Identification and Expression Analysis of the C-TERMINALLY ENCODED PEPTIDE Family in Pisum sativum L.
Abstract
:1. Introduction
2. Results
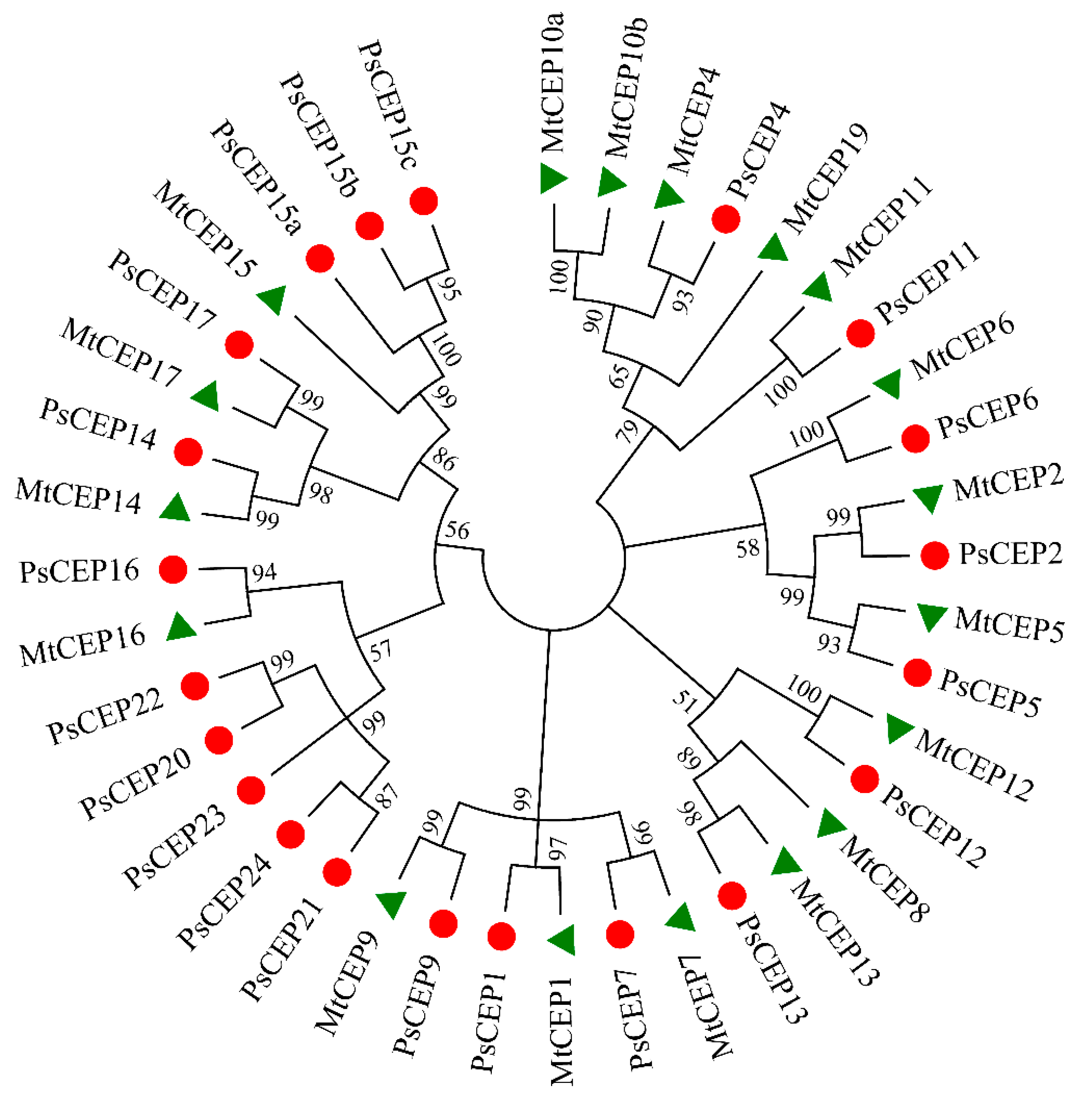
2.1. Identification of the CEP Peptide Family in Pea Genome
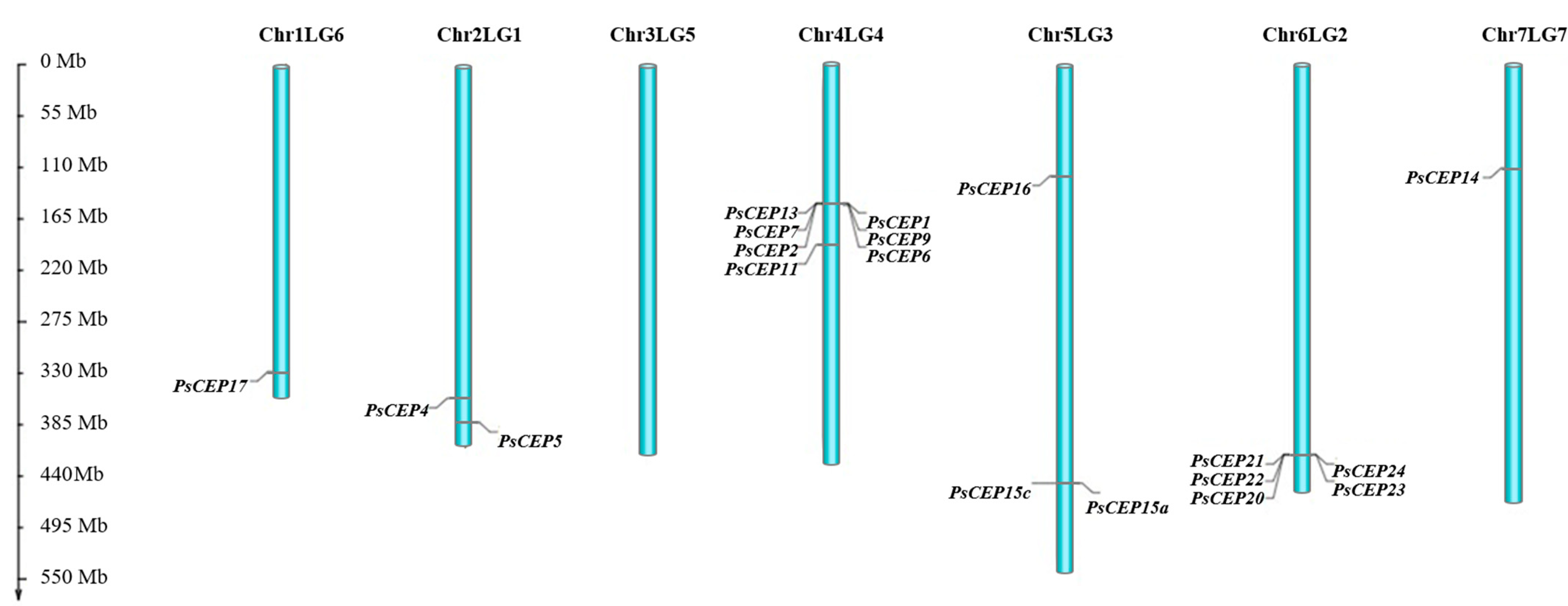
2.2. Chromosome Localization of the PsCEP Genes
2.3. The Structural Elements of the PsCEP Proteins
2.4. Cis-Regulatory Elements in the Promoters of the PsCEP Genes
2.5. Expression Analysis of the PsCEP Genes Based on Transcriptomic Data
2.6. Expression Analysis of the PsCEP Genes in Developing Nodules and in Response to Nitrate Treatment
3. Discussion
4. Materials and Methods
4.1. Identification of the PsCEP Genes and Their Phylogenetic Analysis
4.2. Chromosome Localization of the PsCEP Genes and Microsynteny Analysis
4.3. Promoter Analysis of the PsCEP Genes
4.4. The Structural Characterization of the PsCEP Proteins and Prediction of Physicochemical Properties of CEP Peptides
4.5. Expression Analysis of PsCEP Genes by a qPCR Analysis
4.5.1. Plant Growth Conditions
4.5.2. RNA Extraction and qPCR Analysis
4.6. Expression Analysis of PsCEP Genes Using Transcriptomic Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taleski, M.; Imin, N.; Djordjevic, M.A. CEP Peptide Hormones: Key Players in Orchestrating Nitrogen-Demand Signalling, Root Nodulation, and Lateral Root Development. J. Exp. Bot. 2018, 69, 1829–1836. [Google Scholar] [CrossRef] [Green Version]
- Delay, C.; Imin, N.; Djordjevic, M.A. CEP Genes Regulate Root and Shoot Development in Response to Environmental Cues and Are Specific to Seed Plants. J. Exp. Bot. 2013, 64, 5383–5394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imin, N.; Mohd-Radzman, N.A.; Ogilvie, H.A.; Djordjevic, M.A. The Peptide-Encoding CEP1 Gene Modulates Lateral Root and Nodule Numbers in Medicago Truncatula. J. Exp. Bot. 2013, 64, 5395–5409. [Google Scholar] [CrossRef]
- Mohd-Radzman, N.A.; Binos, S.; Truong, T.T.; Imin, N.; Mariani, M.; Djordjevic, M.A. Novel MtCEP1 Peptides Produced In Vivo Differentially Regulate Root Development in Medicago Truncatula. J. Exp. Bot. 2015, 66, 5289–5300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohyama, K.; Shinohara, H.; Ogawa-Ohnishi, M.; Matsubayashi, Y. A Glycopeptide Regulating Stem Cell Fate in Arabidopsis Thaliana. Nat. Chem. Biol. 2009, 5, 578–580. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Shinohara, H.; Mori, T.; Matsubayashi, Y.; Kawaguchi, M. Root-Derived CLE Glycopeptides Control Nodulation by Direct Binding to HAR1 Receptor Kinase. Nat. Commun. 2013, 4, 2191. [Google Scholar] [CrossRef] [Green Version]
- Tabata, R.; Sumida, K.; Yoshii, T.; Ohyama, K.; Shinohara, H.; Matsubayashi, Y. Perception of Root-Derived Peptides by Shoot LRR-RKs Mediates Systemic N-Demand Signaling. Science 2014, 346, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Roberts, I.; Smith, S.; Stes, E.; De Rybel, B.; Staes, A.; Van De Cotte, B.; Njo, M.F.; Dedeyne, L.; Demol, H.; Lavenus, J. CEP5 and XIP1/CEPR1 Regulate Lateral Root Initiation in Arabidopsis. J. Exp. Bot. 2016, 67, 4889–4899. [Google Scholar] [CrossRef] [Green Version]
- Mohd-Radzman, N.A.; Laffont, C.; Ivanovici, A.; Patel, N.; Reid, D.; Stougaard, J.; Frugier, F.; Imin, N.; Djordjevic, M.A. Different Pathways Act Downstream of the CEP Peptide Receptor CRA2 to Regulate Lateral Root and Nodule Development. Plant Physiol. 2016, 171, 2536–2548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huault, E.; Laffont, C.; Wen, J.; Mysore, K.S.; Ratet, P.; Duc, G.; Frugier, F. Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase. PLoS Genet. 2014, 10, e1004891. [Google Scholar] [CrossRef] [PubMed]
- Laffont, C.; Ivanovici, A.; Gautrat, P.; Brault, M.; Djordjevic, M.A.; Frugier, F. The NIN Transcription Factor Coordinates CEP and CLE Signaling Peptides That Regulate Nodulation Antagonistically. Nat. Commun. 2020, 11, 3167. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Ye, Q.; Chen, H.; Dong, J.; Wang, T. Multigene Editing Reveals That MtCEP1/2/12 Redundantly Control Lateral Root and Nodule Number in Medicago Truncatula. J. Exp. Bot. 2021, 72, 3661–3676. [Google Scholar] [CrossRef] [PubMed]
- Carroll, B.J.; Gresshoff, P.M. Nitrate Inhibition of Nodulation and Nitrogen Fixation in White Clover. Z. Pflanzenphysiol. 1983, 110, 77–88. [Google Scholar] [CrossRef]
- Gautrat, P.; Laffont, C.; Frugier, F. Compact Root Architecture 2 Promotes Root Competence for Nodulation through the MiR2111 Systemic Effector. Curr. Biol. 2020, 30, 1339–1345.e3. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Moreau, C.; Wang, J.; Frugier, F.; Xie, F. NLP1 Binds the CEP1 Signalling Peptide Promoter to Repress Its Expression in Response to Nitrate. New Phytol. 2022, 234, 1547–1552. [Google Scholar] [CrossRef]
- Patel, N.; Mohd-Radzman, N.A.; Corcilius, L.; Crossett, B.; Connolly, A.; Cordwell, S.J.; Ivanovici, A.; Taylor, K.; Williams, J.; Binos, S.; et al. Diverse Peptide Hormones Affecting Root Growth Identified in the Medicago Truncatula Secreted Peptidome. Mol. Cell. Proteom. 2018, 17, 160–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okamoto, S.; Suzuki, T.; Kawaguchi, M.; Higashiyama, T.; Matsubayashi, Y. A Comprehensive Strategy for Identifying Long-Distance Mobile Peptides in Xylem Sap. Plant J. 2015, 84, 611–620. [Google Scholar] [CrossRef] [Green Version]
- Lebedeva, M.A.; Yashenkova, Y.S.; Dodueva, I.E.; Lutova, L.A. Molecular Dialog between Root and Shoot via Regulatory Peptides and Its Role in Systemic Control of Plant Development. Russ. J. Plant Physiol. 2020, 67, 985–1002. [Google Scholar] [CrossRef]
- Boschiero, C.; Dai, X.; Lundquist, P.K.; Roy, S.; Christian de Bang, T.; Zhang, S.; Zhuang, Z.; Torres-Jerez, I.; Udvardi, M.K.; Scheible, W.-R.; et al. MtSSPdb: The Medicago Truncatula Small Secreted Peptide Database. Plant Physiol. 2020, 183, 399–413. [Google Scholar] [CrossRef] [Green Version]
- Kreplak, J.; Madoui, M.-A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A Reference Genome for Pea Provides Insight into Legume Genome Evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Alves-Carvalho, S.; Aubert, G.; Carrère, S.; Cruaud, C.; Brochot, A.-L.; Jacquin, F.; Klein, A.; Martin, C.; Boucherot, K.; Kreplak, J.; et al. Full-Length de Novo Assembly of RNA-Seq Data in Pea (Pisum sativum L.) Provides a Gene Expression Atlas and Gives Insights into Root Nodulation in This Species. Plant J. 2015, 84, 1–19. [Google Scholar] [CrossRef]
- HSDFinder: A BLAST-Based Strategy for Identifying Highly Similar Duplicated Genes in Eukaryotic Genomes. Available online: https://pubmed.ncbi.nlm.nih.gov/36303740/ (accessed on 14 November 2022).
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- PlantRegMap: Charting Functional Regulatory Maps in Plants | Nucleic Acids Research | Oxford Academic. Available online: https://academic.oup.com/nar/article/48/D1/D1104/5614566?login=false (accessed on 13 November 2022).
- Delay, C.; Chapman, K.; Taleski, M.; Wang, Y.; Tyagi, S.; Xiong, Y.; Imin, N.; Djordjevic, M.A. CEP3 Levels Affect Starvation-Related Growth Responses of the Primary Root. J. Exp. Bot. 2019, 70, 4763–4774. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Bisseling, T. Evolution of NIN and NIN-like Genes in Relation to Nodule Symbiosis. Genes 2020, 11, 777. [Google Scholar] [CrossRef] [PubMed]
- Soyano, T.; Shimoda, Y.; Hayashi, M. NODULE INCEPTION Antagonistically Regulates Gene Expression with Nitrate in Lotus Japonicus. Plant Cell Physiol. 2015, 56, 368–376. [Google Scholar] [CrossRef] [Green Version]
- Nishida, H.; Nosaki, S.; Suzuki, T.; Ito, M.; Miyakawa, T.; Nomoto, M.; Tada, Y.; Miura, K.; Tanokura, M.; Kawaguchi, M. Different DNA-Binding Specificities of NLP and NIN Transcription Factors Underlie Nitrate-Induced Control of Root Nodulation. Plant Cell 2021, 33, 2340–2359. [Google Scholar] [CrossRef]
- Zorin, E.A.; Kliukova, M.S.; Afonin, A.M.; Gribchenko, E.S.; Gordon, M.L.; Sulima, A.S.; Zhernakov, A.I.; Kulaeva, O.A.; Romanyuk, D.A.; Kusakin, P.G.; et al. A Variable Gene Family Encoding Nodule-Specific Cysteine-Rich Peptides in Pea (Pisum sativum L.). Front. Plant Sci. 2022, 13, 884726. [Google Scholar] [CrossRef]
- Tsyganov, V.E.; Morzhina, E.V.; Stefanov, S.Y.; Borisov, A.Y.; Lebsky, V.K.; Tikhonovich, I.A. The Pea (Pisum sativum L.) Genes Sym33 and Sym40 Control Infection Thread Formation and Root Nodule Function. Mol. Gen. Genet. 1998, 259, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, E.; Journet, E.-P.; Chabaud, M.; Cosson, V.; Ratet, P.; Duc, G.; Fedorova, E.; Liu, W.; den Camp, R.O.; Zhukov, V.; et al. IPD3 Controls the Formation of Nitrogen-Fixing Symbiosomes in Pea and Medicago spp. MPMI 2011, 24, 1333–1344. [Google Scholar] [CrossRef] [Green Version]
- Streeter, J.; Wong, P.P. Inhibition of Legume Nodule Formation and N2 Fixation by Nitrate. Crit. Rev. Plant Sci. 1988, 7, 1–23. [Google Scholar] [CrossRef]
- Hou, S.; Wang, X.; Chen, D.; Yang, X.; Wang, M.; Turrà, D.; Di Pietro, A.; Zhang, W. The Secreted Peptide PIP1 Amplifies Immunity through Receptor-like Kinase 7. PLoS Pathog. 2014, 10, e1004331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and Sequence Analysis Tools Services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Gribskov, M. Combining Evidence Using p-Values: Application to Sequence Homology Searches. Bioinformatics 1998, 14, 48–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for Occurrences of a given Motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Yang, S.; Liu, J.; Terecskei, K.; Ábrahám, E.; Gombár, A.; Domonkos, Á.; Szűcs, A.; Körmöczi, P.; Wang, T.; et al. Host-Secreted Antimicrobial Peptide Enforces Symbiotic Selectivity in Medicago Truncatula. Proc. Natl. Acad. Sci. USA 2017, 114, 6854–6859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozlowski, L.P. IPC 2.0: Prediction of Isoelectric Point and PKa Dissociation Constants. Nucleic Acids Res. 2021, 49, W285–W292. [Google Scholar] [CrossRef]
- Lullien, V.; Barker, D.G.; de Lajudie, P.; Huguet, T. Plant Gene Expression in Effective and Ineffective Root Nodules of Alfalfa (Medicago sativa). Plant Mol. Biol. 1987, 9, 469–478. [Google Scholar] [CrossRef]
- Sulima, A.S.; Zhukov, V.A.; Kulaeva, O.A.; Vasileva, E.N.; Borisov, A.Y.; Tikhonovich, I.A. New Sources of Sym2A Allele in the Pea (Pisum sativum L.) Carry the Unique Variant of Candidate LysM-RLK Gene LykX. PeerJ 2019, 7, e8070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Gingeras, T.R. Mapping RNA-Seq Reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11.14.1–11.14.19. [Google Scholar] [CrossRef]
- Love, M.; Ahlmann-Eltze, C.; Forbes, K.; Anders, S.; Huber, W.; FP7, R.E.; NHGRI, N. CZI DESeq2: Differential Gene Expression Analysis Based on the Negative Binomial Distribution 2022. Available online: https://github.com/mikelove/DESeq2 (accessed on 5 July 2022).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]











Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebedeva, M.A.; Gancheva, M.S.; Kulaeva, O.A.; Zorin, E.A.; Dobychkina, D.A.; Romanyuk, D.A.; Sulima, A.S.; Zhukov, V.A.; Lutova, L.A. Identification and Expression Analysis of the C-TERMINALLY ENCODED PEPTIDE Family in Pisum sativum L. Int. J. Mol. Sci. 2022, 23, 14875. https://doi.org/10.3390/ijms232314875
Lebedeva MA, Gancheva MS, Kulaeva OA, Zorin EA, Dobychkina DA, Romanyuk DA, Sulima AS, Zhukov VA, Lutova LA. Identification and Expression Analysis of the C-TERMINALLY ENCODED PEPTIDE Family in Pisum sativum L. International Journal of Molecular Sciences. 2022; 23(23):14875. https://doi.org/10.3390/ijms232314875
Chicago/Turabian StyleLebedeva, Maria A., Maria S. Gancheva, Olga A. Kulaeva, Evgeny A. Zorin, Daria A. Dobychkina, Daria A. Romanyuk, Anton S. Sulima, Vladimir A. Zhukov, and Lyudmila A. Lutova. 2022. "Identification and Expression Analysis of the C-TERMINALLY ENCODED PEPTIDE Family in Pisum sativum L." International Journal of Molecular Sciences 23, no. 23: 14875. https://doi.org/10.3390/ijms232314875
APA StyleLebedeva, M. A., Gancheva, M. S., Kulaeva, O. A., Zorin, E. A., Dobychkina, D. A., Romanyuk, D. A., Sulima, A. S., Zhukov, V. A., & Lutova, L. A. (2022). Identification and Expression Analysis of the C-TERMINALLY ENCODED PEPTIDE Family in Pisum sativum L. International Journal of Molecular Sciences, 23(23), 14875. https://doi.org/10.3390/ijms232314875






