Stu-miR827-Targeted StWRKY48 Transcription Factor Negatively Regulates Drought Tolerance of Potato by Increasing Leaf Stomatal Density
Abstract
1. Introduction
2. Results
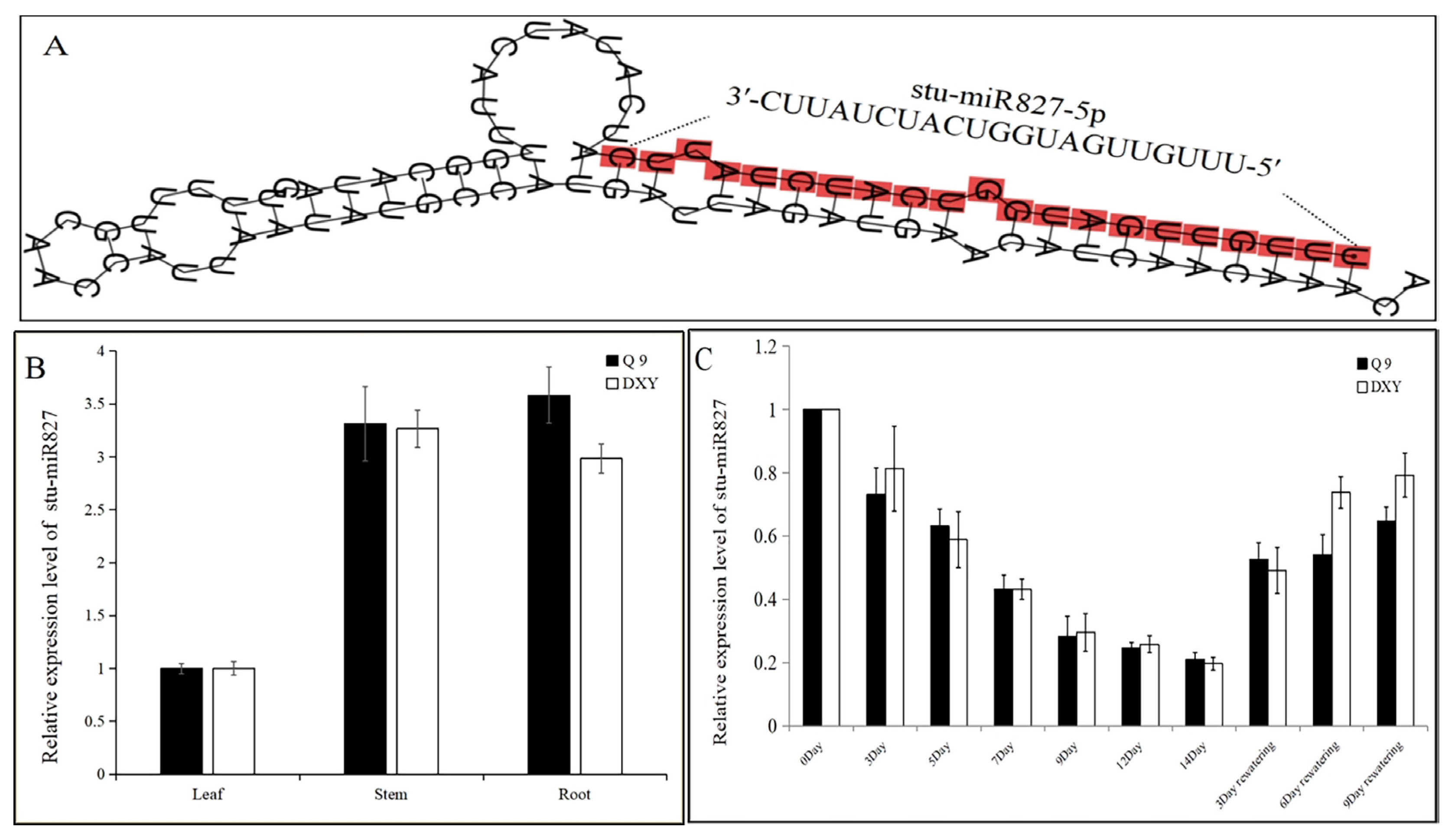
2.1. Identification, Characterization, and Expression Analysis of stu-miR827-5p
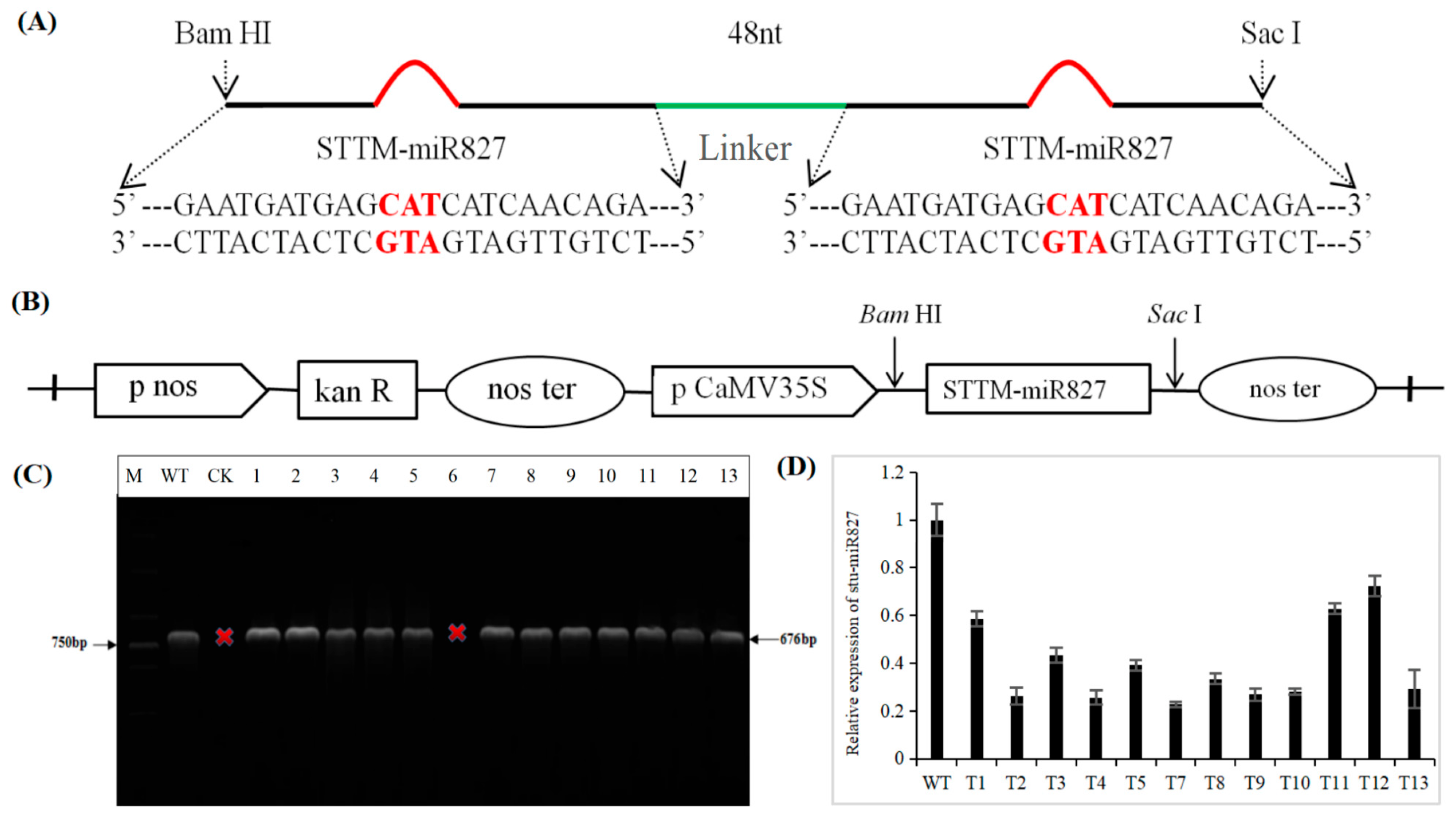
2.2. Generation of Transgenic Potato with Repressed stu-miR827-5p Expression by STTM Approach
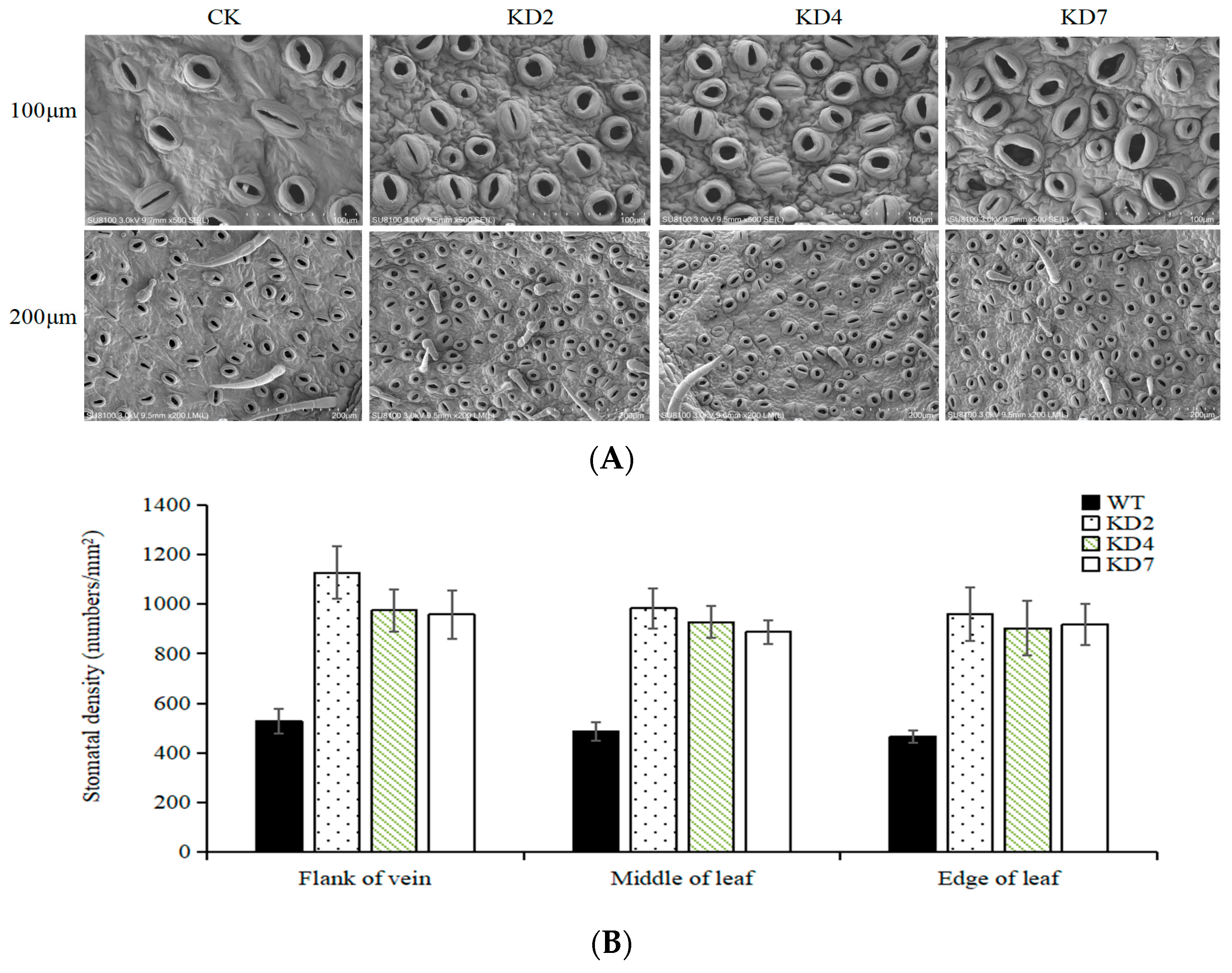
2.3. Suppression of microRNA827-5p Increases Stomatal Density
2.4. Suppression of microRNA827-5p Reduced Drought Tolerance
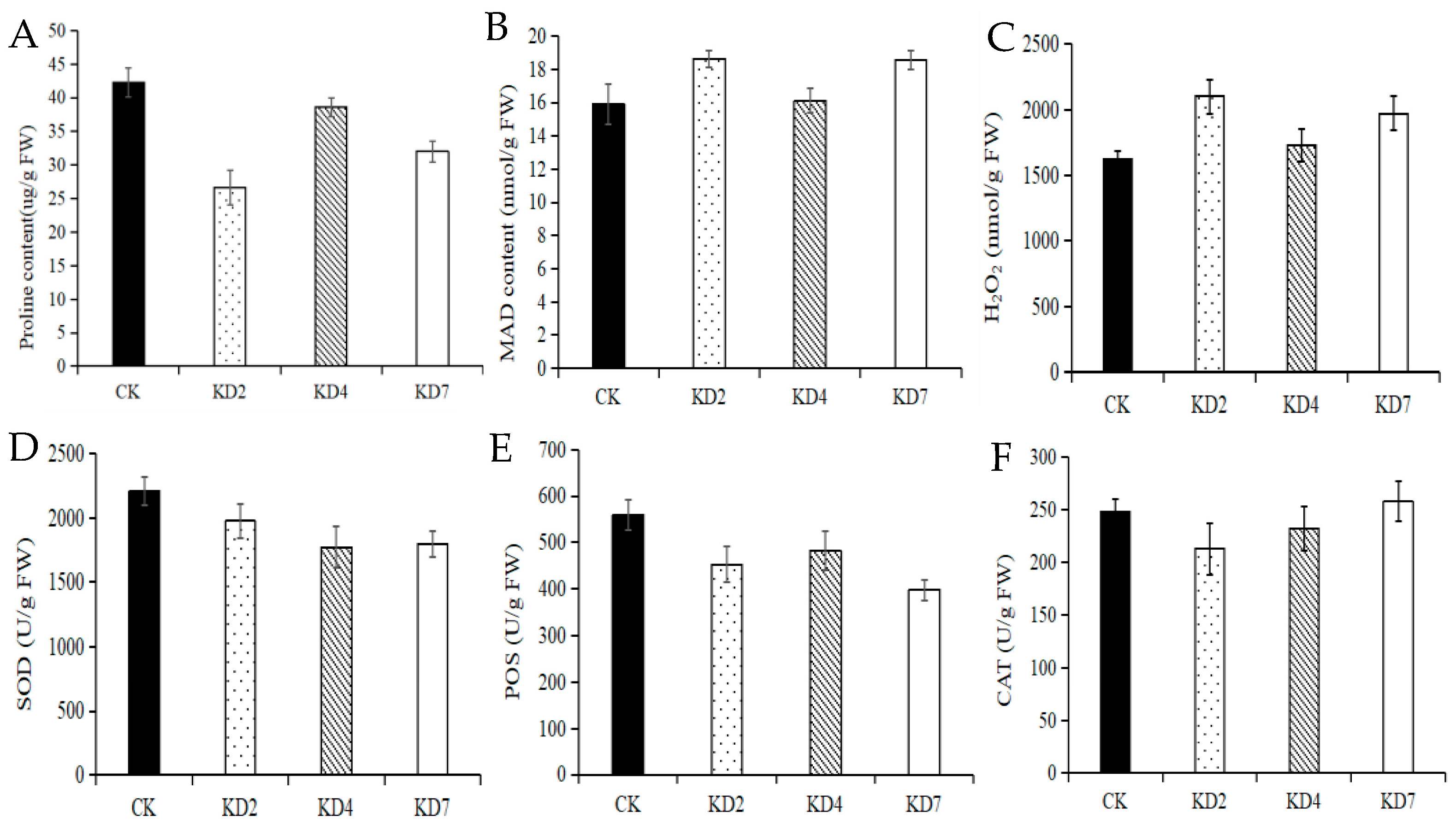
2.5. Suppression of microRNA827-5p Altered Drought-Relevant Physiological Indexes
2.6. Identification of stu-miR827-5p-Targeted StWRKY48 Gene
2.7. Subcellular Localization and Expression Pattern of StWRKY48
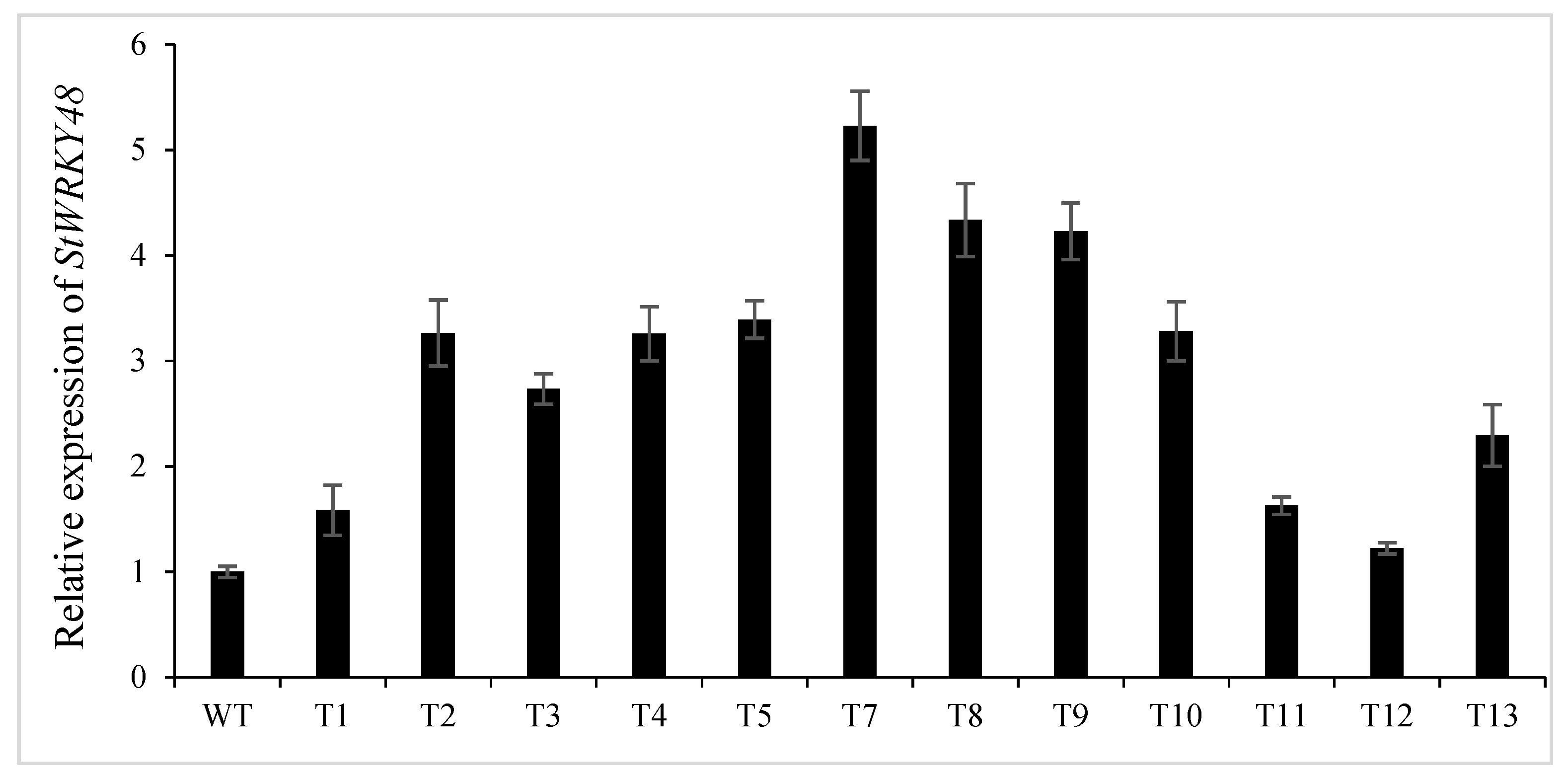
2.8. Knockdown of stu-miR827-5p Increases the Levels of StWRKY48
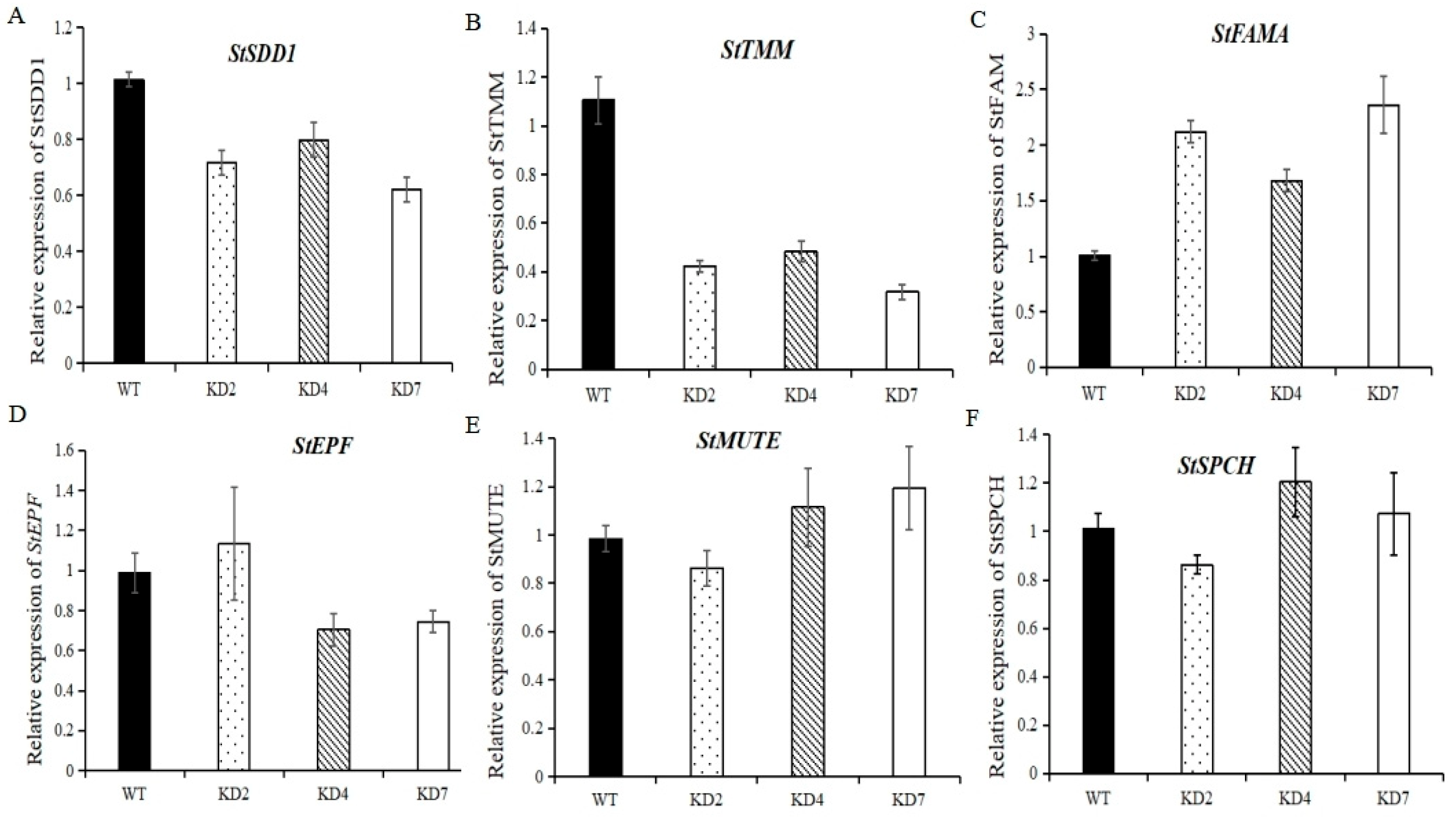
2.9. Suppression of microRNA827-5p Affects the Expression of Stomata Development-Related Genes
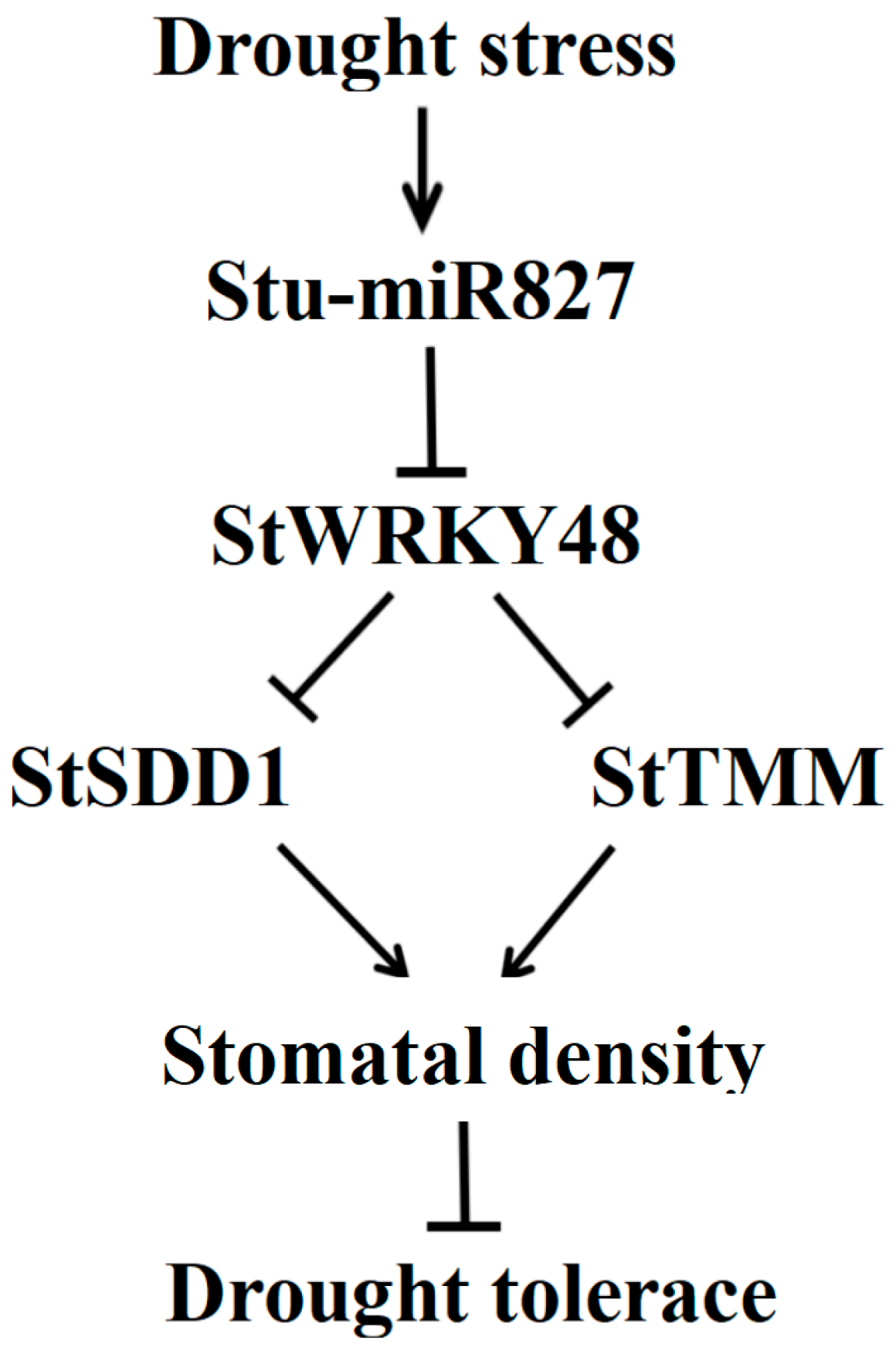
3. Discussion
3.1. Stu-miR827-Mediated StWRKY48 mRNA Cleavage In Vivo
3.2. Stu-miR827 Negatively Regulates Drought Tolerance in Potato
3.3. Stu-miR827 Positively Regulates Stomatal Density by Suppressing the Expression of StSDD1 and StTMM Genes
4. Methods
4.1. The Quantitative Real-Time PCR (qRT-PCR) Analysis
4.2. Construction of Silencing Plasmid of stu-miR827-5p
4.3. Transformation of Potato and Identification of Transgenic Lines
4.4. Analysis of Transgenic Potatoes’ Resistance to Drought Stress
4.5. Measurement of Reactive Oxygen Species (ROS), Proline, and Malonaldehyde (MDA) Levels and Antioxidant Enzyme Activity
4.6. Stomatal Density Measurement
4.7. Prediction and Validation of stu-miR827a-5p Target Genes
4.8. Subcellular Localization Assay
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Le, T.N.; McQueen-Mason, S.J. Desiccation-tolerant plants in dry environments. Life Extrem. Environ. 2006, 5, 269–279. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Dow, G.J.; Bergmann, D.C. Patterning and processes: How stomatal development defines physiological potential. Curr. Opin. Plant Biol. 2014, 21, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.Z.; Zhang, L.; Han, Z.F.; Yang, X.R.; Liu, W.J.; Li, E.T.; Chang, J.B.; Qi, Y.J.; Shpak, E.D.; Chai, J.J. A receptor-like protein acts as a specificity switch for the regulation of stomatal development. Genes Dev. 2017, 31, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Woolfenden, H.C.; Baillie, A.L.; Gray, J.E.; Hobbs, J.K.; Morris, R.J.; Fleming, A.J. Models and mechanisms of stomatal mechanics. Trends Plant Sci. 2018, 23, 822–832. [Google Scholar] [CrossRef]
- McKown, K.H.; Bergmann, D.C. Stomatal development in the grasses: Lessons from models and crops (and crop models). New Phytol. 2020, 227, 1636–1648. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, M.; Zhang, M.; Jiang, W.; Ren, X.; Liang, E.; Zhang, D.; Zhang, C.; Xiao, N.; Li, Y.; et al. A maize phytochrome-interacting factors protein ZmPIF1 enhances drought tolerance by inducing stomatal closure and improves grain yield in Oryza sativa. Plant Biotechnol. J. 2018, 16, 1375–1387. [Google Scholar] [CrossRef]
- Yoo, C.Y.; Pence, H.E.; Jin, J.B.; Miura, K.; Gosney, M.J.; Hasegawa, P.M.; Mickelbart, M.V. The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 2010, 22, 4128–4141. [Google Scholar] [CrossRef]
- Guo, X.Y.; Wang, Y.; Zhao, P.X.; Xu, P.; Yu, G.H.; Zhang, L.Y.; Xiong, Y.; Xiang, C.B. AtEDT1/HDG11 regulates stomatal density and water-use efficiency via ERECTA and E2Fa. New Phytol. 2019, 223, 1478–1488. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Seki, M.; Shinozaki, K.; Yokoyama, S. DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci. 2013, 18, 267–276. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA- binding protein, SPF1, that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef]
- Pnueli, L.; Hallak-Herr, E.; Rozenberg, M. Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J. 2002, 31, 319–330. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef]
- Zhou, Q.; Tian, A.; Zou, H.; Xie, Z.; Lei, G.; Huang, J. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef]
- Qiu, Y.; Yu, D. Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 2009, 65, 35–47. [Google Scholar] [CrossRef]
- Wu, X.; Kishitani, S.; Ito, Y.; Toriyama, K. Accumulation of raffinose in rice seedlings overexpressing OsWRKY11 in relation to desiccation tolerance. Plant Biotechnol. 2009, 26, 431–434. [Google Scholar] [CrossRef]
- Wang, C.; Deng, P.; Chen, L.; Wang, X.; Ma, H.; Hu, W. A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS ONE 2013, 8, e65120. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hu, Y.B.; Wang, H.S.; Feng, S.J.; Zhang, Y.T. Involvement of miR156 in the regulation of vegetative phase change in plants. J. Am. Soc. Hortic. Sci. 2015, 140, 387–395. [Google Scholar] [CrossRef]
- Boccara, M.; Sarazin, A.; Thiébeauld, O.; Jay, F.; Voinnet, O.; Navarro, L.; Colot, V. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 2014, 10, e1003883. [Google Scholar] [CrossRef]
- Alonso-Peral, M.M.; Li, J.; Li, Y.; Allen, R.S.; Schnippenkoetter, W.; Ohms, S.; White, R.G.; Millar, A.A. The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol. 2010, 154, 757–771. [Google Scholar] [CrossRef] [PubMed]
- Marie, T.; Narasimha, R.N.; Mathieu, B.; Stephanie, C.; Suresh, D.; Sajag, A.; Shivaram, P.A.; Oliver, Y.; Senthil, S. Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 2013, 162, 2042–2055. [Google Scholar]
- Zhang, L.; Yao, L.; Zhang, N.; Yang, J.; Zhu, X.; Tang, X.; Calderón-Urrea, A.; Si, H. Lateral root development in potato is mediated by stu-mi164 regulation of NAC transcription factor. Front. Plant Sci. 2018, 29, 383. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef]
- Fujii, H.; Chiou, T.J.; Lin, S.I.; Aung, K.; Zhu, J.K. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 2005, 15, 2038–2043. [Google Scholar] [CrossRef]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef]
- Khraiwesh, B.; Zhu, J.K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta Biochim. Biophys. Acta 2012, 1819, 137–148. [Google Scholar] [CrossRef]
- Sunkar, R.; Li, Y.F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef]
- Bartel, B.; Bartel, D.P. MicroRNAs: At the root of plant development? Plant Physiol. 2003, 132, 709–717. [Google Scholar] [CrossRef]
- Bonnet, E.; Wuyts, J.; Rouze, P.; Van de Peer, Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA 2004, 101, 11511–11516. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Michael, H.; Bu, J.S.; Perry, G.; Peter, L. Characterization of phosphorus-regulated miR399 and miR827 and their isomirs in barley under phosphorus-sufficient and phosphorus-deficient conditions. BMC Plant Biol. 2013, 13, 214. [Google Scholar]
- Zhang, R.; Marshall, D.; Bryan, G.J.; Hornyik, C. Identification and characterization of miRNA transcriptome in potato by high-throughput sequencing. PLoS ONE 2013, 8, e57233. [Google Scholar] [CrossRef]
- Hackenberg, M.; Huang, P.J.; Huang, C.Y.; Shi, B.J.; Gustafson, P.; Langridge, P.A. Comprehensive expression profile of microRNAs and other classes of non-coding small RNAs in barley under phosphorous-deficient and -sufficient conditions. DNA Res. 2013, 20, 109–125. [Google Scholar] [CrossRef]
- Ferdous, J.; Whitford, R.; Nguyen, M.; Brien, C.; Langridge, P.; Tricker, P.J. Drought-inducible expression of Hv-miR827 enhances drought tolerance in transgenic barley. Funct. Integr. Genom. 2017, 17, 279–292. [Google Scholar] [CrossRef]
- Tang, G.; Yan, J.; Gu, Y.; Qiao, M.; Fan, R.; Mao, Y.; Tang, X. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 2012, 58, 118–125. [Google Scholar] [CrossRef]
- Yan, J.; Gu, Y.; Jia, X.; Kang, W.; Pan, S.; Tang, X.; Chen, X.; Tang, G. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 2012, 24, 415–427. [Google Scholar] [CrossRef]
- Guillaumie, S.; Mzid, R.; Méchin, V. The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol. Biol. 2010, 72, 215–234. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Avci, U.; Nakashima, J.; Hahn, M.G.; Chen, F.; Dixon, R.A. Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem bio-mass in dicotyledonous plants. Proc. Natl. Acad. Sci. USA 2010, 107, 22338–22343. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.C.; Dugas, D.V.; Bartel, D.P.; Bartel, B. MicroRNA regulation of NAC domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 2004, 14, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.S.; Xie, Q.; Fei, J.F.; Chua, N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to down regulate auxin signals for Arabidopsis lateral root development. Plant Cell 2009, 17, 1376–1386. [Google Scholar] [CrossRef]
- Wang, J.W.; Wang, L.J.; Mao, Y.B.; Cai, W.J.; Xue, H.W.; Chen, X.Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef]
- Dai, X.; Xu, Y.; Ma, Q. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef]
- Chiou, T.J. The role of microRNAs in sensing nutrient stress. Plant Cell Environ. 2007, 30, 323–332. [Google Scholar] [CrossRef]
- Doerner, P. Phosphate starvation signaling: A threesome controls systemic Pi homeostasis. Curr. Opin. Plant Biol. 2008, 11, 536–540. [Google Scholar] [CrossRef]
- Allen, R.S.; Li, J.Y.; Stahle, M.I.; Dubroue, A.; Gubler, F.; Millar, A.A. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl. Acad. Sci. USA 2007, 104, 16371–16376. [Google Scholar] [CrossRef]
- Vannini, C.; Locatelli, F.; Bracale, M.; Magnani, E.; Marsoni, M.; Osnato, M.; Mattana, M.; Baldoni, E.; Coraggio, I. Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. 2004, 37, 115–127. [Google Scholar] [CrossRef]
- Lu, Y.H.; Sun, R.; Shi, C.; Clark, L.; Li, V.L. Novel and mechanical stress responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 2005, 17, 2186–2203. [Google Scholar] [CrossRef]
- Zhou, M.; Li, D.; Li, Z.; Hu, Q.; Yang, C.; Zhu, L. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 2013, 161, 1375–1391. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Ding, Y.; Tao, Y.; Zhu, C. Emerging roles of microRNAs in the mediation of drought stress response in plants. J. Exp. Bot. 2013, 64, 3077–3086. [Google Scholar] [CrossRef]
- Luan, Y.; Wang, W.; Liu, P. Identification and functional analysis of novel and conserved microRNAs in tomato. Mol. Biol. Rep. 2014, 41, 5385–5394. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 2006, 6, 25. [Google Scholar] [CrossRef]
- Chen, H.; Lai, Z.; Shi, J.; Xiao, Y.; Chen, Z.; Xu, X. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010, 10, 281. [Google Scholar] [CrossRef]
- Li, J.; Besseau, S.; Toronen, P.; Sipari, N.; Kollist, H.; Holm, L. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 2013, 200, 455–472. [Google Scholar] [CrossRef]
- Jiang, Y.J.; Liang, G.; Yu, D.Q. Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 2012, 5, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, R.; Jiang, S.; Kumar, N.; Venkatesh, P.N.; Ramachandran, S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008, 49, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Yang, W.L.; Liu, D.C.; Han, Y.P.; Zhang, A.M.; Li, S.H. Ectopic expression of a grapevine transcription factor VvWRKY11 contributes to osmotic stress tolerance in Arabidopsis. Mol. Biol. Rep. 2011, 38, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, 165–183. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.X.; Miao, Z.Q.; Zhang, J.; Chen, S.Y.; Liu, Q.Q.; Xiang, C.B. Arabidopsis MADS-box factor AGL16 negatively regulates drought resistance via stomatal density and stomatal movement. J. Exp. Bot. 2020, 71, 6092–6106. [Google Scholar] [CrossRef]
- Bussis, D.; von Groll, U.; Fisahn, J.; Altmann, T. Stomatal aperture can compensate altered stomatal density in Arabidopsis thaliana at growth light conditions. Funct. Plant Biol. 2006, 33, 1037–1043. [Google Scholar] [CrossRef]
- Von Groll, U.; Berger, D.; Altmann, T. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 2002, 14, 1527–1539. [Google Scholar] [CrossRef]
- Shpak, E.D.; McAbee, J.M.; Pillitteri, L.J.; Torii, K.U. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 2005, 309, 290–293. [Google Scholar] [CrossRef]
- Lee, J.S.; Kuroha, T.; Hnilova, M.; Khatayevich, D.; Kanaoka, M.M.; McAbee, J.M.; Sarikaya, M.; Tamerler, C.; Torii, K.U. Direct interaction of ligand–receptor pairs specifying stomatal patterning. Genes 2012, 26, 126–136. [Google Scholar] [CrossRef]
- Caine, R.S.; Chater, C.C.; Kamisugi, Y.; Cuming, A.C.; Beerling, D.J.; Gray, J.E.; Fleming, A.J. An ancestral stomatal patterning module revealed in the non-vascular land plant Physcomitrella patens. Development 2016, 143, 3306–3314. [Google Scholar] [CrossRef]
- Olsen, J.L.; Rouze, P.; Verhelst, B.; Lin, Y.C.; Bayer, T.; Collen, J.; Dattolo, E.; De Paoli, E.; Dittami, S.; Maumus, F. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 2016, 530, 331–335. [Google Scholar] [CrossRef]
- Zhang, L.L.; Zhang, R.; Ye, X.; Zheng, X.B.; Tan, B.; Wang, W.; Li, Z.Q.; Li, J.D.; Cheng, J.; Feng, J.C. Overexpressing VvWRKY18 from grapevine reduces the drought tolerance in Arabidopsis by increasing leaf stomatal density. J. Plant Physiol. 2022, 275, 153741. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, N.; Si, H.; Calderón-Urrea, A. Selection and validation of reference genes for RT-qPCR analysis in potato under abiotic stress. Plant Methods 2017, 13, 85. [Google Scholar] [CrossRef]
- Wu, H.J.; Wang, Z.M.; Wang, M.; Wang, X.J. Wide spread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013, 161, 1875–1884. [Google Scholar] [CrossRef]
- Anna, W.; Marcin, P.; Zofifia, S.K. Construction of Artificial miRNAs to prevent drought stress in Solanum tuberosum. In Environmental Responses in Plants; Springer: New York, NY, USA, 2016; Volume 1398, pp. 271–290. [Google Scholar]
- Stentz, F.B.; Umpierrez, G.E.; Cuervo, R.; Kitabchi, A.E. Proinflammatory cytokines, markers of cardiovascular risks, oxidative stress, and lipid peroxidation in patients with hyperglycemic crises. Diabetes 2004, 53, 2079–2086. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Tear, I.D. Rapid determination of free proline for water stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts I Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Li, H. Determination of superoxide dismutase activity by the means of nitroblue tetrazolium. In Principles and Techniques of Plant Physiological Biochemical Experiments; Higher Education Press: Beijing, China, 2000; p. 293. [Google Scholar]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, 155–159. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]

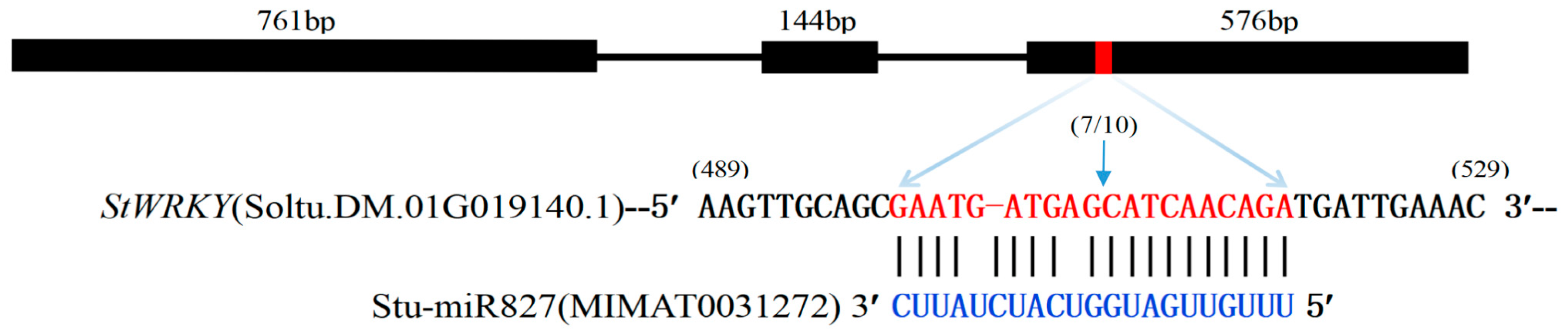

| miRNA | stu-miR827 |
|---|---|
| MS | UUUGUUGAUGGUCAUCUAUUC |
| Gene ID | MI0025949 |
| PS | UUUGUUGAUGGUCAUCUAUUCAUCAUAUCAUUUGGCAUAGUUUUUGCAACCAUUAAUAUGCCAUGAUUAGAUGAACAUCAACAAACA |
| LP (nt) | 87 |
| Families | miR827 |
| NM (nt) | 2 |
| LM (nt) | 21 |
| Side | 5 |
| MFEs (kcal/mol) | −32.05 |
| Chromosome | 12 |
| GenBank | CP047558.1 |
| miRNA | miRNA Sequence | Target Gene ID | Target Annotations |
|---|---|---|---|
| stu-miR827 | UUUGUUGAUGGUCAUCUAUUC | Soltu.DM.01G019140.1 | StWRKY48 |
| Gene | Oligonucleotides Forward (F) and Reverse (R) | Amplicon Length (bp) |
|---|---|---|
| Stu-miR827 | F 5′-TGCCTGGCTCCCTGTATGCCA-3′ | - |
| St18sRNA | R 5′-TTAGAGGAAGGAGAAGTCGTAACAA-3′ | - |
| StWRKY48 | F 5′-TGCTACCACCAAGTTTCCA -3′ | 171 |
| R 5′-CCGAAGAGTAAGAACCGAGG -3′ | ||
| StSDD1 | F 5′-TGAAGGAGAGCCTGTCATAAG -3′ | 111 |
| R 5′-GGCATAGGATGTAACCGATT -3′ | ||
| StEPF | F 5′-AAGACAACATTGGCAAGGG -3′ | 169 |
| R 5′-GCAACAGGACAAGTCTCAGC -3′ | ||
| StTMM | F 5′-GAAATAATGGAGGTGAGCGT -3′ | 137 |
| R 5′-GTTGCTAAACGATGCCTTG -3′ | ||
| StSPCH | F 5′-GACCATCTATCTGTGCTACGC -3′ | 130 |
| R 5′-TTGGCTTCTAAGGATTGGAG -3′ | ||
| StMUTE | F 5′-CGAAAGAGCCTAAGCCCTA -3′ | 130 |
| R 5′-AACATCAGCCACTGGAGAA -3′ | ||
| StFAMA | F 5′-TGAGGAAGTTGAGAGCCAA -3′ | 118 |
| R 5′-CCCTTTGAACATAGGAGCC -3′ | ||
| StEF1α | F 5′- ATTGGAAACGGATATGCTCCA-3′ | 101 |
| R 5′- TCCTTACCTGAACGCCTGTCA-3′ |
| Target Gene | Genomic Locust | Outer Primer | Inner Primer | Size |
|---|---|---|---|---|
| StWRKY48 | Soltu.DM.01G019140.1 | CAGGCTATCAAATGCCGATGA | TGAGCAGAAAGGCAGCATTCAAGACAATC | 397 bp |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Zhang, N.; Bai, J.; Duan, X.; Zhang, L.; Liu, S.; Tang, X.; Jin, X.; Li, S.; Si, H. Stu-miR827-Targeted StWRKY48 Transcription Factor Negatively Regulates Drought Tolerance of Potato by Increasing Leaf Stomatal Density. Int. J. Mol. Sci. 2022, 23, 14805. https://doi.org/10.3390/ijms232314805
Yang J, Zhang N, Bai J, Duan X, Zhang L, Liu S, Tang X, Jin X, Li S, Si H. Stu-miR827-Targeted StWRKY48 Transcription Factor Negatively Regulates Drought Tolerance of Potato by Increasing Leaf Stomatal Density. International Journal of Molecular Sciences. 2022; 23(23):14805. https://doi.org/10.3390/ijms232314805
Chicago/Turabian StyleYang, Jiangwei, Ning Zhang, Jiangping Bai, Xiaoqin Duan, Luhe Zhang, Shengyan Liu, Xun Tang, Xin Jin, Shigui Li, and Huaijun Si. 2022. "Stu-miR827-Targeted StWRKY48 Transcription Factor Negatively Regulates Drought Tolerance of Potato by Increasing Leaf Stomatal Density" International Journal of Molecular Sciences 23, no. 23: 14805. https://doi.org/10.3390/ijms232314805
APA StyleYang, J., Zhang, N., Bai, J., Duan, X., Zhang, L., Liu, S., Tang, X., Jin, X., Li, S., & Si, H. (2022). Stu-miR827-Targeted StWRKY48 Transcription Factor Negatively Regulates Drought Tolerance of Potato by Increasing Leaf Stomatal Density. International Journal of Molecular Sciences, 23(23), 14805. https://doi.org/10.3390/ijms232314805






