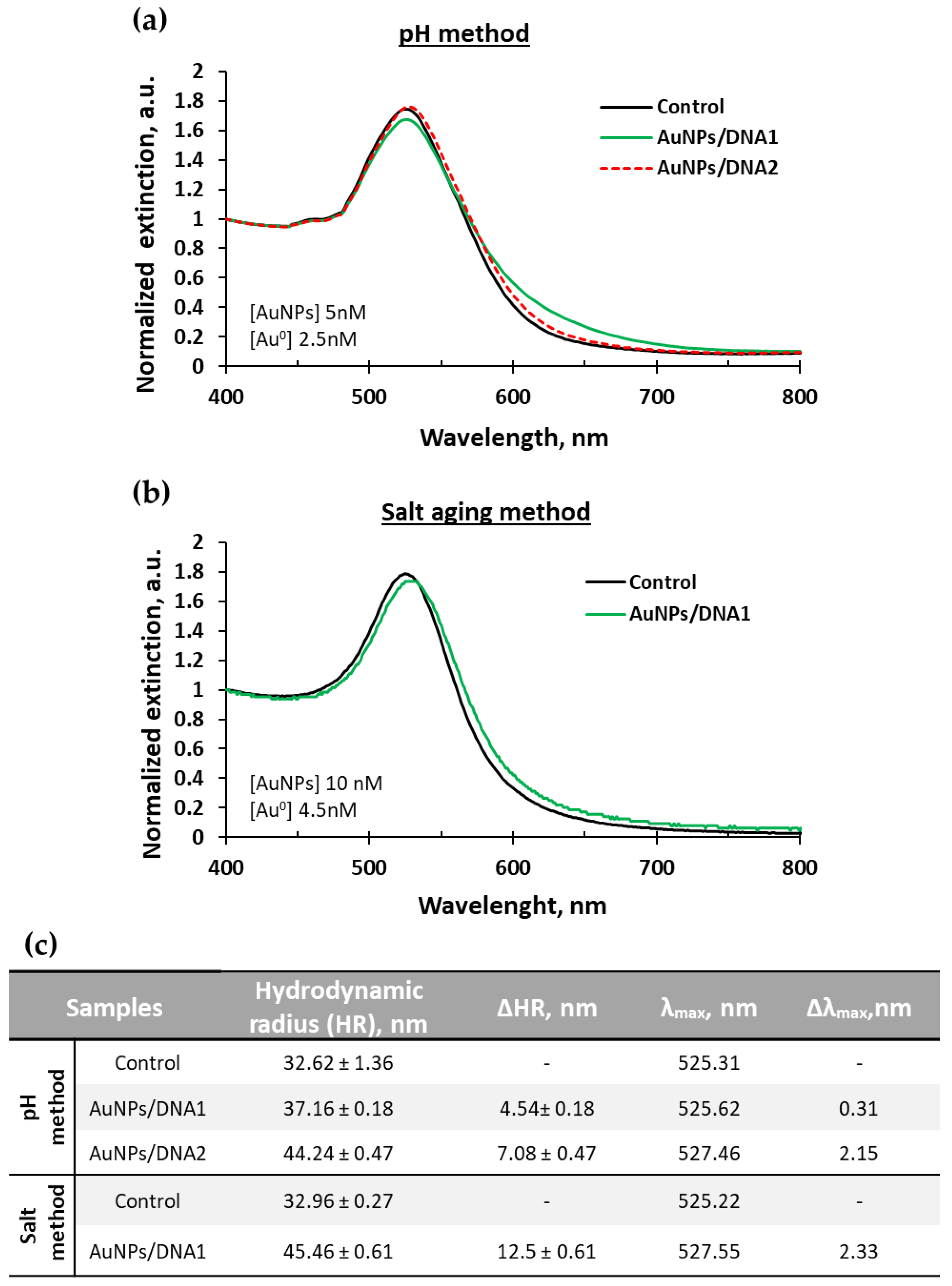

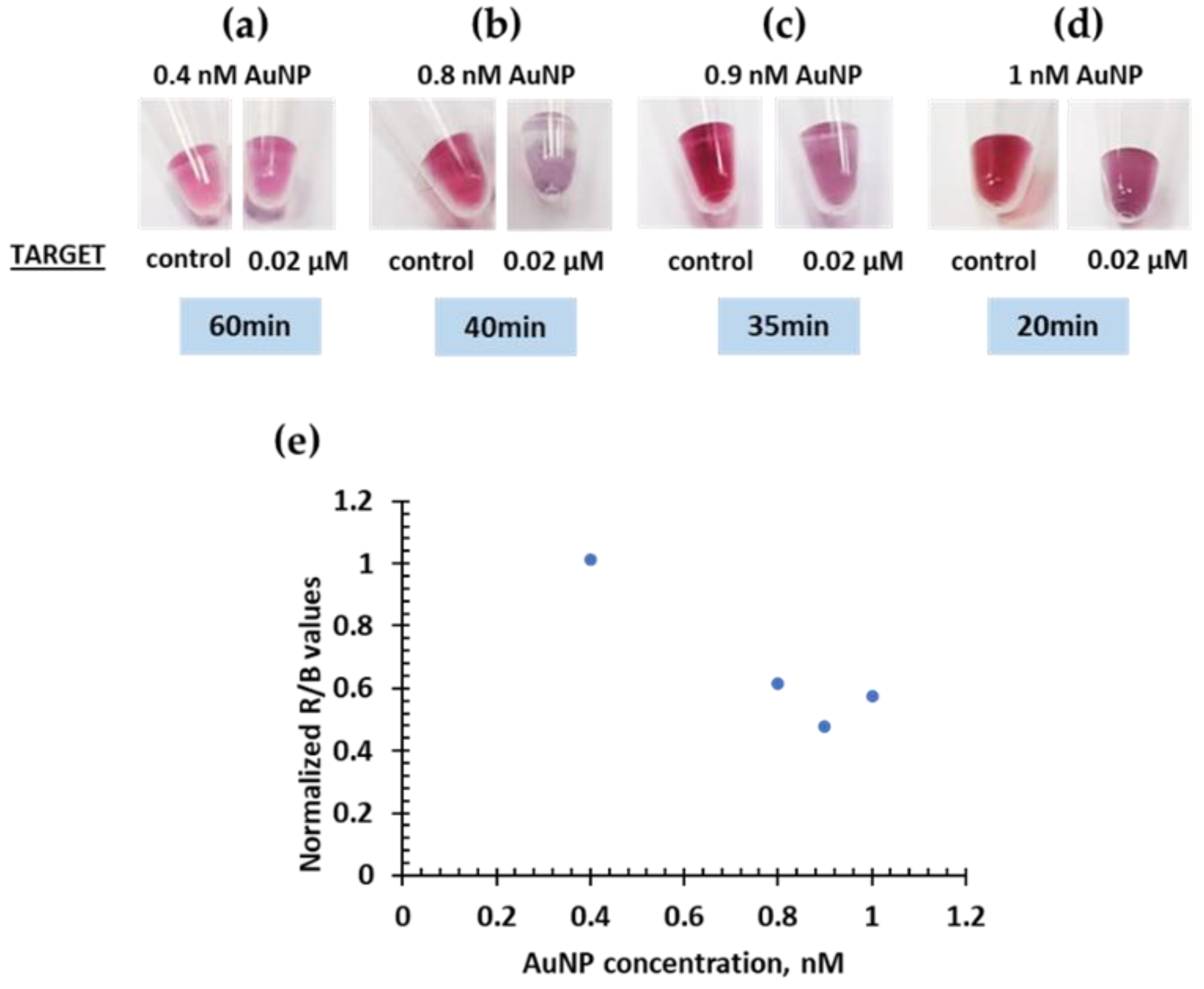
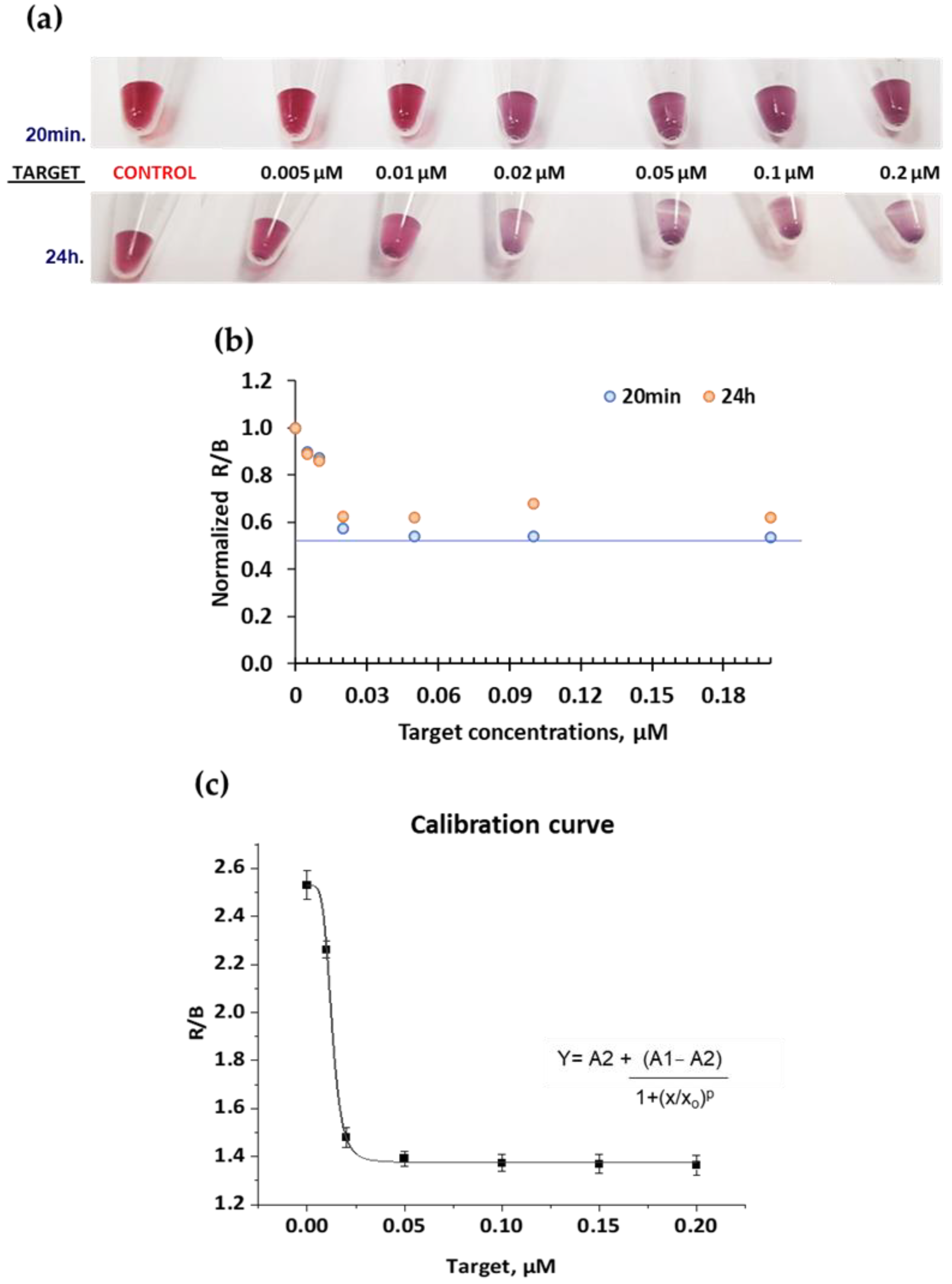
| Real Samples | R/B Values | Nanodrop | Calibration Curve | Culture Quantification |
|---|---|---|---|---|
| DNA Concentration, ng/μL | DNA Concentration, μM | Amoebas/μL | ||
| 1 | 2.30 | 0.85 | 0.010 | 15 |
| 2 | 1.84 | 28.00 | 0.014 | 295 |
| 3 | 1.76 | 42.00 | 0.015 | 776 |
| 4 | 1.69 | 85.00 | 0.015 | 1960 |
| Microorganism | Approach | Sample Type | Measurement | Limit of Detection | Analysis Time | Reference |
|---|---|---|---|---|---|---|
| Sars-CoV | CL * aggregation | Vero cell culture | UV-Vis | 0.18 ng/μL | 10 min | [27] |
| Mycobacterium spp. | NCL * aggregation | Goat feces | Naked eye | 2 ng/μL | 15 min | [44] |
| E. coli | NCL aggregation | Unamplified genomic DNA | Naked eye | ~54 ng | 30 min | [45] |
| Unamplified enzymatic digested genomic DNA | 11.4 ng | |||||
| Salmonella spp. | Surface hybridization | Feces | Naked eye | 5 × 108 CFU | 30 min | [25] |
| Salmonella enterica | CL aggregation | Genomic DNA | Naked eye | 37 fM | 15 min | [46] |
| MERS-CoV | NCL aggregation | Nucleic acids | UV-Vis | 1 pmol/μL | 10 min | [47] |
| Sequence (5′-3′) | |
|---|---|
| Target | GAGCCCACCGCCGCTGCCATCGCGTACGGTCTCGGCAAGAAGTC |
| DNA1 | ATGGCAGCGGCGGTGGGCTCTTTAAAA-SH |
| DNA2 | SH-AAAATTTTGACTTCTTGCCGAGACCGTA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pastrana, C.; Guerreiro, J.R.L.; Elumalai, M.; Carpena-Torres, C.; Crooke, A.; Carracedo, G.; Prado, M.; Huete-Toral, F. Dual-Mode Gold Nanoparticle-Based Method for Early Detection of Acanthamoeba. Int. J. Mol. Sci. 2022, 23, 14877. https://doi.org/10.3390/ijms232314877
Pastrana C, Guerreiro JRL, Elumalai M, Carpena-Torres C, Crooke A, Carracedo G, Prado M, Huete-Toral F. Dual-Mode Gold Nanoparticle-Based Method for Early Detection of Acanthamoeba. International Journal of Molecular Sciences. 2022; 23(23):14877. https://doi.org/10.3390/ijms232314877
Chicago/Turabian StylePastrana, Cristina, J. Rafaela L. Guerreiro, Monisha Elumalai, Carlos Carpena-Torres, Almudena Crooke, Gonzalo Carracedo, Marta Prado, and Fernando Huete-Toral. 2022. "Dual-Mode Gold Nanoparticle-Based Method for Early Detection of Acanthamoeba" International Journal of Molecular Sciences 23, no. 23: 14877. https://doi.org/10.3390/ijms232314877
APA StylePastrana, C., Guerreiro, J. R. L., Elumalai, M., Carpena-Torres, C., Crooke, A., Carracedo, G., Prado, M., & Huete-Toral, F. (2022). Dual-Mode Gold Nanoparticle-Based Method for Early Detection of Acanthamoeba. International Journal of Molecular Sciences, 23(23), 14877. https://doi.org/10.3390/ijms232314877








