Characterization and Genomic Analysis of Escherichia coli O157:H7 Phage UAE_MI-01 Isolated from Birds
Abstract
:1. Introduction
2. Results
2.1. Isolation of the Phage UAE_MI-01 Active against E. coli O157:H7
2.2. Host Range of the Phage UAE_MI-01
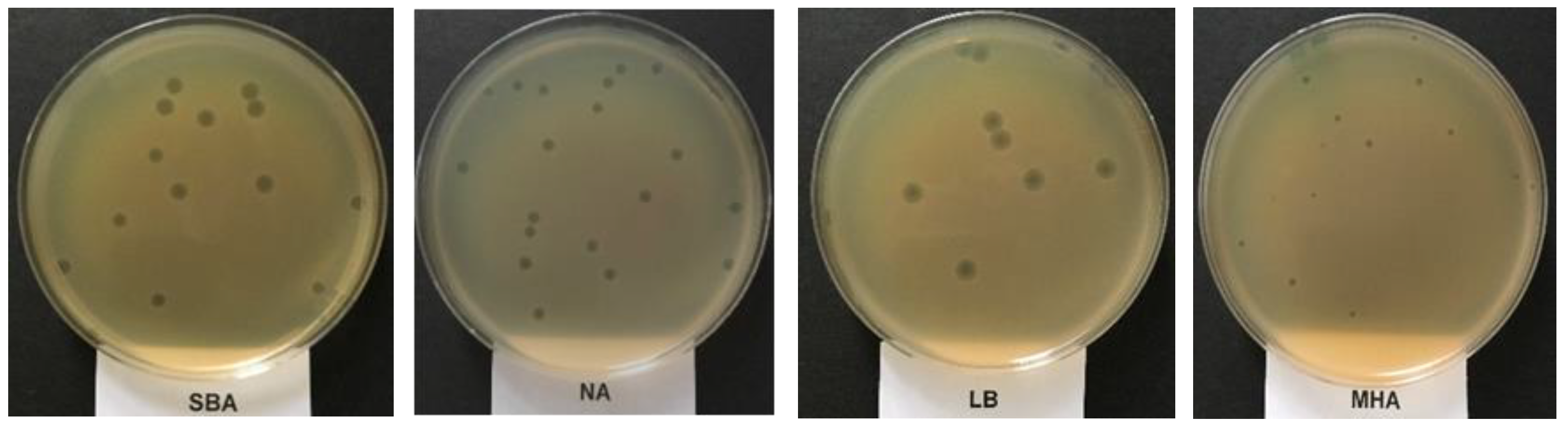
2.3. Plaque’s Morphology and the Determination of the Optimum Media for Phage UAE_MI-01 Propagation Based on Plaque Size
2.4. Effects of Temperature and pH on the Phage UAE_MI-01
2.5. Effects of Chemical Disinfectants on the Phage UAE_MI-01
2.6. Determination of the Adsorption Time, Adsorption Rate Constant, Latent Period and Burst Size of the Phage UAE_MI-01
2.7. Electron Microscopy of the Phage UAE_MI-01
2.8. Genomic Analysis and Bioinformatics of the Phage UAE_MI-01
3. Discussion
4. Materials and Methods
4.1. Media
4.2. Cultivation of the Propagation Host E. coli O157:H7
4.3. Isolation, Purification and Propagation of the Phage UAE_MI-01 using E. coli O157:H7 as the Propagation Host
4.4. Determination of the Host Range of the Phage UAE_MI-01
4.5. Plaque Morphology of the Phage UAE_MI-01 and Determination of the Optimum Media Based on Plaque Size
4.6. Evaluation of Physical and Chemical Agents on the Stability of the Phage UAE_MI-01
4.7. One-Step-Growth Curve to Determine the Adsorption Time, Adsorption Rate Constant, Latent Period and Burst Size of the Phage UAE_MI-01
4.8. Examination of the Phage UAE_MI-01 Using TEM
4.9. Isolation of the Phage UAE_MI-01 DNA
4.10. Genome Sequencing of the Phage UAE_MI-01
4.11. Bioinformatics of the Phage UAE_MI-01
4.12. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kadariya, J.; Smith, T.C.; Thapaliya, D. Staphylococcus aureus and staphylococcal food-borne disease: An ongoing challenge in public health. BioMed Res. Int. 2014, 2014, 827965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, S.P. Foodborne pathogens and disease special issue on the national and international pulse net network. Foodborne Pathog. Dis. 2019, 16, 439–440. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Lin, C.-W.; Wang, J.; Oh, D.H. Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omer, M.K.; Alvarez-Ordonez, A.; Prieto, M.; Skjerve, E.; Asehun, T.; Alvseike, O.A. A Systematic review of bacterial foodborne outbreaks related to red meat and meat products. Foodborne Pathog. Dis. 2018, 15, 598–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Authority, E.F.S.; Prevention, E.C. for D.; Control (ECDC) the European union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 2015, 13, 4329. [Google Scholar] [CrossRef]
- Coulombe, G.; Catford, A.; Martinez-Perez, A.; Buenaventura, E. Outbreaks of Escherichia coli O157:H7 infections linked to romaine lettuce in Canada from 2008 to 2018: An analysis of food safety context. J. Food Prot. 2020, 83, 1444–1462. [Google Scholar] [CrossRef]
- Berg, J.; McAllister, T.I.M.; Bach, S.; Stilborn, R.; Hancock, D.; LeJEUNE, J. Escherichia coli O157: H7 excretion by commercial feedlot cattle fed either barley-or corn-based finishing diets. J. Food Prot. 2004, 67, 666–671. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7. [Google Scholar] [CrossRef]
- Sperandio, V.; Nguyen, Y. Enterohemorrhagic E. coli (EHEC) pathogenesis. Front. Cell. Infect. Microbiol. 2012, 2, 90. [Google Scholar] [CrossRef] [Green Version]
- Pham-Khanh, N.H.; Sunahara, H.; Yamadeya, H.; Sakai, M.; Nakayama, T.; Yamamoto, H.; Miyanaga, K.; Kamei, K. Isolation, Characterization, and complete genome sequence of a tequatrovirus phage, Escherichia phage KIT03, which simultaneously infects Escherichia coli O157: H7 and Salmonella enterica. Curr. Microbiol. 2019, 76, 1130–1137. [Google Scholar] [CrossRef]
- Thomas, M.K.; Murray, R.; Flockhart, L.; Pintar, K.; Fazil, A.; Nesbitt, A.; Marshall, B.; Tataryn, J.; Pollari, F. Estimates of foodborne illness–related hospitalizations and deaths in Canada for 30 specified pathogens and unspecified agents. Foodborne Pathog. Dis. 2015, 12, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Arshadi, N.; Mousavi Gargari, S.L.; Amani, J.; Nazarian, S. Immunogenicity of inactivated Escherichia coli O157:H7 with Stx2B microparticle in mice. Iran. J. Basic Med. Sci. 2022, 25, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Bell, B.P.; Griffin, P.M.; Lozano, P.; Christie, D.L.; Kobayashi, J.M.; Tarr, P.I. Predictors of hemolytic uremic syndrome in children during a large outbreak of Escherichia coli O157: H7 infections. Pediatrics 1997, 100, e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavia, A.T.; Nichols, C.R.; Green, D.P.; Tauxe, R.V.; Mottice, S.; Greene, K.D.; Wells, J.G.; Siegler, R.L.; Brewer, E.D.; Hannon, D. Hemolytic-uremic syndrome during an outbreak of Escherichia coli O157: H7 infections in institutions for mentally retarded persons: Clinical and epidemiologic observations. J. Pediatr. 1990, 116, 544–551. [Google Scholar] [CrossRef]
- Carter, A.O.; Borczyk, A.A.; Carlson, J.A.; Harvey, B.; Hockin, J.C.; Karmali, M.A.; Krishnan, C.; Korn, D.A.; Lior, H. A severe outbreak of Escherichia coli O157: H7–associated hemorrhagic colitis in a nursing home. N. Engl. J. Med. 1987, 317, 1496–1500. [Google Scholar] [CrossRef]
- Wong, C.S.; Jelacic, S.; Habeeb, R.L.; Watkins, S.L.; Tarr, P.I. The Risk of the hemolytic–uremic syndrome after antibiotic treatment of Escherichia coli O157: H7 infections. N. Engl. J. Med. 2000, 342, 1930–1936. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Ku, H.-J.; Lee, D.-H.; Kim, Y.-T.; Shin, H.; Ryu, S.; Lee, J.-H. Characterization, and genomic study of the novel bacteriophage HY01 infecting both Escherichia coli O157: H7 and Shigella flexneri: Potential as a biocontrol agent in food. PLoS ONE 2016, 11, e0168985. [Google Scholar] [CrossRef] [Green Version]
- Fernández, L.; Gutiérrez, D.; García, P.; Rodríguez, A. The perfect bacteriophage for therapeutic applications—A quick guide. Antibiotics 2019, 8, 126. [Google Scholar] [CrossRef] [Green Version]
- Abdelkader, K.; Gerstmans, H.; Saafan, A.; Dishisha, T.; Briers, Y. The preclinical and clinical progress of bacteriophages and their lytic enzymes: The parts are easier than the whole. Viruses 2019, 11, 96. [Google Scholar] [CrossRef] [Green Version]
- Janczuk-Richter, M.; Marinović, I.; NIEDZIÓLKA-Jönsson, J.; Szot-Karpińska, K. Recent applications of bacteriophage-based electrodes: A mini-review. Electrochem. Commun. 2019, 99, 11–15. [Google Scholar] [CrossRef]
- Wittebole, X.; De Roock, S.; Opal, S.M. A Historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 2014, 5, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Borysowski, J.; Miedzybrodzki, R.; Gorski, A. Phage Therapy: Current Research and Applications; Caister Academic Press: Norfolk, UK, 2014; p. 406. [Google Scholar]
- Soffer, N.; Abuladze, T.; Woolston, J.; Li, M.; Hanna, L.F.; Heyse, S.; Charbonneau, D.; Sulakvelidze, A. Bacteriophages safely reduce Salmonella contamination in pet food and raw pet food ingredients. Bacteriophage 2016, 6, e1220347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abuladze, T.; Li, M.; Menetrez, M.Y.; Dean, T.; Senecal, A.; Sulakvelidze, A. Bacteriophages reduce experimental contamination of hard surfaces, tomato, spinach, broccoli, and ground beef by Escherichia coli O157: H7. Appl. Environ. Microbiol. 2008, 74, 6230–6238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Górski, A.; Międzybrodzki, R.; Węgrzyn, G.; Jończyk-Matysiak, E.; Borysowski, J.; Weber-Dąbrowska, B. Phage therapy: Current status and perspectives. Med. Res. Rev. 2020, 40, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Litt, P.K.; Jaroni, D. Isolation and Physiomorphological characterization of Escherichia coli O157: H7-infecting bacteriophages recovered from beef cattle operations. Int. J. Microbiol. 2017, 2017, 7013236. [Google Scholar] [CrossRef] [Green Version]
- Raya, R.R.; Varey, P.; Oot, R.A.; Dyen, M.R.; Callaway, T.R.; Edrington, T.S.; Kutter, E.M.; Brabban, A.D. Isolation and characterization of a new t-even bacteriophage, cev1, and determination of its potential to reduce Escherichia coli O157:H7 Levels in Sheep. Appl. Environ. Microbiol. 2006, 72, 6405–6410. [Google Scholar] [CrossRef] [Green Version]
- Sultan-Alolama, M.I.; El-Tarabily, K.A.; Vijayan, R. Complete genome sequence of Escherichia coli O157: H7 phage UAE_MI-01, isolated from bird feces. Microbiol. Resour. Announc. 2021, 10, e00348-21. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Waddell, T.; Meng, J.; Franklin, K.; Ackermann, H.-W.; Ahmed, R.; Mazzocco, A.; Yates, J.; Lingohr, E.J.; Johnson, R.P. The Host-range, genomics and proteomics of Escherichia coli O157: H7 bacteriophage rV5. Virol. J. 2013, 10, 76. [Google Scholar] [CrossRef] [Green Version]
- Callaway, T.R.; Edrington, T.S.; Brabban, A.D.; Anderson, R.C.; Rossman, M.L.; Engler, M.J.; Carr, M.A.; Genovese, K.J.; Keen, J.E.; Looper, M.L. Bacteriophage isolated from feedlot cattle can reduce Escherichia coli O157: H7 populations in ruminant gastrointestinal tracts. Foodborne Pathog. Dis. 2008, 5, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Morita, M.; Tanji, Y.; Mizoguchi, K.; Akitsu, T.; Kijima, N.; Unno, H. Characterization of a virulent bacteriophage specific for Escherichia coli O157: H7 and analysis of its cellular receptor and two tail fiber genes. FEMS Microbiol. Lett. 2002, 211, 77–83. [Google Scholar] [CrossRef]
- Lee, Y.-D.; Park, J.-H. Characterization and application of phages isolated from sewage for reduction of Escherichia coli O157: H7 in biofilm. LWT-Food Sci. Technol. 2015, 60, 571–577. [Google Scholar] [CrossRef]
- Park, M.; Lee, J.-H.; Shin, H.; Kim, M.; Choi, J.; Kang, D.-H.; Heu, S.; Ryu, S. Characterization, and comparative genomic analysis of a novel bacteriophage, sfp10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157: H7. Appl. Environ. Microbiol. 2012, 78, 58–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.-H.; Adeyemi, D.E.; Park, M.-K. Characterization of a new and efficient polyvalent phage infecting Escherichia coli O157: H7, Salmonella spp., and Shigella Sonnei. Microorganisms 2021, 9, 2105. [Google Scholar] [CrossRef]
- Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Grabski, M.; Topka, G.; Dydecka, A.; Kosznik-Kwaśnicka, K.; Lukasz, G.; Jurczak-Kurek, A.; Wolkowicz, T. Characterization of a bacteriophage, VB_Eco4M-7, that effectively infects many Escherichia coli O157 strains. Sci. Rep. 2020, 10, 3743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhang, C.; Wei, F.; Yu, F.; Zhao, Z. Bactericidal activity of a holin-endolysin system derived from Vibrio alginolyticus phage HH109. Microb. Pathog. 2021, 159, 105135. [Google Scholar] [CrossRef]
- Wang, I.-N.; Smith, D.L.; Young, R. Holins: The protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, Y.; Xu, X.; Ahmed, T.; Yang, Y.; Loh, B.; Leptihn, S.; Yan, C.; Chen, J.; Li, B. The holin-endolysin lysis system of the op2-like phage x2 infecting Xanthomonas oryzae Pv. oryzae. Viruses 2021, 13, 1949. [Google Scholar] [CrossRef]
- Li, S.; Liu, L.; Zhu, J.; Zou, L.; Li, M.; Cong, Y.; Rao, X.; Hu, X.; Zhou, Y.; Chen, Z. Characterization and genome sequencing of a novel coliphage isolated from engineered Escherichia coli. Intervirology 2010, 53, 211–220. [Google Scholar] [CrossRef]
- Davis, J.E.; Sinsheimer, R.L. The replication of bacteriophage ms2: 1. transfer of parental nucleic acid to progeny phage. J. Mol. Biol. 1963, 6, 203–207. [Google Scholar] [CrossRef]
- Green, M.R.; Hughes, H.; Sambrook, J.; MacCallum, P. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012; p. 2028. [Google Scholar]
- Vidaver, A.K.; Koski, R.K.; Van Etten, J.L. Bacteriophage Φ6: A lipid-containing virus of Pseudomonas phaseolicola. J. Virol. 1973, 11, 799–805. [Google Scholar] [CrossRef]
- Williams, S.T.; Wellington, E.M.H.; Tipler, L.S. The taxonomic implications of the reactions of representative nocardia strains to actinophage. Microbiology 1980, 119, 173–178. [Google Scholar] [CrossRef] [Green Version]
- Brownell, G.H.; Adams, J.N.; Bradley, S.G. Growth and characterization of nocardiophages for Nocardia canicruria and Nocardia erythropolis mating types. Microbiology 1967, 47, 247–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowding, J.E. Characterization of a bacteriophage virulent for Streptomyces coelicolor A3 (2). Microbiology 1973, 76, 163–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sykes, I.K.; Lanning, S.; Williams, S.T. The Effect of pH on soil actinophage. Microbiology 1981, 122, 271–280. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, H.-W.; Heldal, M. Basic electron microscopy of aquatic viruses. Man. Aquat. Viral Ecol. 2010, 18, 182–192. [Google Scholar]
- Brum, J.R.; Steward, G.F. Morphological characterization of viruses in the stratified water column of alkaline, hypersaline mono lake. Microb. Ecol. 2010, 60, 636–643. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. ProgressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [Green Version]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Mead, P.S.; Slutsker, L.; Dietz, V.; McCaig, L.F.; Bresee, J.S.; Shapiro, C.; Griffin, P.M.; Tauxe, R.V. Food-related Illness and death in the United States. Emerg. Infect. Dis. 1999, 5, 607. [Google Scholar] [CrossRef] [PubMed]
- Gambushe, S.M.; Zishiri, O.T.; El Zowalaty, M.E. Review of Escherichia Coli O157: H7 prevalence, pathogenicity, heavy metal and antimicrobial resistance, African perspective. Infect. Drug Resist. 2022, 15, 4645–4673. [Google Scholar] [CrossRef] [PubMed]





| Bacteria | Sensitivity to Phage UAE_MI-01 |
|---|---|
| Escherichia coli O157:H7 NCTC 12900 | + |
| Escherichia coli ATCC 25922 | − |
| Escherichia coli ATCC 8739 | + |
| Escherichia coli ESBL-producing (Patient isolate) | − |
| Escherichia coli ATCC 15223 | + |
| Escherichia coli ATCC 23227 | − |
| Escherichia coli ATCC 9637 | − |
| Escherichia coli ATCC 35218 | − |
| Escherichia coli ATCC 23224 | − |
| Bacillus subtilis ATCC 6051 | − |
| Pseudomonas aeruginosa ATCC 25668 | − |
| Pseudomonas aeruginosa ATCC 27853 | − |
| Methicillin-resistant Staphylococcus aureus (Patient isolate) | − |
| Staphylococcus aureus ATCC 6358 | − |
| Staphylococcus aureus ATCC 29213 | − |
| Staphylococcus epidermidis ATCC 12228 | − |
| Staphylococcus saprophyticus ATCC-BAA 750 | − |
| Streptococcus pyogenes ATCC 19615 | − |
| Enterococcus faecalis ATCC 51299 | − |
| Enterococcus faecalis (Patient isolate) | − |
| Enterococcus casseliflavus (Patient isolate) | − |
| Enterobacter aerogenes ATCC 13018 | − |
| Enterobacter hormaechei (Patient isolate) | − |
| Klebsiella pneumonia ESBL-producing ATCC 700603 | − |
| Klebsiella pneumonia KPC 2 +ve (Patient isolate) | − |
| Haemophilus influenzae ATCC 9007 | − |
| Stenotrophomonas maltophilia ATCC 17666 | − |
| Salmonella enterica ATCC 14028 | − |
| Salmonella sp. (Patient isolate) | − |
| Proteus vulgaris ATCC 29905 | − |
| Mycobacterium smegmatis ATCC 607 | − |
| Type of Medium | Soyabean Casein Digest Agar | Nutrient Agar | Luria-Bertani Agar | Muller Hinton Agar |
|---|---|---|---|---|
| (log10 pfu mL−1) | 6.59 ± 0.21 a | 7.81 ± 0.11 b | 5.53 ± 0.39 c | 6.56 ± 0.17 a |
| Temperature/Time | −20 °C (log10 pfu mL−1) | 4 °C (log10 pfu mL−1) | 25 °C (log10 pfu mL−1) | 45 °C (log10 pfu mL−1) | 55 °C (log10 pfu mL−1) | 65 °C (log10 pfu mL−1) | 75 °C (log10 pfu mL-1) | 100 °C (log10 pfu mL−1) |
|---|---|---|---|---|---|---|---|---|
| 15 min | 7.48 ± 0.10 Aa | 7.51 ± 0.16 Aa | 7.46 ± 0.13 Aa | 7.49 ± 0.14 Aa | 7.43 ± 0.18 Aa | 7.47 ± 0.19 Aa | 3.29 ± 0.09 Ab | 0.00 ± 0.00 Ac |
| 30 min | 7.49 ± 0.15 Aa | 7.53 ± 0.16 Aa | 7.42 ± 0.15 Aa | 7.48 ± 0.21 Aa | 7.46 ± 0.12 Aa | 7.43 ± 0.26 Aa | 0.00 ± 0.00 Bb | 0.00 ± 0.00 Ab |
| Temperature | 0:00 Time (log10 pfu mL−1) | After 1 Month (log10 pfu mL−1) | After 5 Months (log10 pfu mL−1) |
|---|---|---|---|
| −20 °C | 7.49 ± 0.07 Aa | 7.42 ± 0.14 Aa | 7.10 ± 0.15 Ab |
| 4 °C | 7.48 ± 0.15 Aa | 7.39 ± 0.15 Aa | 7.47 ± 0.11 Ba |
| 25 °C | 7.41 ± 0.18 Aa | 7.44 ± 0.17 Aa | 6.79 ± 0.28 Cb |
| pH Value | 3 | 4 | 7 | 9 | 10 |
|---|---|---|---|---|---|
| (log10 pfu mL−1) | 0.00 ± 0.00 a | 5.72 ± 0.12 b | 7.79 ± 0.11 c | 7.76 ± 0.15 c | 7.78 ± 0.16 c |
| Disinfectant | Initial Titer (log10 pfu mL−1) | After 2 min (log10 pfu mL−1) | After 3 min (log10 pfu mL−1) | Percentage of Survivors (%) |
|---|---|---|---|---|
| Ethanol 70% | 7.48 ± 0.11 Aa | 6.62 ± 0.06 Ab | 4.57 ± 0.11 Ac | 0.13% ± 0.02 A |
| Liquid hand wash soap | 7.44 ± 0.18 Aa | 7.11 ± 0.07 Bb | 0.00 ± 0.00 Bc | 0.00% ± 0.00 B |
| Sodium hypochlorite 2% | 7.51 ± 0.15 Aa | 6.09 ± 0.10 Cb | 0.00 ± 0.00 Bc | 0.00% ± 0.00 B |
| Commercial disinfectant 20% | 7.45 ± 0.10 Aa | 7.09 ± 0.06 Bb | 7.06 ± 0.13 Cb | 66% ± 0.21 C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sultan-Alolama, M.I.; Amin, A.; El-Tarabily, K.A.; Vijayan, R. Characterization and Genomic Analysis of Escherichia coli O157:H7 Phage UAE_MI-01 Isolated from Birds. Int. J. Mol. Sci. 2022, 23, 14846. https://doi.org/10.3390/ijms232314846
Sultan-Alolama MI, Amin A, El-Tarabily KA, Vijayan R. Characterization and Genomic Analysis of Escherichia coli O157:H7 Phage UAE_MI-01 Isolated from Birds. International Journal of Molecular Sciences. 2022; 23(23):14846. https://doi.org/10.3390/ijms232314846
Chicago/Turabian StyleSultan-Alolama, Mohamad Ismail, Amr Amin, Khaled A. El-Tarabily, and Ranjit Vijayan. 2022. "Characterization and Genomic Analysis of Escherichia coli O157:H7 Phage UAE_MI-01 Isolated from Birds" International Journal of Molecular Sciences 23, no. 23: 14846. https://doi.org/10.3390/ijms232314846
APA StyleSultan-Alolama, M. I., Amin, A., El-Tarabily, K. A., & Vijayan, R. (2022). Characterization and Genomic Analysis of Escherichia coli O157:H7 Phage UAE_MI-01 Isolated from Birds. International Journal of Molecular Sciences, 23(23), 14846. https://doi.org/10.3390/ijms232314846







