H2S Enhanced the Tolerance of Malus hupehensis to Alkaline Salt Stress through the Expression of Genes Related to Sulfur-Containing Compounds and the Cell Wall in Roots
Abstract
1. Introduction
2. Results
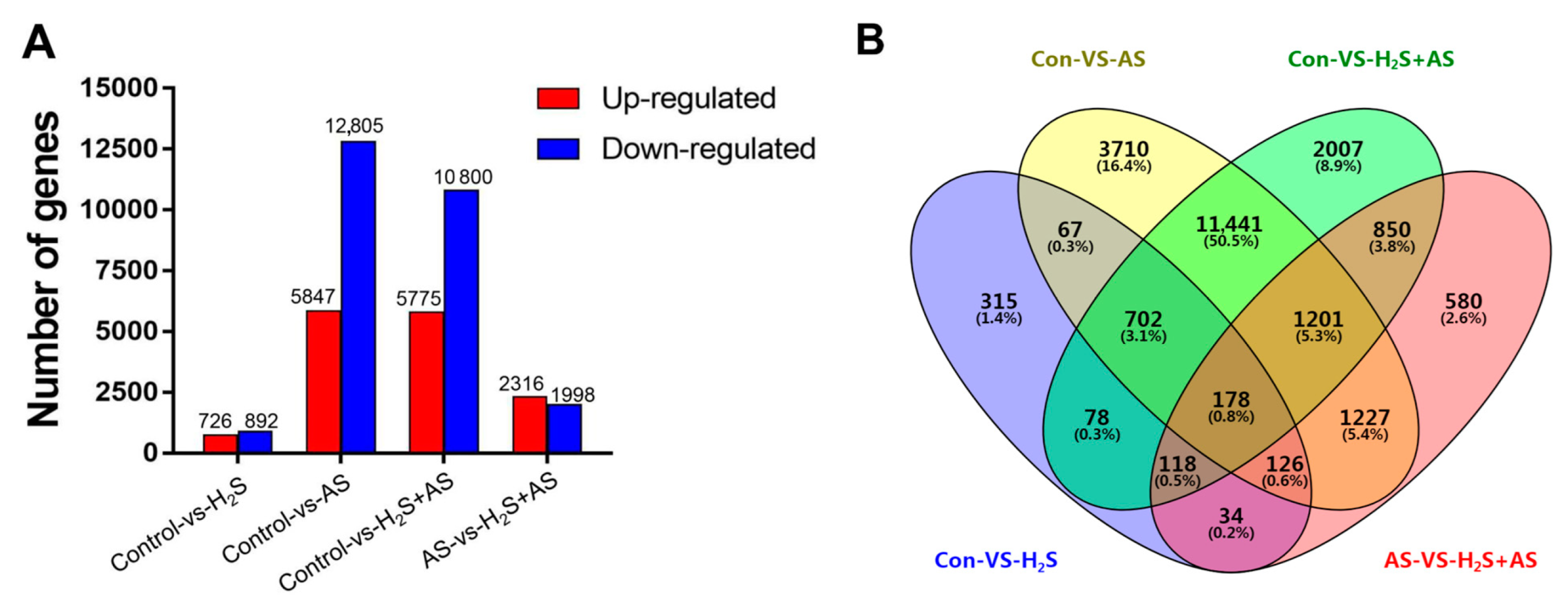
2.1. Analysis of DEGs in the Roots of M. hupehensis in Response to H2S and Alkaline Salt Stress
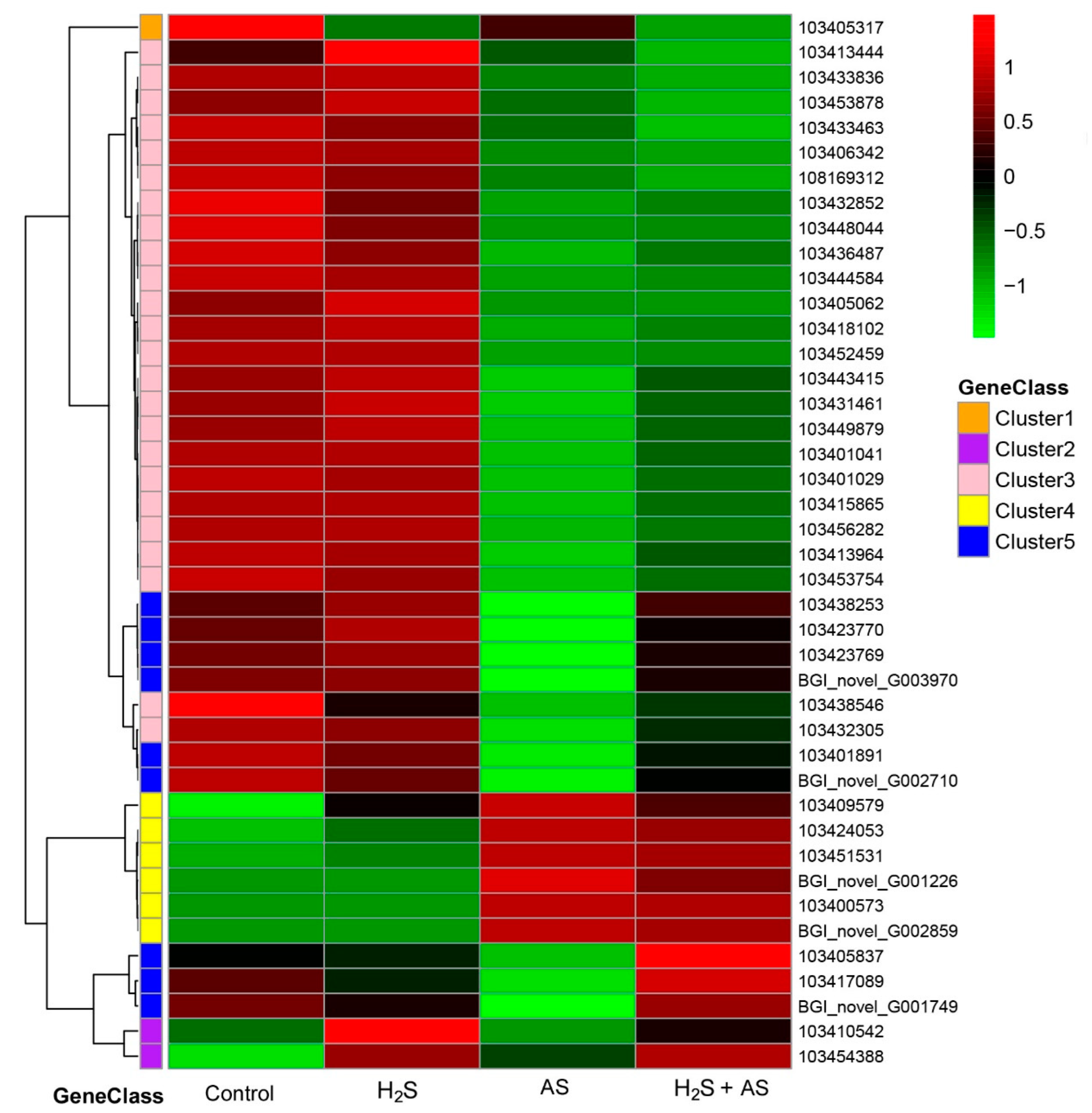
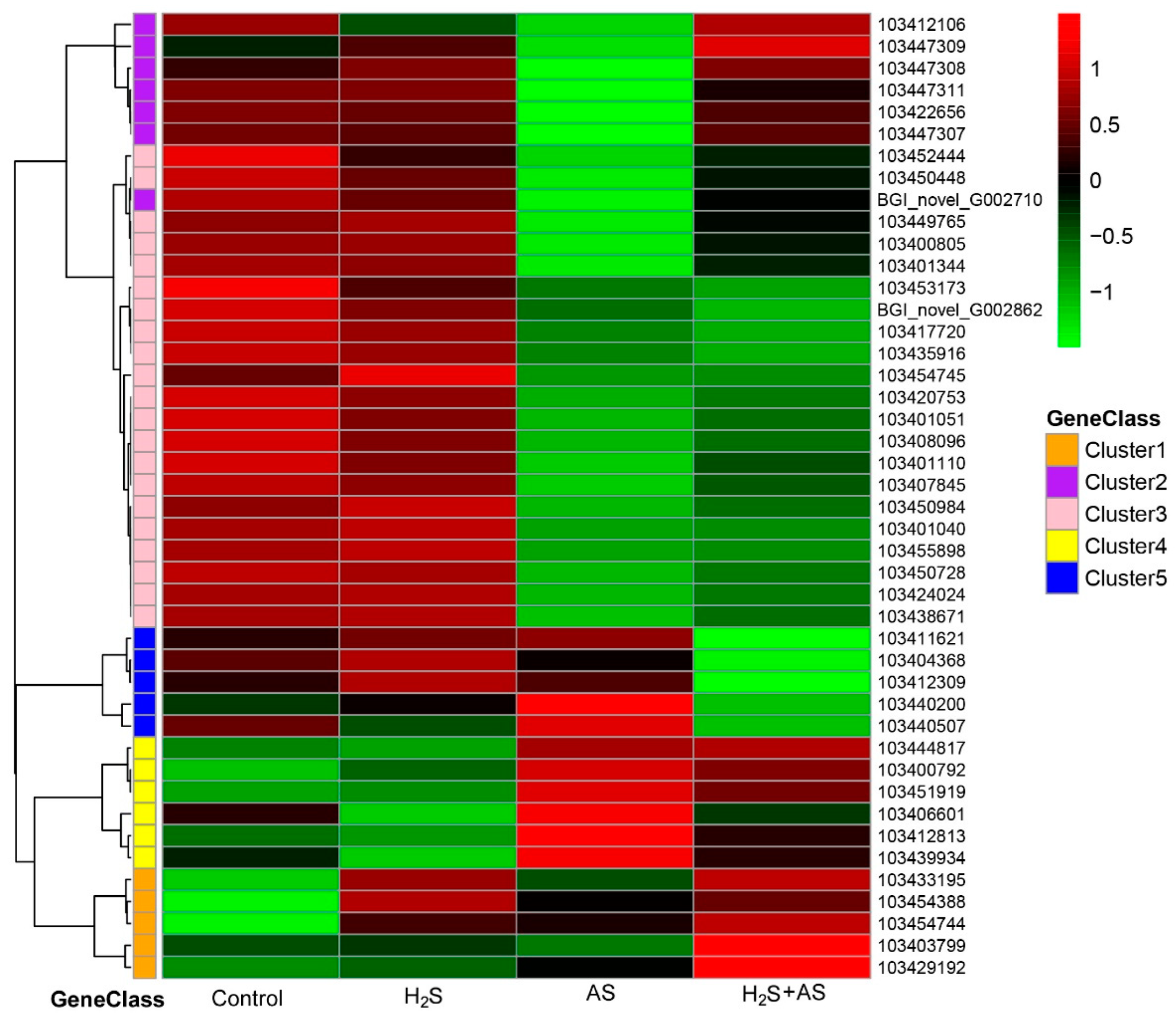
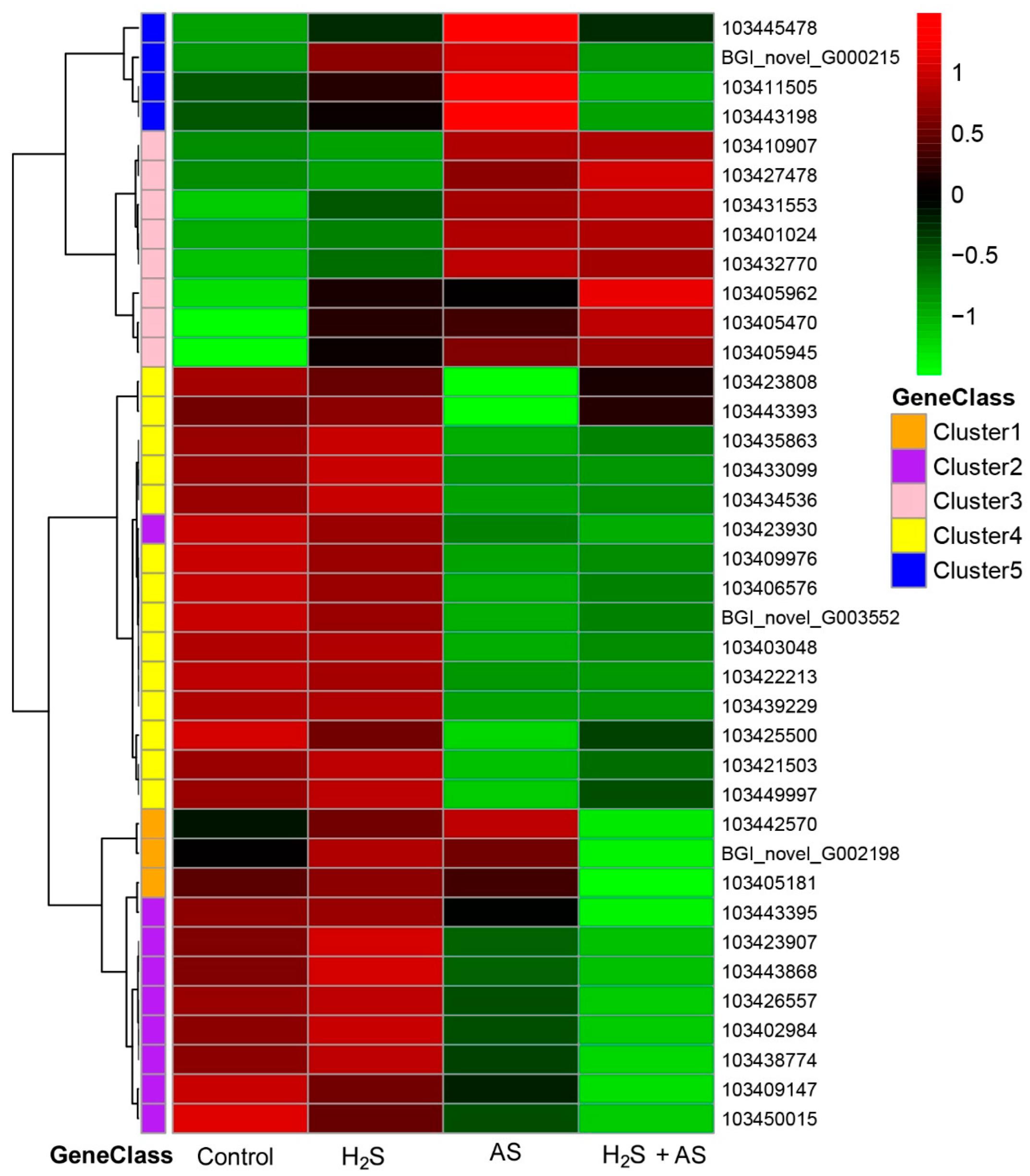
2.2. H2S Treatment Changed the DEG Expression Pattern of M. hupehensis Roots under Alkaline Salt Stress
2.3. GO Term and KEGG Pathway Enrichment Analysis of DEGs between H2S and Alkaline Salt Stress
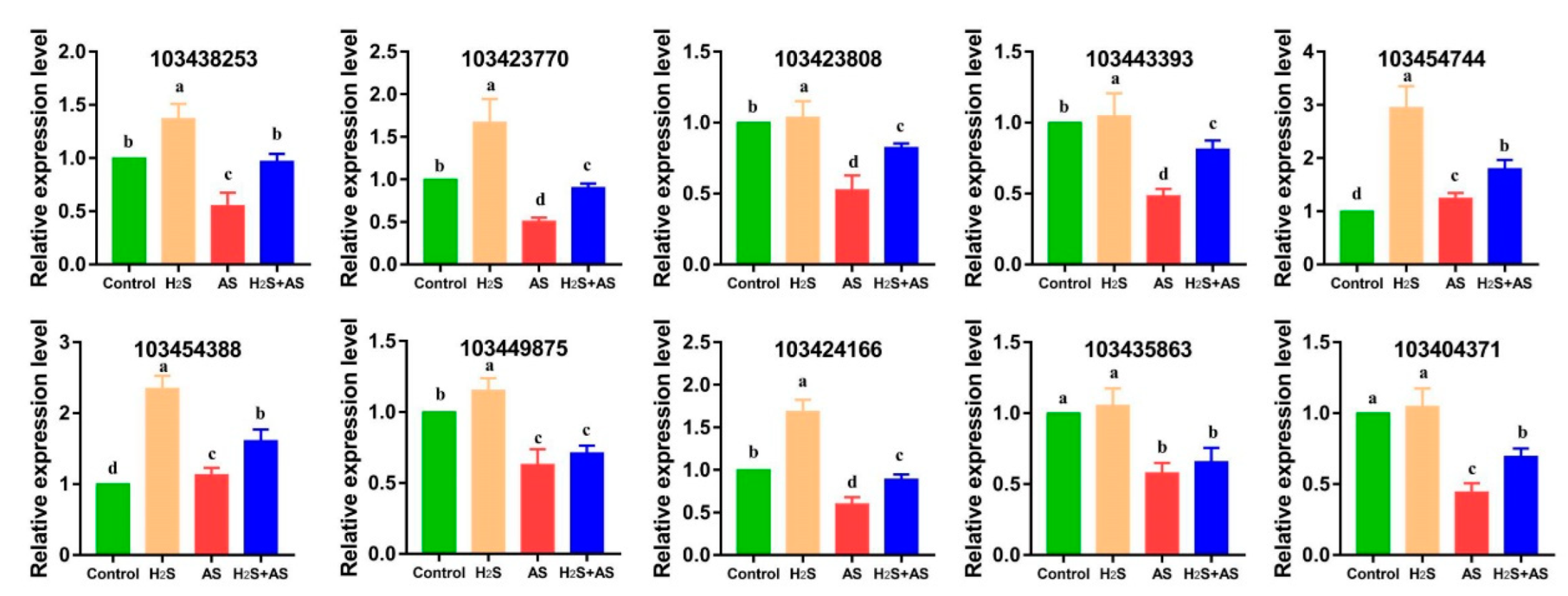
2.4. Transcriptome Analysis and PCR Validation in Response to H2S and Alkaline Salt Stress
3. Discussion
3.1. H2S-Mediated Antioxidant System and S-Containing Compounds Alleviate Alkaline Salts in M. hupehensis Roots
3.2. Role of Cell Wall Metabolism–Related Genes in H2S Alleviating Alkaline Salt Stress
3.3. H2S Induces Soluble Sugar and Osmoprotectant-Regulation-Related Genes in Response to Alkaline Salt Stress
4. Methods and Materials
4.1. Treatment of Plant Materials
4.2. RNA Extraction and cDNA Library Preparation for Transcriptome Sequencing
4.3. RNA-Seq Read Mapping, Assembly, and Annotation of the Transcriptome
4.4. Differentially Expressed Gene Annotation and Analysis
4.5. Analysis and Validation of qRT-PCR
4.6. Experimental Design and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kawanabe, S.; Zhu, T.C. Degeneration and conservation of aneurolepidium chinense grassland in Northern China. J. Jpn. Grassland Sci. 1991, 37, 91–99. [Google Scholar]
- Shi, D.C.; Wang, D.L. Effects of various salt-alkaline mixed stresses on Aneurolepidium chinense (Trin.) Kitag. Plant Soil 2005, 271, 15–26. [Google Scholar] [CrossRef]
- Yang, C.W.; Chong, J.N.; Li, C.Y.; Kim, C.M.; Shi, D.C.; Wang, D.L. Osmotic adjustment and ion balance traits of an alkali resistant halophyte Kochia sieversiana during adaptation to salt and alkali conditions. Plant Soil 2007, 294, 263–276. [Google Scholar] [CrossRef]
- Yu, S.; Yu, L.H.; Hou, Y.L.; Zhang, Y.F.; Guo, W.; Xue, Y.W. Contrasting effects of NaCl and NaHCO3 stresses on seed germination, seedling growth, photosynthesis, and osmoregulators of the common bean (Phaseolus vulgaris L.). Agronomy 2019, 9, 19. [Google Scholar] [CrossRef]
- Liu, P.; Zhu, Y.X.; Liu, H.; Liang, Z.J.; Zhang, M.Y.; Zou, C.Y.; Yuan, G.S.; Gao, S.B.; Pan, G.T.; Shen, Y.O.; et al. Acombination of a genome-wide association study and a transcriptome analysis reveals circRNAs as new regulators involved in the response to salt stress in maize. Int. J. Mol. Sci. 2022, 23, 9755. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.L.; Zhang, R.X.; Yuan, H.Y.; Wang, M.M.; Yang, H.Y. Root Damage under alkaline stress is associated with reactive oxygen species accumulation in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1580. [Google Scholar] [CrossRef]
- Li, H.; Shi, J.Y.; Wang, Z.P.; Zhang, W.W.; Yang, H.Q. H2S pretreatment mitigates the alkaline salt stress on Malus hupehensis roots by regulating Na+/K+ homeostasis and oxidative stress. Plant Physiol. Bioch. 2020, 156, 233–241. [Google Scholar] [CrossRef]
- Capula-Rodríguez, R.; Valdez-Aguilar, L.A.; Cartmill, D.L.; Cartmill, A.D.; Alia-Tejacal, I. Supplementary calcium and potassium improve the response of tomato (Solanum lycopersicum L.) to simultaneous alkalinity, salinity, and boron stress. Commun. Soil Sci. Plant Anal. 2016, 47, 505–511. [Google Scholar]
- Zhang, K.H.; Tang, J.R.; Wang, Y.; Kang, H.Y.; Zeng, J. The tolerance to salinealkaline stress was dependent on the roots in wheat. Physiol. Mol. Biol. Plants 2020, 26, 947–954. [Google Scholar] [CrossRef]
- Zhang, P.; Duo, T.Q.; Wang, F.D.; Zhang, X.Z.; Yang, Z.Z.; Hu, G.F. De novo transcriptome in roots of switchgrass (Panicum virgatum L.) reveals gene expression dynamic and act network under alkaline salt stress. BMC Genom. 2021, 22, 82. [Google Scholar] [CrossRef]
- An, P.; Li, X.; Zheng, Y.; Matsuura, A.; Abe, J.; Eneji, A.E.; Tanimoto, E.; Inanaga, S. Effects of NaCl on root growth and cell wall composition of two soya bean cultivars with contrasting salt tolerance. J. Agron. Crop Sci. 2014, 200, 212–218. [Google Scholar] [CrossRef]
- Xu, P.P.; Fang, S.; Chen, H.Y.; Cai, W.M. The brassinosteroid-responsive xyloglucan endotransglucosylase/hydrolase 19 (XTH19) and XTH23 genes are involved in lateral root development under salt stress in Arabidopsis. Plant J. 2020, 104, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.H.; Li, J.; Ma, C.H.; Yang, Q.H.; Liu, C.J.; Feng, B.L. Salt-tolerant broomcorn millet (Panicum miliaceum L.) resists salt stress via modulation of cell wall biosynthesis and Na+ balance. Land Degrad. Dev. 2020, 32, 1–15. [Google Scholar] [CrossRef]
- Rausch, T.; Wachter, A. Sulfur metabolism: A versatile platform for launching defence operations. Trends Plant Sci. 2005, 10, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.G.; Min, X.; Zhou, Z.H. Hydrogen sulfide: A signal molecule in plant cross-adaptation. Front. Plant Sci. 2016, 7, 1621. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.L.; Tian, Y.; Li, L.; Yu, M.; Hou, R.P.; Ren, X.M. H2S alleviates salinity stress in cucumber by maintaining the Na+/K+ balance and regulating H2S metabolism and oxidative stress response. Front. Plant Sci. 2019, 10, 678. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Saegusa, D.; Fujita, M.; Tran, L.S. Hydrogen sulfide regulates salt tolerance in rice by maintaining Na+/K+ balance, mineral homeostasis and oxidative metabolism under excessive salt Stress. Front. Plant Sci. 2015, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.W.; Mao, Y.; Zhou, H.; Li, F.; Wu, M.Z.; Zhang, J. Endogenous hydrogen sulfifide enhances salt tolerance by coupling the reestablishment of redox homeostasis and preventing salt-induced K+ loss in seedlings of Medicago sativa. Plant Sci. 2014, 225, 117–129. [Google Scholar] [CrossRef]
- Deng, Y.Q.; Bao, J.; Yuan, F.; Liang, X.; Feng, Z.T.; Wang, B.S. Exogenous hydrogen sulfide alleviates salt stress in wheat seedlings by decreasing Na+ content. Plant Growth Regul. 2016, 79, 391–399. [Google Scholar] [CrossRef]
- Ziogas, V.; Molassiotis, A.; Fotopoulos, V.; Tanou, G. Hydrogen sulfide: A potent tool in postharvest fruit biology and possible mechanism of action. Front. Plant Sci. 2018, 9, 1375. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.F.; Jin, Y.H.; Huang, W.; Sun, Q.; Liu, F.; Huang, X.Z. Full-length transcriptome sequences of ephemeral plant Arabidopsis pumila provides insight into gene expression dynamics during continuous salt stress. BMC Genom. 2018, 19, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.H.; Wu, T.T.; Zhao, X.; Wang, Z.Q.; Chen, Y. Comparative physiological and full-length transcriptome analyses reveal the molecular mechanism of melatoninmediated salt tolerance in okra (Abelmoschus esculentus L.). BMC Plant Biol. 2021, 21, 180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.W.; Song, J.F.; Yue, S.Q.; Duan, K.X.; Yang, H.Q. MhMAPK4 from Malus hupehensis Rehd. decreases cell death in tobacco roots by controlling Cd2+ uptake. Ecotox. Environ. Saf. 2019, 168, 230–240. [Google Scholar] [CrossRef]
- Cai, Z.C.; Wang, C.C.; Chen, C.H.; Zou, L.S.; Yin, S.X.; Liu, S.J.; Yuan, J.H.; Wu, N.; Liu, X.H. Comparative transcriptome analysis reveals variations of bioactive constituents in Lonicera japonica flowers under salt stress. Plant Physiol. Bioch. 2022, 15, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.C.; Wang, M.; Ren, T.T.; Li, K.Y.; Li, Y.Q.; Marowa, P.; Zhang, C.S. Comparative transcriptome analysis reveals the molecular mechanism of salt tolerance in Apocynum venetum. Plant Physiol. Bioch. 2021, 167, 816–830. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.Q.; Duan, K.X.; Zhang, W. Biology and physiology of Malus hupehensis for the apogamic plant resource. Acta. Hortic. 2008, 769, 441–447. [Google Scholar] [CrossRef]
- Zhu, J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Wei, T.L.; Wang, Y.; Xie, Z.Z.; Guo, D.Y.; Chen, C.W.; Fan, Q.J.; Deng, X.D.; Liu, J.H. Enhanced ROS scavenging and sugar accumulation contribute to drought tolerance of naturally occurring autotetraploids in Poncirus trifoliata. Plant Biotechnol. J. 2018, 17, 1394–1407. [Google Scholar] [CrossRef] [PubMed]
- Droux, M. Sulfur assimilation and the role of sulfur in plant metabolism: A survey. Photosynth. Res. 2004, 79, 331–348. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, M.; Droux, M. Synthesis of the sulfur amino acids: Cysteine and methionine. Photosynth. Res. 2005, 86, 345–362. [Google Scholar] [CrossRef] [PubMed]
- Kopriva, S.; Rennenberg, H. Control of sulphate assimilation and glutathione synthesis: Interaction with N and C metabolism. J. Exp. Bot. 2004, 55, 1831–1842. [Google Scholar] [CrossRef]
- Kopriva, S. Regulation of sulfate assimilation in Arabidopsis and beyond. Ann. Bot. 2006, 97, 479–495. [Google Scholar] [CrossRef]
- Romero, L.C.; Domínguez-Solis, J.R.; Gutierrez-Alcala, G.; Gotor, C. Salt regulation of O-acetylserine(thiol)lyase in Arabidopsis thaliana and increased tolerance in yeast. Plant Physiol. Biochem. 2001, 39, 643–647. [Google Scholar] [CrossRef]
- Amir, R. Current understanding of the factors regulating methionine content in vegetative tissues of higher plants. Amino Acids. 2010, 39, 917–931. [Google Scholar] [CrossRef]
- Nazar, R.; Iqbal, N.; Masood, A.; Syeed, S.; Khan, N.A. Understanding the significance of sulfur in improving salinity tolerance in plants. Environ. Exp. Bot. 2011, 70, 80–87. [Google Scholar] [CrossRef]
- Jaleel, C.A.; Manivannan, P.; Lakshmanan, G.M.; Sridharan, R.; Panneerselvam, R. NaCl as a physiological modulator of proline metabolism and antioxidant potential in Phyllanthus amarus. C. R. Biol. 2007, 330, 806–813. [Google Scholar] [CrossRef]
- Reiter, W.D. Biosynthesis and properties of the plant cell wall. Curr. Opin. Plant Biol. 2002, 5, 536–542. [Google Scholar] [CrossRef]
- Chai, L.; Wang, J.M.; Fan, Z.Y.; Liu, Z.B.; Li, X.F.; Yang, Y. Ascorbate peroxidase gene from Brassica napus enhances salt and drought tolerances in Arabidopsis thaliana. Afr. J. Biotechnol. 2011, 10, 18085–18091. [Google Scholar]
- Choi, H.W.; Hwang, B.K. The pepper extracellular peroxidase CaPO2 is required for salt, drought and oxidative stress tolerance as well as resistance to fungal pathogens. Planta 2012, 235, 1369–1382. [Google Scholar] [CrossRef] [PubMed]
- Maia, J.M.; Ferreira-Silva, S.L.; Voigt, E.L.; de Macedo, C.E.C.; Ponte, L.F.A.; Silveira, J.A.G. Activities of antioxidant enzymes and root growth inhibition in cowpea seedlings exposed to different salt levels. Acta Bot. Bras. 2012, 26, 2. [Google Scholar]
- Kumar, D.; Singh, P.; Yusuf, M.A.; Upadhyaya, C.P.; Roy, S.D.; Hohn, T.; Sarin, N.B. The Xerophyta viscosa Aldose Reductase (ALDRXV4) Confers Enhanced Drought and Salinity Tolerance to Transgenic Tobacco Plants by Scavenging Methylglyoxal and Reducing the Membrane Damage. Mol. Biotechnol. 2013, 54, 292–303. [Google Scholar] [CrossRef]
- Fry, S.C. Cross-linking of matrix polymers in the growing cell walls of angiosperms. Ann. Rev. Plant Physiol. 1986, 37, 165–186. [Google Scholar] [CrossRef]
- Hu, L.X.; Li, H.Y.; Chen, L.; Lou, Y.H.; Amombo, E. RNA-seq for gene identification and transcript profiling in relation to root growth of bermudagrass (Cynodon dactylon) under salinity stress. BMC Genom. 2015, 16, 575. [Google Scholar] [CrossRef]
- Zhong, H.; Läuchli, A. Changes of cell wall composition and polymer size in primary roots of cotton seedlings under high salinity. J. Exp. Bot. 1993, 44, 773–778. [Google Scholar] [CrossRef]
- Buchanan, B.; Gruissem, W.; Jones, R. Biochemistry and Molecular Biology of Plants; John Wiley & Sons: Hoboken, NJ, USA, 2005. [Google Scholar]
- Muller, B.; Reymond, M.; Tardieu, F. The elongation rate at the base of a maize leaf shows an invariant pattern during both the steady-stage elongation and the establishment of the elongation zone. J. Exp. Bot. 2001, 52, 1259–1268. [Google Scholar] [CrossRef]
- Reidy, B.; Nösberger, J.; Fleming, A. Differential expression of XET-related genes in the leaf elongation zone of F. pratensis. J. Exp. Bot. 2001, 52, 1847–1856. [Google Scholar] [CrossRef][Green Version]
- Levy, S.; York, W.S.; Stuike-prill, R.; Meyer, B.; Staehelin, L.A. Simulations of the static and dynamic molecular conformations of xyloglucan. The role of the fucosylated sidechain in surface-specific sidechain folding. Plant J. 1991, 1, 195–215. [Google Scholar] [CrossRef]
- Du, J.; Guo, S.R.; Sun, J.; Shu, S.R. Proteomic and physiological analyses reveal the role of exogenous spermidine on cucumber roots in response to Ca(NO3)2 stress. Plant Mol. Biol. 2018, 97, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Walias, F.J.; García, M.; Moreno, M.; Giannoukos, I.; González, N.; Sanz-García, E.; Necira, K.; Canto, T.; Tenllado, F. Transgenerational tolerance to salt and osmotic stresses induced by plant virus infection. Int. J. Mol. Sci. 2022, 23, 12497. [Google Scholar] [CrossRef] [PubMed]
- Dahro, B.; Wang, F.; Peng, T.; Liu, J.H. PtrA/NINV, an alkaline/neutral invertase gene of Poncirus trifoliata, confers enhanced tolerance to multiple abiotic stresses by modulating ROS levels and maintaining photosynthetic efficiency. BMC Plant Biol. 2016, 16, 76. [Google Scholar] [CrossRef]
- Wei, T.L.; Wang, Y.; Liu, J.H. Comparative transcriptome analysis reveals synergistic and disparate defense pathways in the leaves and roots of trifoliate orange (Poncirus trifoliata) autotetraploids with enhanced salt tolerance. Hortic. Res.-Engl. 2020, 7, 88. [Google Scholar] [CrossRef]
- Rady, M.M.; Elrys, A.S.; Abo El-Maati, M.F.; Desoky, E.S.M. Interplaying roles of silicon and proline effectively improve salt and cadmium stress tolerance in Phaseolus vulgaris plant. Plant Physiol. Biochem. 2019, 139, 558–5680. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.M.F.; Ali, E.F. Evaluation of proline functions in saline conditions. Phytochemistry 2017, 140, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Broyart, C.; Rabanal, F.A.; Jonak, C. The redox-sensitive chloroplast trehalose-6-phosphate phosphatase AtTPPD regulates salt stress tolerance. Antioxidants Redox Signal. 2014, 21, 1289–1304. [Google Scholar] [CrossRef]
- Gao, Y.; Yang, X.; Yang, X.; Zhao, T.; An, X.; Chen, Z. Characterization and expression pattern of the trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase gene families in Populus. Int. J. Biol. Macromol. 2021, 187, 9–23. [Google Scholar] [CrossRef]
- Garg, A.K.; Kim, J.K.; Owens, T.G.; Ranwala, A.P.; Choi, Y.D.; Kochian, L.V.; Wu, R.J. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. USA 2002, 99, 15898–15903. [Google Scholar] [CrossRef]
- Ge, L.F.; Chao, D.Y.; Shi, M.; Zhu, M.Z.; Gao, J.P.; Lin, H.X. Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 2008, 228, 191–201. [Google Scholar] [CrossRef]
- Li, H.; Yu, T.T.; Ning, Y.S.; Li, H.; Zhang, W.W.; Yang, H.Q. Hydrogen sulfide alleviates alkaline salt stress by regulating the expression of microRNAs in Malus hupehensis Rehd. roots. Front. Plant Sci. 2021, 12, 663519. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Mao, X.Z.; Cai, T.; Olyarchuk, J.G.; Wei, L.P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
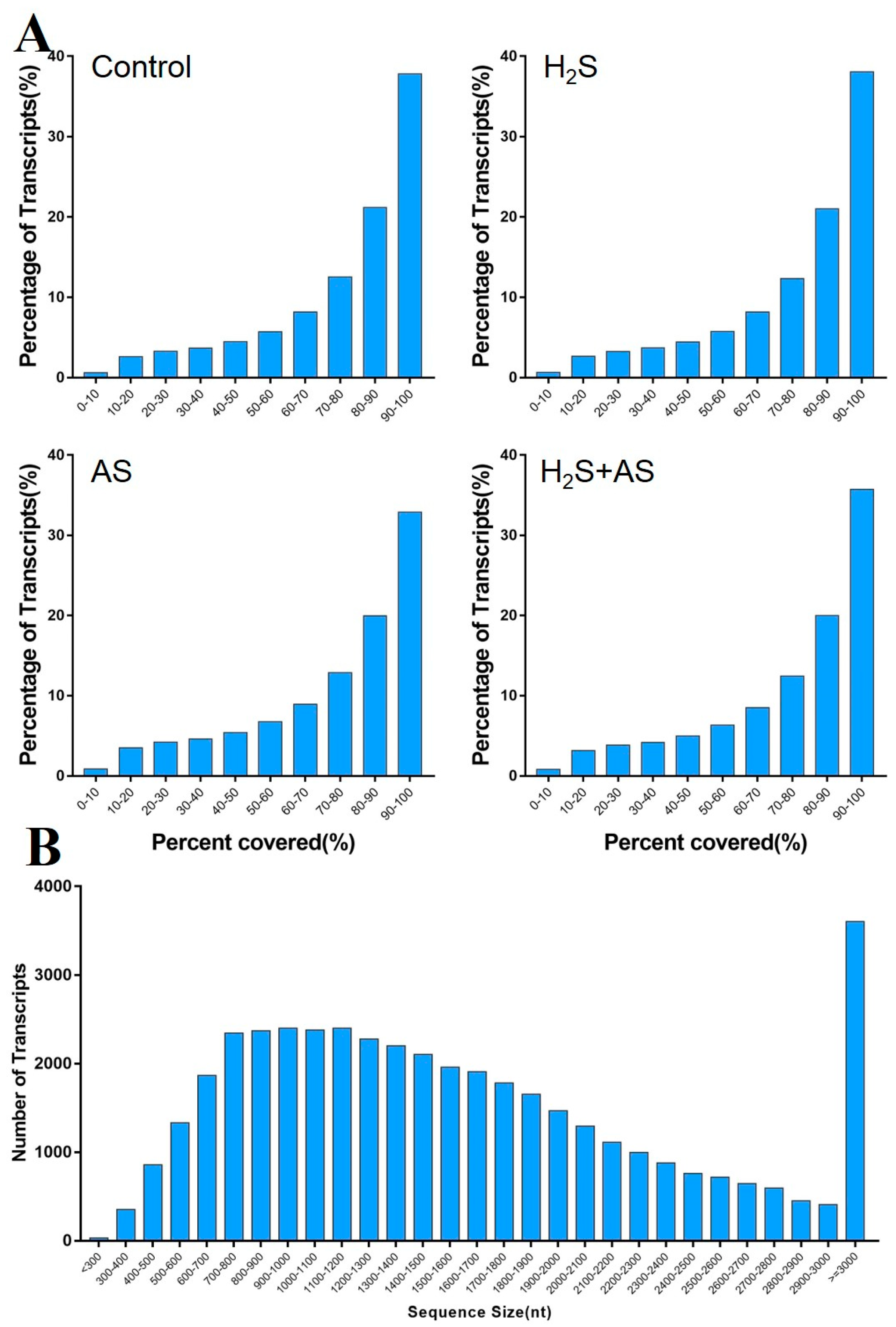



| Sample | Raw Reads | Clean Reads (%) | Clean Bases | N (%) | Adapter (%) | Low Quality (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|---|---|---|
| Control-1 | 45.09 M | 40.84 M (90.58%) | 6.13 | 97,530 (0.22%) | 2,061,610 (4.57%) | 2,086,720 (4.63%) | 96.61% | 87.44% |
| Control-2 | 42.2 M | 38.23 M (90.5%) | 5.73 | 89,922 (0.21%) | 1,862,696 (4.41%) | 2,061,138 (4.88%) | 96.62% | 87.53% |
| Control-3 | 43.77 M | 39.38 M (89.98%) | 5.91 | 93,516 (0.21%) | 2,109,890 (4.82%) | 2,182,808 (4.99%) | 96.6% | 87.47% |
| H2S-1 | 47.19 M | 42.2 M (89.4%) | 6.33 | 106,196 (0.23%) | 2,533,522 (5.37%) | 2,348,632 (4.98%) | 96.61% | 87.5% |
| H2S-2 | 46.48 M | 41.74 M (89.43%) | 6.26 | 100,562 (0.22%) | 2,337,408 (5.03%) | 2,302,112 (4.95%) | 96.61% | 87.53% |
| H2S-3 | 47.02 M | 41.85 M (89.8%) | 6.28 | 102,490 (0.22%) | 2,621,430 (5.58%) | 2,440,370 (5.19%) | 96.53% | 87.23% |
| H2S + AS-1 | 46.29 M | 41.82 M (89.02%) | 6.27 | 100,578 (0.22%) | 2,083,188 (4.5%) | 2,282,972 (4.93%) | 96.64% | 87.56% |
| H2S + AS-2 | 47.33 M | 42.71 M (90.35%) | 6.41 | 109,606 (0.23%) | 2,246,082 (4.73%) | 2,265,452 (4.79%) | 96.71% | 87.8% |
| H2S + AS-3 | 49.02 M | 43.78 M (90.24%) | 6.57 | 108,808 (0.22%) | 2,832,818 (5.78%) | 2,305,498 (4.7%) | 96.69% | 87.75% |
| AS-1 | 47.94 M | 42.63 M (89.3%) | 6.39 | 105,086 (0.22%) | 2,797,352 (5.84%) | 2,407,260 (5.02%) | 96.61% | 87.52% |
| AS-2 | 47.35 M | 42.12 M (88.97%) | 6.32 | 100,798 (0.21%) | 3,043,710 (6.43%) | 2,078,552 (4.39%) | 96.8% | 88.12% |
| AS-3 | 42.15 M | 37.82 M (89.71%) | 5.67 | 90,538 (0.21%) | 2,118,994 (5.03%) | 2,127,618 (5.05%) | 96.55% | 87.31% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Zhang, W.; Han, M.; Song, J.; Ning, Y.; Yang, H. H2S Enhanced the Tolerance of Malus hupehensis to Alkaline Salt Stress through the Expression of Genes Related to Sulfur-Containing Compounds and the Cell Wall in Roots. Int. J. Mol. Sci. 2022, 23, 14848. https://doi.org/10.3390/ijms232314848
Li H, Zhang W, Han M, Song J, Ning Y, Yang H. H2S Enhanced the Tolerance of Malus hupehensis to Alkaline Salt Stress through the Expression of Genes Related to Sulfur-Containing Compounds and the Cell Wall in Roots. International Journal of Molecular Sciences. 2022; 23(23):14848. https://doi.org/10.3390/ijms232314848
Chicago/Turabian StyleLi, Huan, Weiwei Zhang, Mengyuan Han, Jianfei Song, Yuansheng Ning, and Hongqiang Yang. 2022. "H2S Enhanced the Tolerance of Malus hupehensis to Alkaline Salt Stress through the Expression of Genes Related to Sulfur-Containing Compounds and the Cell Wall in Roots" International Journal of Molecular Sciences 23, no. 23: 14848. https://doi.org/10.3390/ijms232314848
APA StyleLi, H., Zhang, W., Han, M., Song, J., Ning, Y., & Yang, H. (2022). H2S Enhanced the Tolerance of Malus hupehensis to Alkaline Salt Stress through the Expression of Genes Related to Sulfur-Containing Compounds and the Cell Wall in Roots. International Journal of Molecular Sciences, 23(23), 14848. https://doi.org/10.3390/ijms232314848







