HDAC3 Knockdown Dysregulates Juvenile Hormone and Apoptosis-Related Genes in Helicoverpa armigera
Abstract
:1. Introduction
2. Results
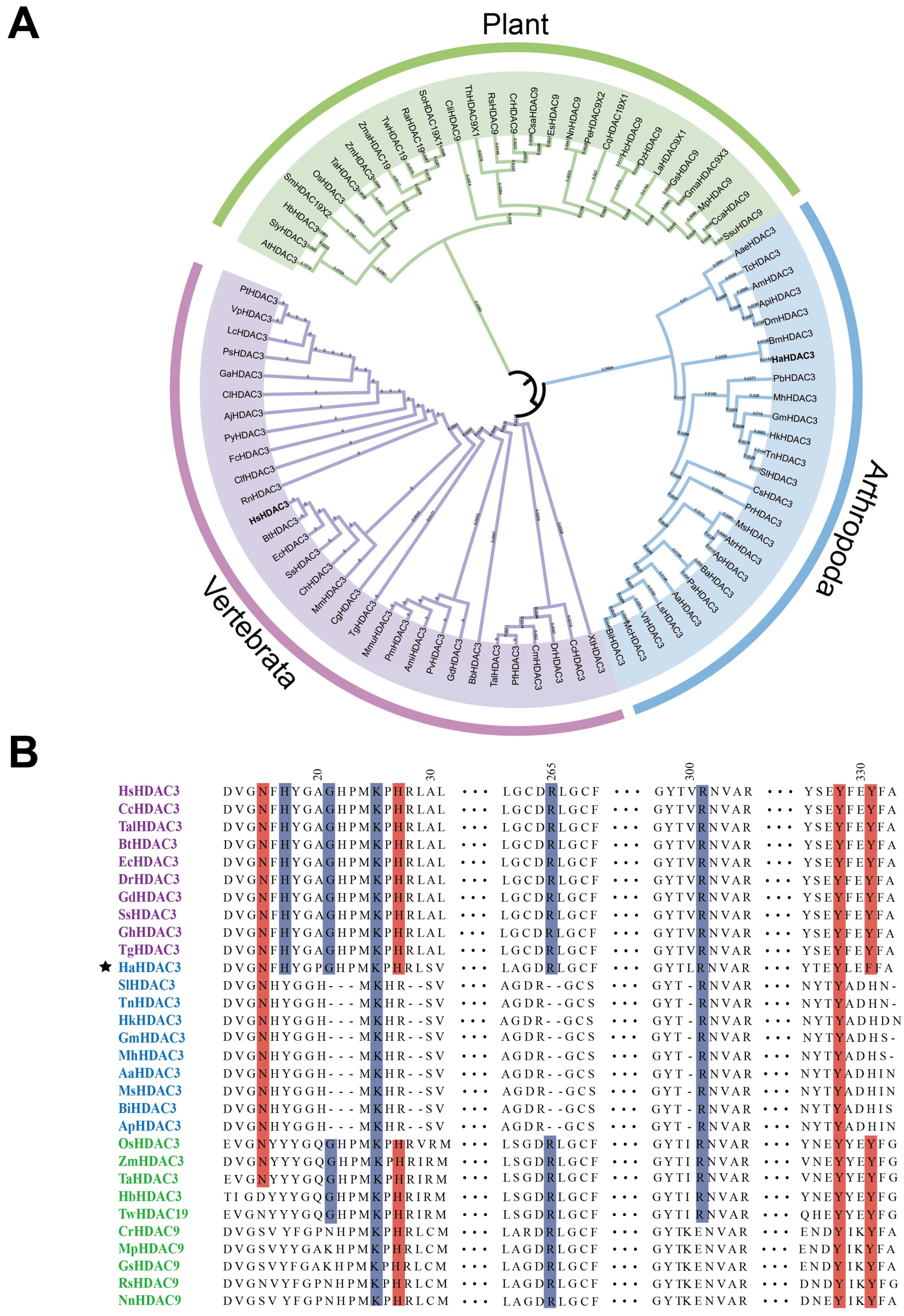
2.1. Construction of HDAC3 Phylogenetic Trees
2.2. Analysis of HDAC3 Expression in Various Tissues and Developmental Stages
2.3. Subcellular HaHDAC3 Localization
2.4. HDAC3 Is Required for Larval, Pupal, and Adult Survival
2.5. HDAC3 Knockdown Affects Histone H3 Acetylation
2.6. Knockdown of HDAC3 Suppresses the Expression of Genes Involved in JH Action and Promotes the Expression of Genes to Apoptosis Pathway
3. Discussion
4. Materials and Methods
4.1. Phylogenetic Tree Construction and Sequence Alignment
4.2. Insect Rearing and Feeding Experiments
4.3. Analysis of HaHDAC3 Expression and Localization in Sf9 Cells
4.4. Expression Pattern Analysis
4.5. Small Interfering RNA (siRNA) Synthesis and Microinjection
4.6. Protein Extraction
4.7. Western Blot Analysis
4.8. Transcriptome Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, H.; Shi, Y.; Wang, L.; Liu, S.; Wu, S.; Yang, Y.; Feyereisen, R.; Wu, Y. CYP6AE gene cluster knockout in Helicoverpa armigera reveals role in detoxification of phytochemicals and insecticides. Nat. Commun. 2018, 9, 4820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerns, D.D.; Yang, F.; Kerns, D.L.; Stewart, S.D. Evaluation of Bt resistance in Helicoverpa zea (Lepidoptera: Noctuidae) strains using various Bt cotton plant tissues. Pest Manag. Sci. 2022, 78, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Tavernarakis, N.; Wang, S.L.; Dorovkov, M.; Ryazanov, A.; Driscoll, M. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat. Genet. 2000, 24, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.Y.; Vander Meer, R.K. Phenotypic Effects of PBAN RNAi Using Oral Delivery of dsRNA to Corn Earworm (Lepidoptera: Noctuidae) and Tobacco Budworm Larvae. J. Econ. Entomol. 2019, 112, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Zeng, H.; Zhang, Y.; Xu, D.; Qiu, D. Silencing the HaHR3 gene by transgenic plant-mediated RNAi to disrupt Helicoverpa armigera development. Int. J. Biol. Sci. 2013, 9, 370–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grishok, A.; Tabara, H.; Mello, C.C.; Grishok, A.; Tabara, H.; Mello, C.C. Genetic Requirements for Inheritance of RNAi in C. elegans. Science 2016, 287, 2494–2497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.; Xu, S.; Lu, H.; Li, F.; Li, S.; Chang, L.; Heckel, D.G.; Bock, R.; Zhang, J. Resistance to RNA interference by plant-derived double-stranded RNAs but not plant-derived short interfering RNAs in Helicoverpa armigera. Plant Cell Environ. 2022, 45, 1930–1941. [Google Scholar] [CrossRef]
- Shukla, J.N.; Kalsi, M.; Sethi, A.; Narva, K.E.; Fishilevich, E.; Singh, S.; Mogilicherla, K.; Palli, S.R. Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 2016, 13, 656–669. [Google Scholar] [CrossRef]
- Kim, Y.J.; Wang, R.; Gao, L.; Li, D.; Xu, C.; Mang, H.; Jeon, J.; Chen, X.; Zhong, X.; Kwak, J.M.; et al. POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 14858–14863. [Google Scholar] [CrossRef] [Green Version]
- Backs, J.; Olson, E.N. Control of cardiac growth by histone acetylation/deacetylation. Circ. Res. 2006, 98, 15–24. [Google Scholar] [CrossRef]
- Watson, P.J.; Fairall, L.; Santos, G.M.; Schwabe, J.W.R. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 2012, 481, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Yiew, K.H.; Chatterjee, T.K.; Hui, D.Y.; Weintraub, N.L. Histone deacetylases and cardiometabolic diseases. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1914–1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, M.; Zhao, Q.; Zhang, H.; Chen, Y.; Yuan, Z.; Xu, Y.; Zhang, M. Proteomic Analysis of HDAC3 Selective Inhibitor in the Regulation of Inflammatory Response of Primary Microglia. Neural Plast. 2017, 2017, 6237351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Tong, Q.; Zhang, Z.; Wang, S.; Zheng, Y.; Liu, Q.; Qian, L.B.; Chen, S.Y.; Sun, J.; Cai, L. Inhibition of HDAC3 prevents diabetic cardiomyopathy in OVE26 mice via epigenetic regulation of DUSP5-ERK1/2 pathway. Clin. Sci. 2017, 131, 1841–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emmett, M.J.; Lazar, M.A. Integrative regulation of physiology by histone deacetylase 3. Nat. Rev. Mol. Cell Biol. 2019, 20, 102–115. [Google Scholar] [CrossRef]
- Yang, W.M.; Yao, Y.L.; Sun, J.M.; Davie, J.R.; Seto, E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem. 1997, 272, 28001–28007. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Kalkum, M.; Chait, B.T.; Roeder, R.G. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 2002, 9, 611–623. [Google Scholar] [CrossRef]
- Bhaskara, S.; Chyla, B.J.; Amann, J.M.; Knutson, S.K.; Cortez, D.; Sun, Z.W.; Hiebert, S.W. Deletion of Histone Deacetylase 3 Reveals Critical Roles in S Phase Progression and DNA Damage Control. Mol. Cell 2008, 30, 61–72. [Google Scholar] [CrossRef] [Green Version]
- Guenther, M.G.; Lane, W.S.; Fischle, W.; Verdin, E.; Lazar, M.A.; Shiekhattar, R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000, 14, 1048–1057. [Google Scholar] [CrossRef]
- Gryder, B.E.; Wu, L.; Woldemichael, G.M.; Pomella, S.; Quinn, T.R.; Park, P.M.C.; Cleveland, A.; Stanton, B.Z.; Song, Y.; Rota, R.; et al. Chemical genomics reveals histone deacetylases are required for core regulatory transcription. Nat. Commun. 2019, 10, 3004. [Google Scholar] [CrossRef]
- Zhu, C.C.; Bornemann, D.J.; Zhitomirsky, D.; Miller, E.L.; O’Connor, M.B.; Simon, J.A. Drosophila histone deacetylase-3 controls imaginal disc size through suppression of apoptosis. PLoS Genet. 2008, 4, e1000009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foglietti, C.; Filocamo, G.; Cundari, E.; De Rinaldis, E.; Lahm, A.; Cortese, R.; Steinkühler, C. Dissecting the biological functions of Drosophila histone deacetylases by RNA interference and transcriptional profiling. J. Biol. Chem. 2006, 281, 17968–17976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- George, S.; Palli, S.R. Histone Deacetylase 11 Knockdown Blocks Larval Development and Metamorphosis in the Red Flour Beetle, Tribolium castaneum. Front. Genet. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- George, S.; Gaddelapati, S.C.; Palli, S.R. Histone deacetylase 1 suppresses Krüppel homolog 1 gene expression and influences juvenile hormone action in Tribolium castaneum. Proc. Natl. Acad. Sci. USA 2019, 116, 17759–17764. [Google Scholar] [CrossRef] [Green Version]
- Chandra, S.; Albishi, N.M.; Kumar, R.; Reddy, S. Juvenile hormone-induced histone deacetylase 3 suppresses apoptosis to maintain larval midgut in the yellow fever mosquito. Proc. Natl. Acad. Sci. USA 2022, 119, e2118871119. [Google Scholar] [CrossRef]
- Malvaez, M.; McQuown, S.C.; Rogge, G.A.; Astarabadi, M.; Jacques, V.; Carreiro, S.; Rusche, J.R.; Wood, M.A. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc. Natl. Acad. Sci. USA 2013, 110, 2647–2652. [Google Scholar] [CrossRef] [Green Version]
- Chi, Z.; Chen, S.; Xu, T.; Zhen, W.; Yu, W.; Jiang, D.; Guo, X.; Wang, Z.; Zhang, K.; Li, M.; et al. Histone Deacetylase 3 Couples Mitochondria to Drive IL-1β-Dependent Inflammation by Configuring Fatty Acid Oxidation. Mol. Cell 2020, 80, 43–58.e7. [Google Scholar] [CrossRef]
- Kayukawa, T.; Nagamine, K.; Ito, Y.; Nishita, Y.; Ishikawa, Y.; Shinoda, T. Krüppel homolog 1 inhibits insect metamorphosis via direct transcriptional repression of broad-complex, a pupal specifier gene. J. Biol. Chem. 2016, 291, 1751–1762. [Google Scholar] [CrossRef] [Green Version]
- Staley, B.K.; Irvine, K.D. Hippo signaling in Drosophila: Recent advances and insights. Dev. Dyn. 2012, 241, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Bao, Y.; Hata, Y.; Ikeda, M.; Withanage, K. Mammalian Hippo pathway: From development to cancer and beyond. J. Biochem. 2011, 149, 361–379. [Google Scholar] [CrossRef]
- Guo, C.; Gow, C.H.; Li, Y.; Gardner, A.; Khan, S.; Zhang, J. Regulated clearance of histone deacetylase 3 protects independent formation of nuclear receptor corepressor complexes. J. Biol. Chem. 2012, 287, 12111–12120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, S.H.; Liao, X.; Weiss, R.E.; Lazar, M.A. The interaction between nuclear receptor corepressor and histone deacetylase 3 regulates both positive and negative thyroid hormone action in vivo. Mol. Endocrinol. 2010, 24, 1359–1367. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Hsieh, J. HDAC3 controls gap 2/mitosis progression in adult neural stem/progenitor cells by regulating CDK1 levels. Proc. Natl. Acad. Sci. USA 2014, 111, 13541–13546. [Google Scholar] [CrossRef] [Green Version]
- Ishii, S.; Kurasawa, Y.; Wong, J.; Yu-Lee, L.Y. Histone deacetylase 3 localizes to the mitotic spindle and is required for kinetochore-microtubule attachment. Proc. Natl. Acad. Sci. USA 2008, 105, 4179–4184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rundlett, S.E.; Carmen, A.A.; Kobayashi, R.; Bavykin, S.; Turner, B.M.; Grunstein, M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 1996, 93, 14503–14508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Wharton, W.; Yuan, Z.; Tsai, S.-C.; Olashaw, N.; Seto, E. Activation of the Growth-Differentiation Factor 11 Gene by the Histone Deacetylase (HDAC) Inhibitor Trichostatin A and Repression by HDAC3. Mol. Cell. Biol. 2004, 24, 5106–5118. [Google Scholar] [CrossRef] [Green Version]
- Ozawa, T.; Mizuhara, T.; Arata, M.; Shimada, M.; Niimi, T.; Okada, K.; Okada, Y.; Ohta, K. Histone deacetylases control module-specific phenotypic plasticity in beetle weapons. Proc. Natl. Acad. Sci. USA 2016, 113, 15042–15047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kayukawa, T.; Jouraku, A.; Ito, Y.; Shinoda, T. Molecular mechanism underlying juvenile hormone-mediated repression of precocious larval-adult metamorphosis. Proc. Natl. Acad. Sci. USA 2017, 114, 1057–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konopova, B.; Smykal, V.; Jindra, M. Common and distinct roles of juvenile hormone signaling genes in metamorphosis of holometabolous and hemimetabolous insects. PLoS ONE 2011, 6, e28728. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, Y.; Truman, J.W.; Riddiford, L.M. The role of broad in the development of Tribolium castaneum: Implications for the evolution of the holometabolous insect pupa. Development 2008, 135, 569–577. [Google Scholar] [CrossRef]
- Zhou, X.; Riddiford, L.M. Broad specifies pupal development and mediates the ‘status quo’ action of juvenile hormone on the pupal-adult transformation in Drosophila and Manduca. Development 2002, 129, 2259–2269. [Google Scholar] [CrossRef] [PubMed]
- Konopova, B.; Jindra, M. Broad-Complex acts downstream of Met in juvenile hormone signaling to coordinate primitive holometabolan metamorphosis. Development 2008, 135, 559–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minakuchi, C.; Namiki, T.; Shinoda, T. Krüppel homolog 1, an early juvenile hormone-response gene downstream of Methoprene-tolerant, mediates its anti-metamorphic action in the red flour beetle Tribolium castaneum. Dev. Biol. 2009, 325, 341–350. [Google Scholar] [CrossRef] [Green Version]
- Aishwarya, S.; Ambikai, G.; Lori, A. Epigenetic regulation of transcription in Drosophila Aishwarya. Front. Biosci. 2012, 12, 909–937. [Google Scholar]
- Walshe, D.P.; Lehane, S.M.; Lehane, M.J.; Haines, L.R. Prolonged gene knockdown in the tsetse fly Glossina by feeding double stranded RNA. Insect Mol. Biol. 2009, 18, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef]
- Mao, Y.B.; Cai, W.J.; Wang, J.W.; Hong, G.J.; Tao, X.Y.; Wang, L.J.; Huang, Y.P.; Chen, X.Y. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007, 25, 1307–1313. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Krieg, R.C.; Dong, Y.; Schwamborn, K.; Knuechel, R. Protein quantification and its tolerance for different interfering reagents using the BCA-method with regard to 2D SDS PAGE. J. Biochem. Biophys. Methods 2005, 65, 13–19. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, H.; Xu, Z.; Li, W.; Cai, C.; Wang, W.; Ge, P.; Jia, X.; Li, Y.; Ding, T.; Ma, W.; et al. HDAC3 Knockdown Dysregulates Juvenile Hormone and Apoptosis-Related Genes in Helicoverpa armigera. Int. J. Mol. Sci. 2022, 23, 14820. https://doi.org/10.3390/ijms232314820
Chang H, Xu Z, Li W, Cai C, Wang W, Ge P, Jia X, Li Y, Ding T, Ma W, et al. HDAC3 Knockdown Dysregulates Juvenile Hormone and Apoptosis-Related Genes in Helicoverpa armigera. International Journal of Molecular Sciences. 2022; 23(23):14820. https://doi.org/10.3390/ijms232314820
Chicago/Turabian StyleChang, Huimin, Zhenlu Xu, Wenkang Li, Chenggu Cai, Wenjing Wang, Pengliang Ge, Xue Jia, Yingge Li, Tianze Ding, Wei Ma, and et al. 2022. "HDAC3 Knockdown Dysregulates Juvenile Hormone and Apoptosis-Related Genes in Helicoverpa armigera" International Journal of Molecular Sciences 23, no. 23: 14820. https://doi.org/10.3390/ijms232314820
APA StyleChang, H., Xu, Z., Li, W., Cai, C., Wang, W., Ge, P., Jia, X., Li, Y., Ding, T., Ma, W., Banaei-Moghaddam, A. M., Mo, H., & Ren, M. (2022). HDAC3 Knockdown Dysregulates Juvenile Hormone and Apoptosis-Related Genes in Helicoverpa armigera. International Journal of Molecular Sciences, 23(23), 14820. https://doi.org/10.3390/ijms232314820





