Discovery of a Novel Bloom’s Syndrome Protein (BLM) Inhibitor Suppressing Growth and Metastasis of Prostate Cancer
Abstract
:1. Introduction
2. Results
2.1. BLM Is Highly Expressed in PCa and Its Expression Correlates with Poor Survival
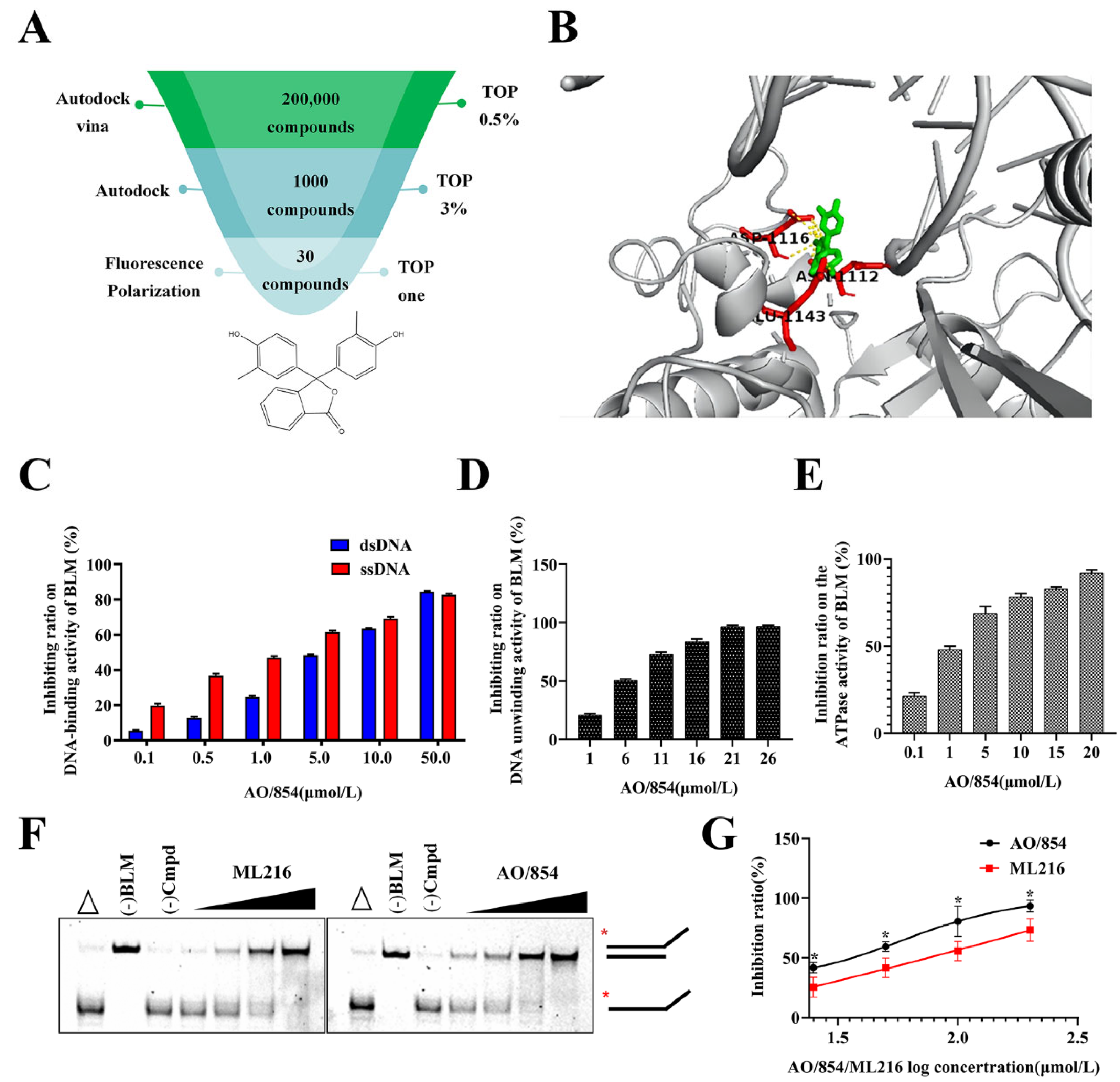
2.2. A High-Throughput Screen for Inhibitors of BLM
2.3. AO/854 Is a Potent Small Molecule Inhibitor of BLM
2.4. AO/854 Represses Cell Proliferation and Migration in PCa Cells
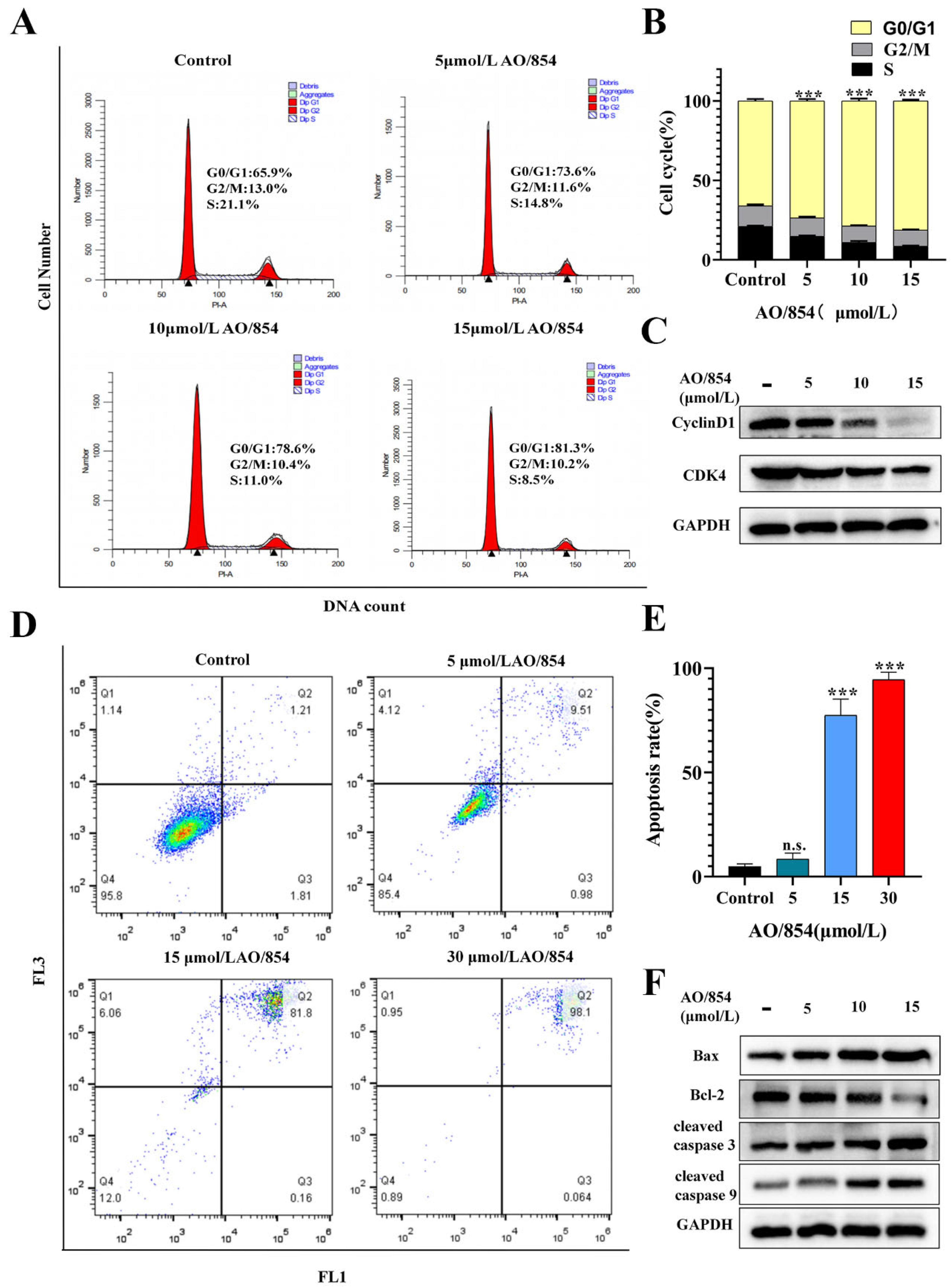
2.5. Flow Cytometry Cell Cycle Arrest and Apoptosis Analysis of AO/854
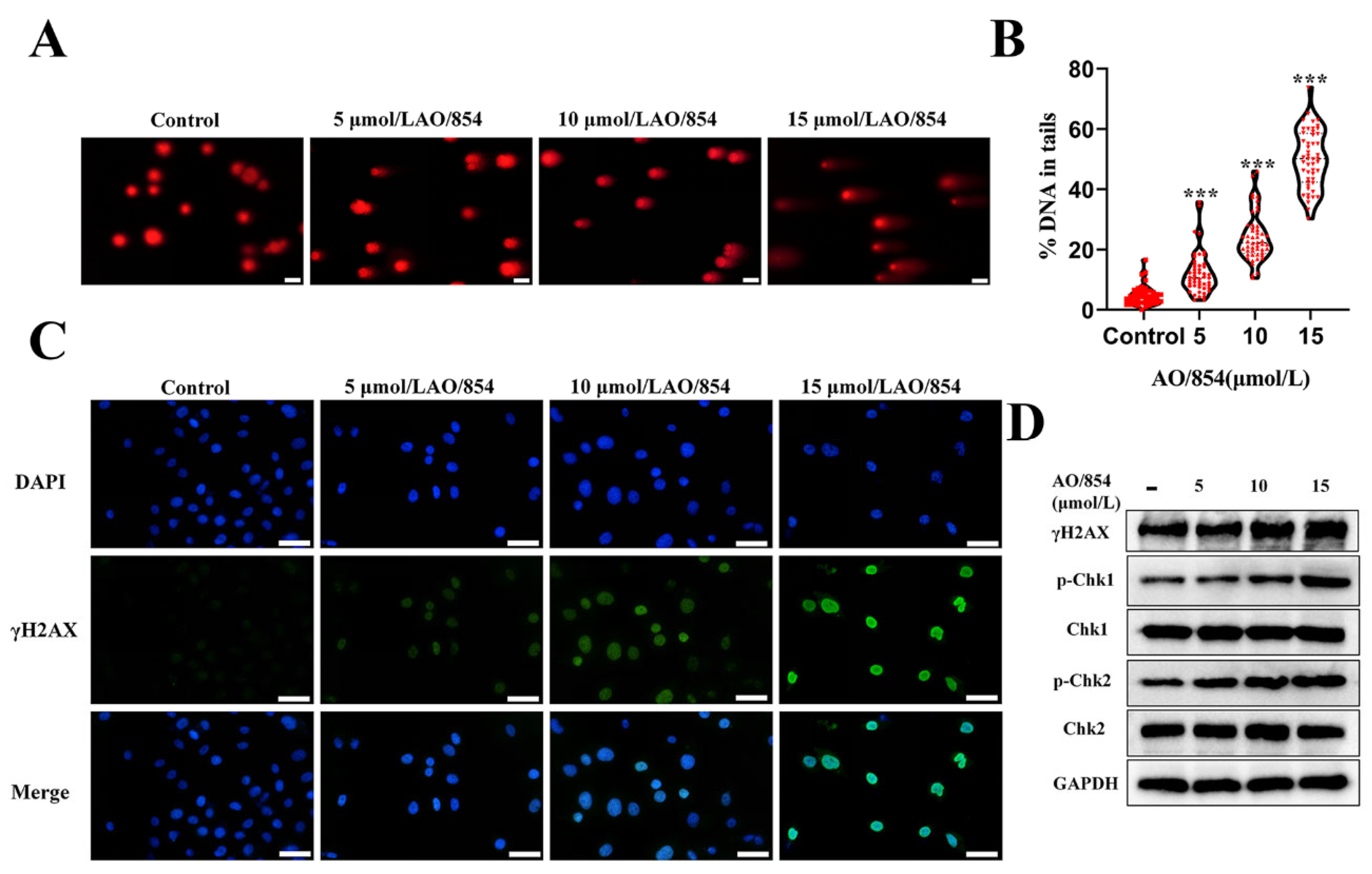
2.6. DNA Damage Is Observed in PC3 Cells Surviving AO/854 Treatment
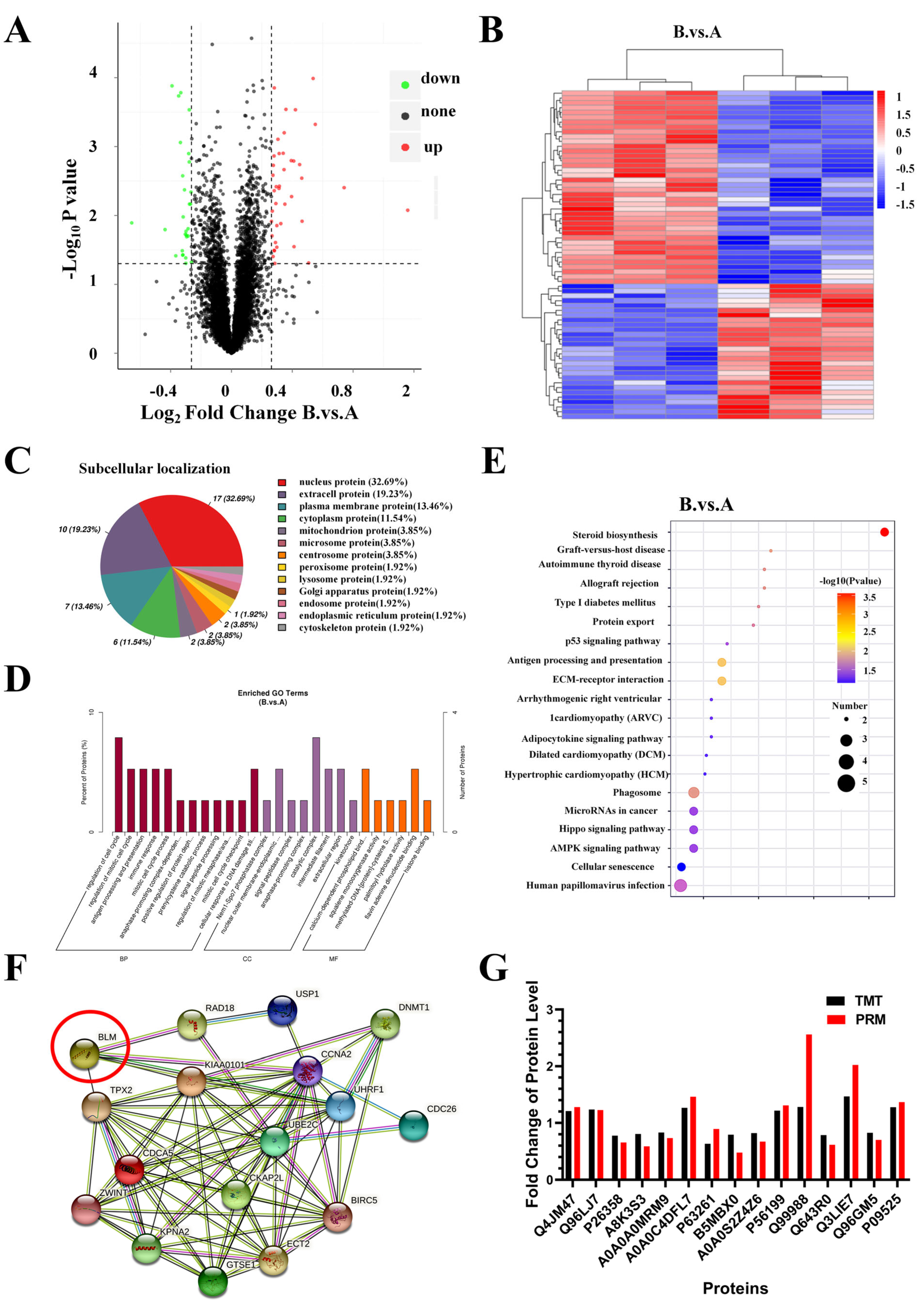
2.7. Identification and Bioinformatics Analysis of Differently Expressed Proteins (DEPs) in PC3 Cells after AO/854 Treatment
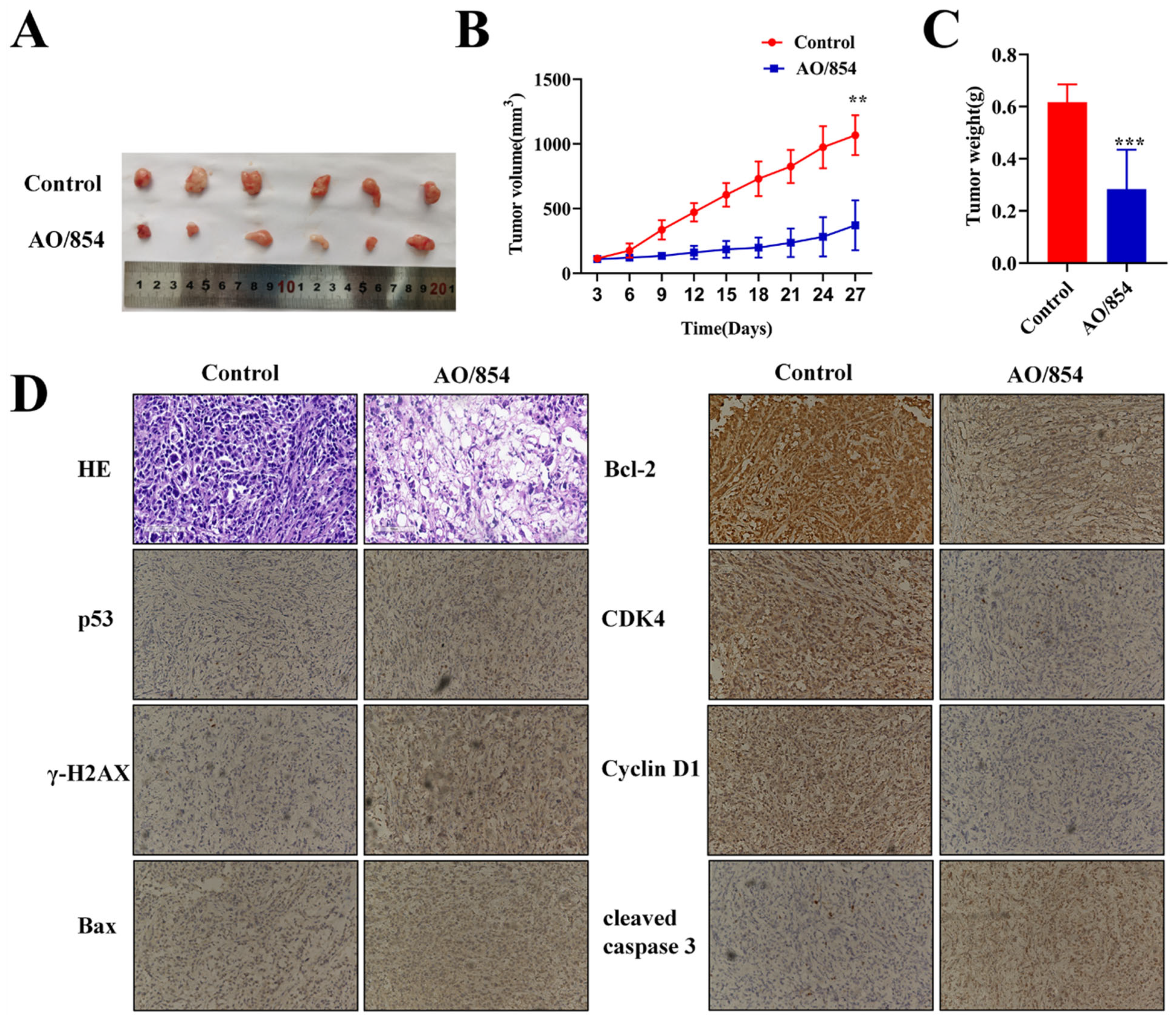
2.8. AO/854-Suppressed Tumour Growth in PC3 Xenograft Model
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. Cell Culture and Animals
4.3. Plasmids, Antibodies and Reagents
4.4. Bioinformatics Analysis
4.5. Immunohistochemical (IHC) Staining and Immunofluorescence (IF) Analysis
4.6. Western Blotting (WB)
4.7. Molecular Docking Study
4.8. Expression and Purification of the BLM642−1290 Helicase
4.9. Fluorescence Polarization Assay
4.10. ATPase Activity Analysis Assay
4.11. Electrophoretic Mobility Shift Assay (EMSA)
4.12. Unwinding Activity of Intracellular Assay
4.13. Circular Dichroism (CD) Assay
4.14. Cell Counting Kit-8 (CCK8) Drug Sensitivity Assessment and Cell Colony Formation Assay
4.15. Cell Cycle and Apoptosis Analysis
4.16. Transwell Assay and Scratch Wound-Healing Assay
4.17. Alkaline Comet Assay
4.18. Nude Mouse Tumor Xenograft Model
4.19. Quantitative Proteomics and Parallel Reaction Monitoring (PRM)
4.20. Statistics and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Sandhu, S.; Moore, C.M.; Chiong, E.; Beltran, H.; Bristow, R.G.; Williams, S.G. Prostate cancer. Lancet 2021, 398, 1075–1090. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Parker, J.; Crawley, D.; Garmo, H.; Lindahl, B.; Styrke, J.; Adolfsson, J.; Lambe, M.; Stattin, P.; Van Hemelrijck, M.; Beckmann, K. Use of Warfarin or Direct Oral Anticoagulants and Risk of Prostate Cancer in PCBaSe: A Nationwide Case-Control Study. Front. Oncol. 2020, 10, 571838. [Google Scholar] [CrossRef] [PubMed]
- Genkinger, J.M.; Wu, K.; Wang, M.; Albanes, D.; Black, A.; van den Brandt, P.A.; Burke, K.A.; Cook, M.B.; Gapstur, S.M.; Giles, G.G.; et al. Measures of body fatness and height in early and mid-to-late adulthood and prostate cancer: Risk and mortality in The Pooling Project of Prospective Studies of Diet and Cancer. Ann. Oncol. 2020, 31, 103–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labbé, D.P.; Zadra, G.; Yang, M.; Reyes, J.M.; Lin, C.Y.; Cacciatore, S.; Ebot, E.M.; Creech, A.L.; Giunchi, F.; Fiorentino, M.; et al. High-fat diet fuels prostate cancer progression by rewiring the metabolome and amplifying the MYC program. Nat. Commun. 2019, 10, 4358. [Google Scholar] [CrossRef] [Green Version]
- Baumgart, S.J.; Nevedomskaya, E.; Haendler, B. Dysregulated Transcriptional Control in Prostate Cancer. Int. J. Mol. Sci. 2019, 20, 2883. [Google Scholar] [CrossRef] [Green Version]
- Angeles, A.K.; Bauer, S.; Ratz, L.; Klauck, S.M.; Sültmann, H. Genome-Based Classification and Therapy of Prostate Cancer. Diagnostics 2018, 8, 62. [Google Scholar] [CrossRef] [Green Version]
- Antonarakis, E.S.; Piulats, J.M.; Gross-Goupil, M.; Goh, J.; Ojamaa, K.; Hoimes, C.J.; Vaishampayan, U.; Berger, R.; Sezer, A.; Alanko, T.; et al. Pembrolizumab for Treatment-Refractory Metastatic Castration-Resistant Prostate Cancer: Multicohort, Open-Label Phase II KEYNOTE-199 Study. J. Clin. Oncol. 2020, 38, 395–405. [Google Scholar] [CrossRef]
- Chen, M.; Wan, L.; Zhang, J.; Zhang, J.; Mendez, L.; Clohessy, J.G.; Berry, K.; Victor, J.; Yin, Q.; Zhu, Y.; et al. Deregulated PP1α phosphatase activity towards MAPK activation is antagonized by a tumor suppressive failsafe mechanism. Nat. Commun. 2018, 9, 159. [Google Scholar] [CrossRef]
- Gravis, G.; Boher, J.M.; Chen, Y.H.; Liu, G.; Fizazi, K.; Carducci, M.A.; Oudard, S.; Joly, F.; Jarrard, D.M.; Soulie, M.; et al. Burden of Metastatic Castrate Naive Prostate Cancer Patients, to Identify Men More Likely to Benefit from Early Docetaxel: Further Analyses of CHAARTED and GETUG-AFU15 Studies. Eur. Urol. 2018, 73, 847–855. [Google Scholar] [CrossRef]
- Huang, Q.; Ma, B.; Su, Y.X.; Chan, K.; Qu, H.; Huang, J.Y.; Wang, D.; Qiu, J.; Liu, H.; Yang, X.; et al. miR-197-3p Represses the Proliferation of Prostate Cancer by Regulating the VDAC1/AKT/β-catenin Signaling Axis. Int. J. Biol. Sci. 2020, 16, 1417–1426. [Google Scholar] [CrossRef]
- Hong, Y.; Sonneville, R.; Wang, B.; Scheidt, V.; Meier, B.; Woglar, A.; Demetriou, S.; Labib, K.; Jantsch, V.; Gartner, A. LEM-3 is a midbody-tethered DNA nuclease that resolves chromatin bridges during late mitosis. Nat. Commun. 2018, 9, 728. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.Q.; Hou, X.M.; Li, M.; Dou, S.X.; Xi, X.G. BLM unfolds G-quadruplexes in different structural environments through different mechanisms. Nucleic Acids Res. 2015, 43, 4614–4626. [Google Scholar] [CrossRef] [Green Version]
- Xue, C.; Daley, J.M.; Xue, X.; Steinfeld, J.; Kwon, Y.; Sung, P.; Greene, E.C. Single-molecule visualization of human BLM helicase as it acts upon double- and single-stranded DNA substrates. Nucleic Acids Res. 2019, 47, 11225–11237. [Google Scholar] [CrossRef]
- Croteau, D.L.; Popuri, V.; Opresko, P.L.; Bohr, V.A. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014, 83, 519–552. [Google Scholar] [CrossRef] [Green Version]
- Crickard, J.B.; Xue, C.; Wang, W.; Kwon, Y.; Sung, P.; Greene, E.C. The RecQ helicase Sgs1 drives ATP-dependent disruption of Rad51 filaments. Nucleic Acids Res. 2019, 47, 4694–4706. [Google Scholar] [CrossRef]
- Lu, H.; Davis, A.J. Human RecQ Helicases in DNA Double-Strand Break Repair. Front. Cell Dev. Biol. 2021, 9, 640755. [Google Scholar] [CrossRef]
- Panier, S.; Maric, M.; Hewitt, G.; Mason-Osann, E.; Gali, H.; Dai, A.; Labadorf, A.; Guervilly, J.H.; Ruis, P.; Segura-Bayona, S.; et al. SLX4IP Antagonizes Promiscuous BLM Activity during ALT Maintenance. Mol. Cell 2019, 76, 27–43.e11. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, V.; Agarwal, H.; Priya, S.; Batra, H.; Modi, P.; Pandey, M.; Saha, D.; Raghavan, S.C.; Sengupta, S. MRN complex-dependent recruitment of ubiquitylated BLM helicase to DSBs negatively regulates DNA repair pathways. Nat. Commun. 2018, 9, 1016. [Google Scholar] [CrossRef]
- Chang, E.Y.; Novoa, C.A.; Aristizabal, M.J.; Coulombe, Y.; Segovia, R.; Chaturvedi, R.; Shen, Y.; Keong, C.; Tam, A.S.; Jones, S.J.M.; et al. RECQ-like helicases Sgs1 and BLM regulate R-loop-associated genome instability. J. Cell Biol. 2017, 216, 3991–4005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Hu, J.; Li, Y.; Bi, L.; Guo, L.; Jia, X.; Zhang, X.; Li, D.; Hou, X.M.; Modesti, M.; et al. Bloom Syndrome Helicase Compresses Single-Stranded DNA into Phase-Separated Condensates. Angew. Chem. Int. Ed. Engl. 2022, 61, e202209463. [Google Scholar] [PubMed]
- Brosh, R.M., Jr.; Matson, S.W. History of DNA Helicases. Genes 2020, 11, 255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Lv, H.C.; Lv, W.H.; Shi, M.; Zhang, M.M.; Luan, M.W.; Zhu, H.; Zhang, R.; Jiang, Y. Genome-wide haplotype association study identifies BLM as a risk gene for prostate cancer in Chinese population. Tumor Biol. 2015, 36, 2703–2707. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.S.; Feng, S.J.; Xie, D.W.; Feng, D.L.; Jiang, Y.H.; Zhang, X.D. RecQ helicase BLM regulates prostate cancer cell proliferation and apoptosis. Oncol. Lett. 2017, 14, 4206–4212. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, G.H.; Dexheimer, T.S.; Rosenthal, A.S.; Chu, W.K.; Singh, D.K.; Mosedale, G.; Bachrati, C.Z.; Schultz, L.; Sakurai, M.; Savitsky, P.; et al. A small molecule inhibitor of the BLM helicase modulates chromosome stability in human cells. Chem. Biol. 2013, 20, 55–62. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.X.; Zhang, Z.L.; Yin, Q.K.; Tu, J.L.; Wang, J.E.; Xu, Y.H.; Rao, Y.; Ou, T.M.; Huang, S.L.; Li, D.; et al. Design, Synthesis, and Evaluation of New Quinazolinone Derivatives that Inhibit Bloom Syndrome Protein (BLM) Helicase, Trigger DNA Damage at the Telomere Region, and Synergize with PARP Inhibitors. J. Med. Chem. 2020, 63, 9752–9772. [Google Scholar] [CrossRef]
- Yin, Q.K.; Wang, C.X.; Wang, Y.Q.; Guo, Q.L.; Zhang, Z.L.; Ou, T.M.; Huang, S.L.; Li, D.; Wang, H.G.; Tan, J.H.; et al. Discovery of Isaindigotone Derivatives as Novel Bloom’s Syndrome Protein (BLM) Helicase Inhibitors That Disrupt the BLM/DNA Interactions and Regulate the Homologous Recombination Repair. J. Med. Chem. 2019, 62, 3147–3162. [Google Scholar] [CrossRef]
- Zhang, W.M.; Yang, S.; Liu, J.H.; Bao, L.C.; Lu, H.; Li, H.; Pan, W.D.; Jiao, Y.C.; He, Z.X.; Liu, J. Screening antiproliferative drug for breast cancer from bisbenzylisoquinoline alkaloid tetrandrine and fangchinoline derivatives by targeting BLM helicase. BMC Cancer 2019, 19, 1009. [Google Scholar] [CrossRef]
- Rausch, C.; Zhang, P.; Casas-Delucchi, C.S.; Daiß, J.L.; Engel, C.; Coster, G.; Hastert, F.D.; Weber, P.; Cardoso, M.C. Cytosine base modifications regulate DNA duplex stability and metabolism. Nucleic Acids Res. 2021, 49, 12870–12894. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Kaur, E.; Agrawal, R.; Sengupta, S. Functions of BLM Helicase in Cells: Is It Acting Like a Double-Edged Sword? Front. Genet. 2021, 12, 634789. [Google Scholar] [CrossRef]
- Alzahrani, F.A.; Ahmed, F.; Sharma, M.; Rehan, M.; Mahfuz, M.; Baeshen, M.N.; Hawsawi, Y.; Almatrafi, A.; Alsagaby, S.A.; Kamal, M.A.; et al. Investigating the pathogenic SNPs in BLM helicase and their biological consequences by computational approach. Sci. Rep. 2020, 10, 12377. [Google Scholar] [CrossRef]
- Newman, J.A.; Savitsky, P.; Allerston, C.K.; Bizard, A.H.; Özer, Ö.; Sarlós, K.; Liu, Y.; Pardon, E.; Steyaert, J.; Hickson, I.D.; et al. Crystal structure of the Bloom’s syndrome helicase indicates a role for the HRDC domain in conformational changes. Nucleic Acids Res. 2015, 43, 5221–5235. [Google Scholar] [CrossRef]
- Chen, K.; Xu, H.Q.; Zhao, J.F. Bloom Syndrome Protein Activates AKT and PRAS40 in Prostate Cancer Cells. Oxid. Med. Cell. Longev. 2019, 2019, 3685817. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Wang, Y.; Wang, L.; Wang, Q.; Du, L.Q.; Fan, S.; Liu, Q.; Li, L. Accumulation and Phosphorylation of RecQ-Mediated Genome Instability Protein 1 (RMI1) at Serine 284 and Serine 292 during Mitosis. Int. J. Mol. Sci. 2015, 16, 26395–26405. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, A.; Singh, T.R.; Ali, A.M.; Wahengbam, K.; Meetei, A.R. Monopolar spindle 1 (MPS1) protein-dependent phosphorylation of RecQ-mediated genome instability protein 2 (RMI2) at serine 112 is essential for BLM-Topo III α-RMI1-RMI2 (BTR) protein complex function upon spindle assembly checkpoint (SAC) activation during mitosis. J. Biol. Chem. 2013, 288, 33500–33508. [Google Scholar]
- Balbo Pogliano, C.; Ceppi, I.; Giovannini, S.; Petroulaki, V.; Palmer, N.; Uliana, F.; Gatti, M.; Kasaciunaite, K.; Freire, R.; Seidel, R.; et al. The CDK1-TOPBP1-PLK1 axis regulates the Bloom’s syndrome helicase BLM to suppress crossover recombination in somatic cells. Sci. Adv. 2022, 8, eabk0221. [Google Scholar] [CrossRef]
- Mendez-Dorantes, C.; Tsai, L.J.; Jahanshir, E.; Lopezcolorado, F.W.; Stark, J.M. BLM has Contrary Effects on Repeat-Mediated Deletions, based on the Distance of DNA DSBs to a Repeat and Repeat Divergence. Cell Rep. 2020, 30, 1342–1357.e4. [Google Scholar] [CrossRef]
- Patel, D.S.; Misenko, S.M.; Her, J.; Bunting, S.F. BLM helicase regulates DNA repair by counteracting RAD51 loading at DNA double-strand break sites. J. Cell Biol. 2017, 216, 3521–3534. [Google Scholar] [CrossRef] [Green Version]
- Shastri, V.M.; Subramanian, V.; Schmidt, K.H. A novel cell-cycle-regulated interaction of the Bloom syndrome helicase BLM with Mcm6 controls replication-linked processes. Nucleic Acids Res. 2021, 49, 8699–8713. [Google Scholar] [CrossRef] [PubMed]
- Bononi, A.; Goto, K.; Ak, G.; Yoshikawa, Y.; Emi, M.; Pastorino, S.; Carparelli, L.; Ferro, A.; Nasu, M.; Kim, J.H.; et al. Heterozygous germline BLM mutations increase susceptibility to asbestos and mesothelioma. Proc. Natl. Acad. Sci. USA 2020, 117, 33466–33473. [Google Scholar] [CrossRef] [PubMed]
- Crocetto, F.; Barone, B.; Caputo, V.F.; Fontana, M.; de Cobelli, O.; Ferro, M. BRCA Germline Mutations in Prostate Cancer: The Future Is Tailored. Diagnostics 2021, 11, 908. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.W.; Fugger, K.; West, S.C. Unresolved recombination intermediates lead to ultra-fine anaphase bridges, chromosome breaks and aberrations. Nat. Cell Biol. 2018, 20, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Maity, J.; Horibata, S.; Zurcher, G.; Lee, J.M. Targeting of RecQ Helicases as a Novel Therapeutic Strategy for Ovarian Cancer. Cancers 2022, 14, 1219. [Google Scholar] [CrossRef]
- Kong, Y.; Xu, C.; Sun, X.; Sun, H.; Zhao, X.; He, N.; Ji, K.; Wang, Q.; Du, L.; Wang, J.; et al. BLM helicase inhibition synergizes with PARP inhibition to improve the radiosensitivity of olaparib resistant non-small cell lung cancer cells by inhibiting homologous recombination repair. Cancer Biol. Med. 2021, 18, 1150–1171. [Google Scholar] [CrossRef]
- Shi, T.; van Soest, D.M.K.; Polderman, P.E.; Burgering, B.M.T.; Dansen, T.B. DNA damage and oxidant stress activate p53 through differential upstream signaling pathways. Free Radic. Biol. Med. 2021, 172, 298–311. [Google Scholar] [CrossRef]
- Lindström, M.S.; Bartek, J.; Maya-Mendoza, A. p53 at the crossroad of DNA replication and ribosome biogenesis stress pathways. Cell Death Differ. 2022, 29, 972–982. [Google Scholar] [CrossRef]
- Carroll, A.G.; Voeller, H.J.; Sugars, L.; Gelmann, E.P. p53 oncogene mutations in three human prostate cancer cell lines. Prostate 1993, 23, 123–134. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.Z.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]





| Feature | N (60) | Immunohistochemical Score of BLM | χ2 | p Value | |
|---|---|---|---|---|---|
| <8 (%) | ≥8 (%) | ||||
| Age(years) | 60 | 17 | 43 | 0.219 | 0.640 |
| ≤70 | 24 | 6 (25.0) | 18 (75.0) | ||
| >70 | 36 | 11 (32.4) | 25 (67.6) | ||
| Clinical stage | 60 | 17 | 43 | 10.506 | 0.001 * |
| I–II | 20 | 11 (55.0) | 9 (45.0) | ||
| III–IV | 40 | 6 (15.0) | 34 (85.0) | ||
| Gleason score | 60 | 17 | 43 | 2.694 | 0.101 |
| <7 | 5 | 3 (60.0) | 2 (40.0) | ||
| ≥7 | 55 | 14 (25.5) | 41 (74.5) | ||
| Gleason grade | 60 | 17 | 43 | 9.877 | 0.002 * |
| 1–3 | 15 | 9 (60.0) | 6 (40.0) | ||
| 4–5 | 45 | 8 (17.8) | 37 (82.2) | ||
| N-regional lymph nodes | 60 | 17 | 43 | 1.927 | 0.165 |
| N0 | 43 | 10 (23.3) | 33 (76.7) | ||
| N1 | 17 | 7 (41.2) | 10 (58.8) | ||
| M-distant metastasis | 60 | 17 | 43 | 0.410 | 0.522 |
| M0 | 50 | 15 (30.0) | 35 (70.0) | ||
| M1 | 10 | 2 (20.0) | 8 (80.0) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.-Y.; Xu, H.-Q.; Zhao, J.-F.; Ruan, Y.; Chen, B. Discovery of a Novel Bloom’s Syndrome Protein (BLM) Inhibitor Suppressing Growth and Metastasis of Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 14798. https://doi.org/10.3390/ijms232314798
Ma X-Y, Xu H-Q, Zhao J-F, Ruan Y, Chen B. Discovery of a Novel Bloom’s Syndrome Protein (BLM) Inhibitor Suppressing Growth and Metastasis of Prostate Cancer. International Journal of Molecular Sciences. 2022; 23(23):14798. https://doi.org/10.3390/ijms232314798
Chicago/Turabian StyleMa, Xiao-Yan, Hou-Qiang Xu, Jia-Fu Zhao, Yong Ruan, and Bin Chen. 2022. "Discovery of a Novel Bloom’s Syndrome Protein (BLM) Inhibitor Suppressing Growth and Metastasis of Prostate Cancer" International Journal of Molecular Sciences 23, no. 23: 14798. https://doi.org/10.3390/ijms232314798
APA StyleMa, X.-Y., Xu, H.-Q., Zhao, J.-F., Ruan, Y., & Chen, B. (2022). Discovery of a Novel Bloom’s Syndrome Protein (BLM) Inhibitor Suppressing Growth and Metastasis of Prostate Cancer. International Journal of Molecular Sciences, 23(23), 14798. https://doi.org/10.3390/ijms232314798







