Increased Expression of the RBPMS Splice Variants Inhibits Cell Proliferation in Ovarian Cancer Cells
Abstract
1. Introduction
2. Results
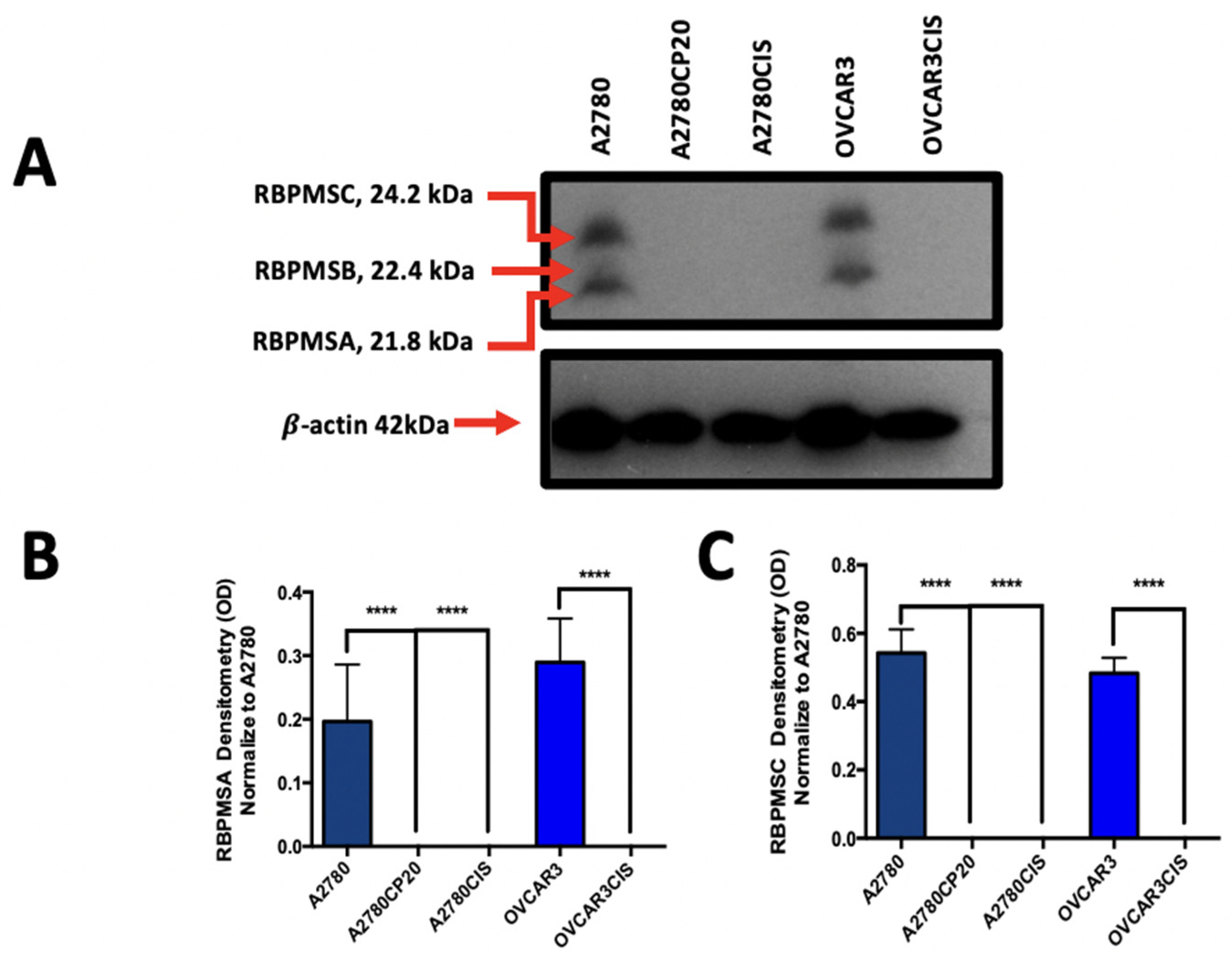
2.1. RBPMSA and RBPMSC Protein Levels Are Reduced in Cisplatin Resistance Ovarian Cancer Cell Lines
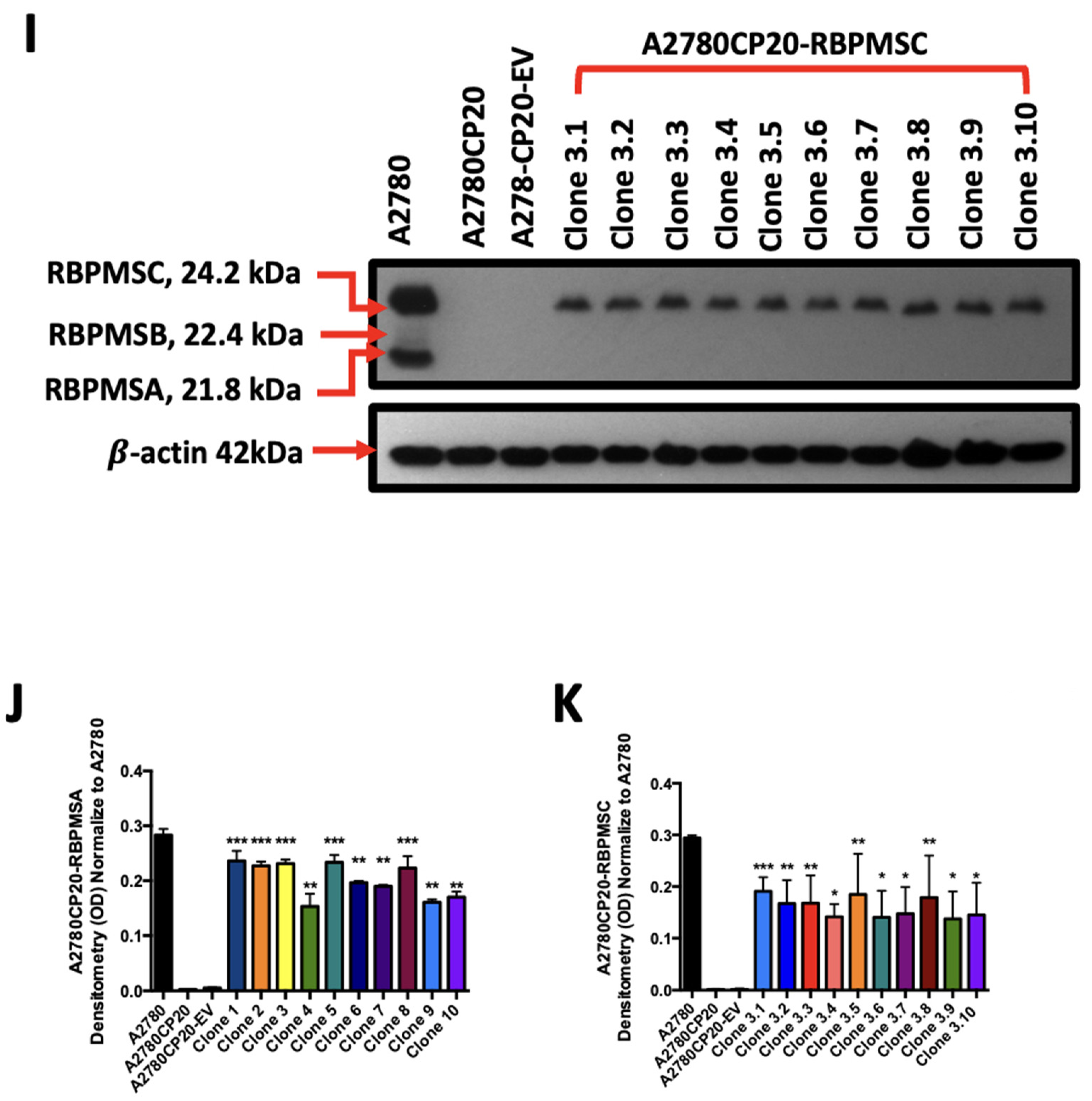
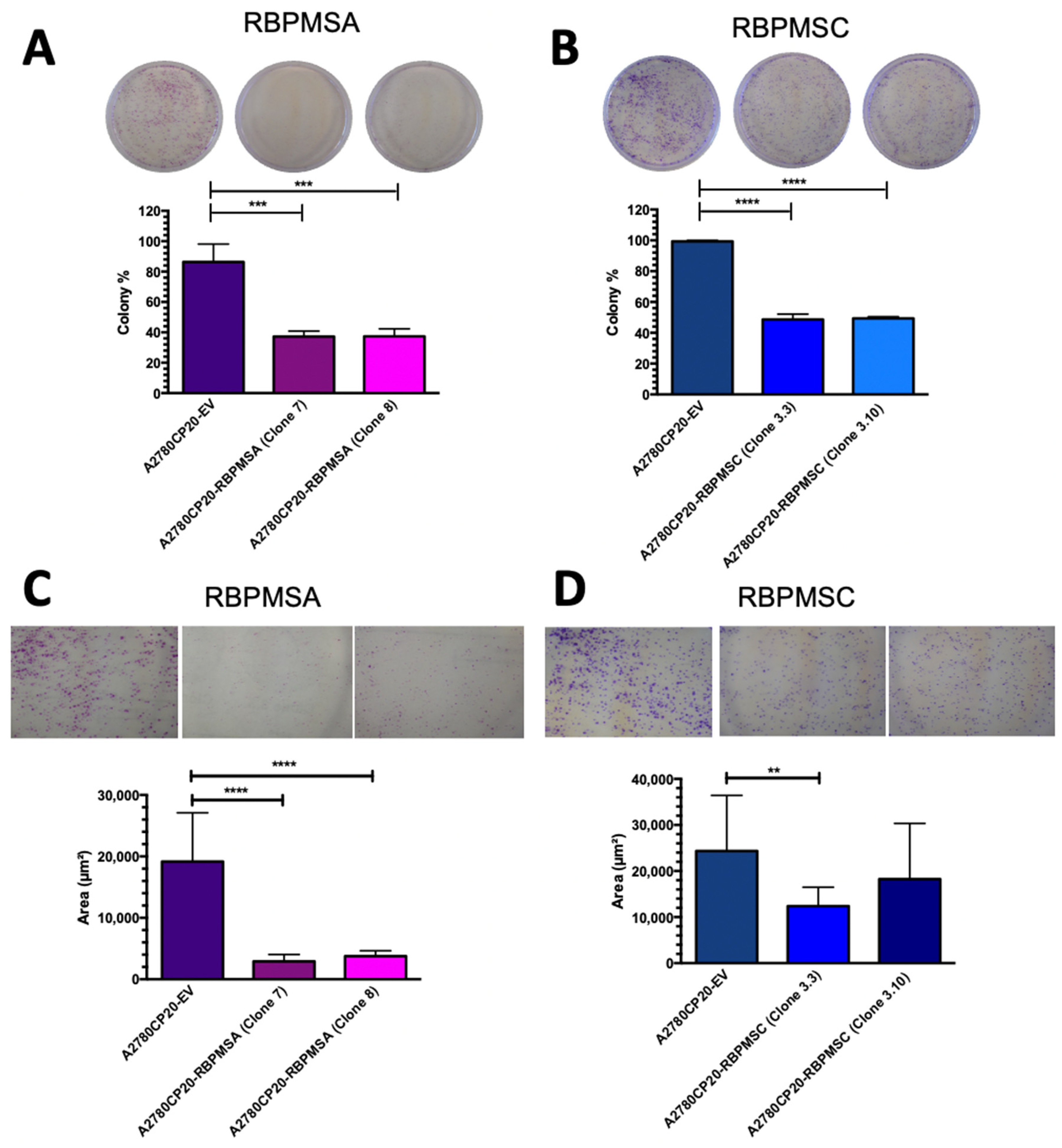
2.2. Overexpression of RBPMSA and RBPMSC Decreased Cell Growth and Proliferation of Cisplatin Resistant Ovarian Cancer Cells
2.3. RBPMSA Overexpression Increased the Sensitivity of Ovarian Cancer Cells to Cisplatin Treatment
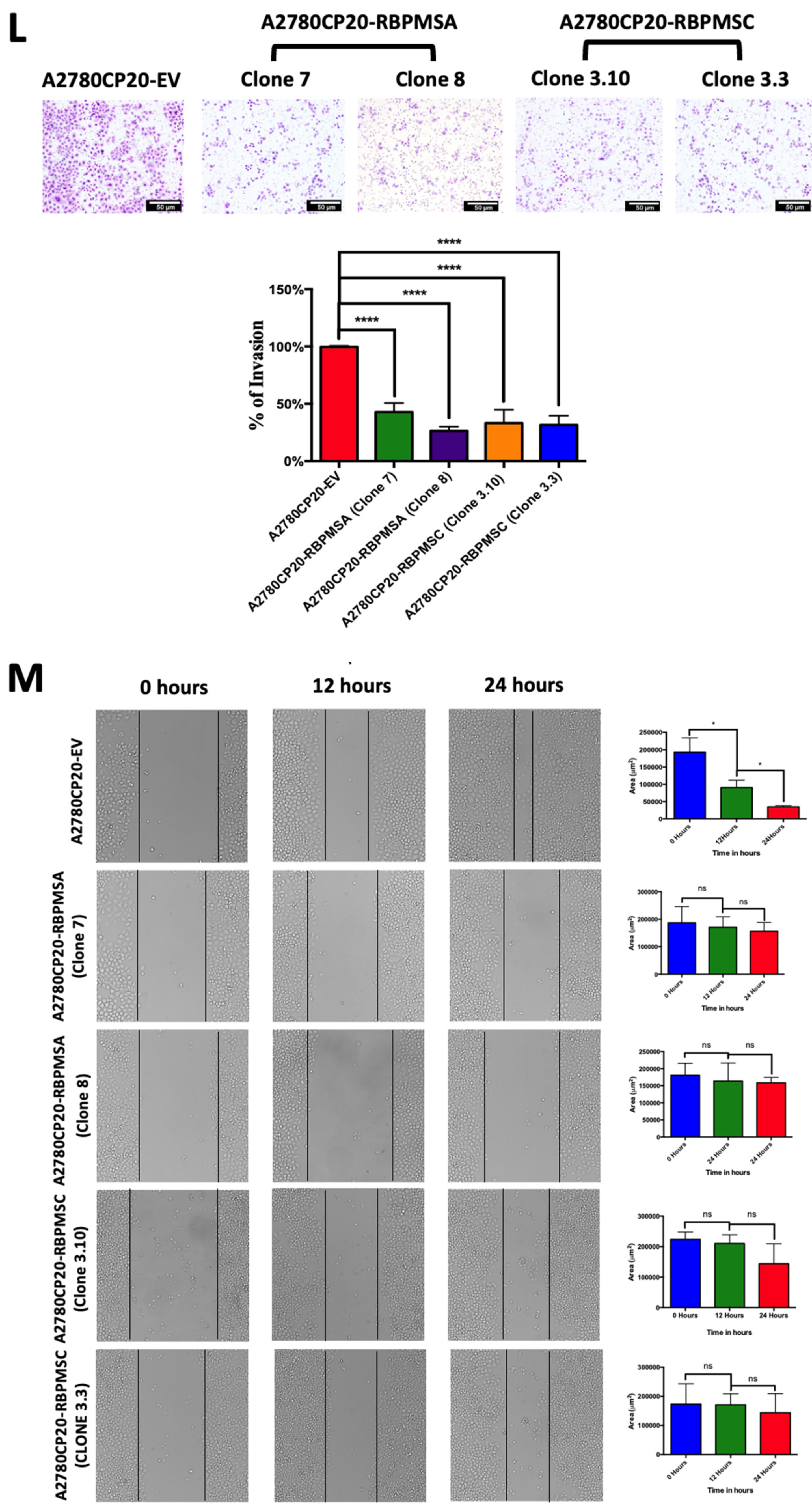
2.4. RBPMSA and RBPMSC Overexpression Decreased the Migration and the Invasion Ability of Ovarian Cancer Cells
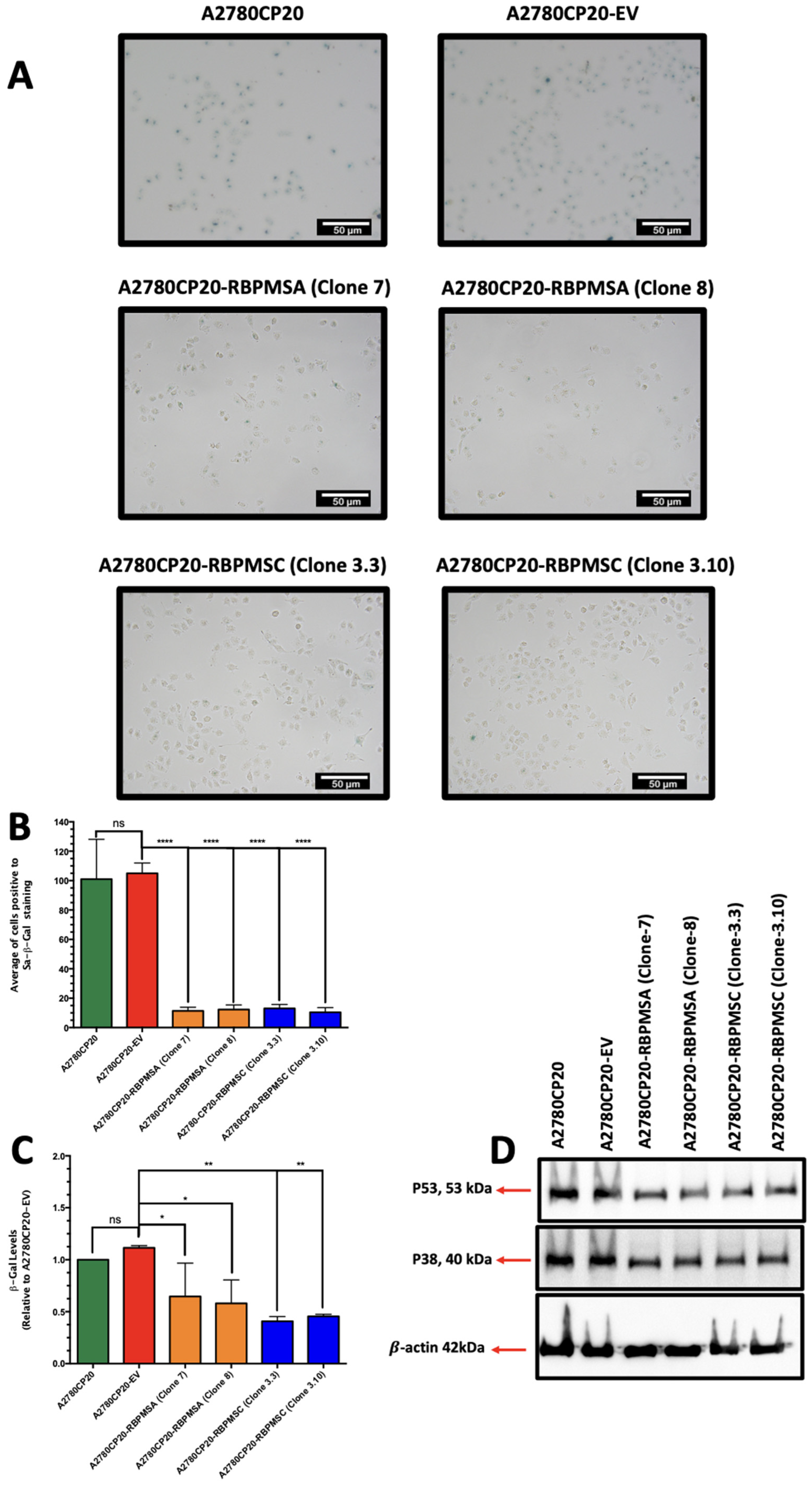
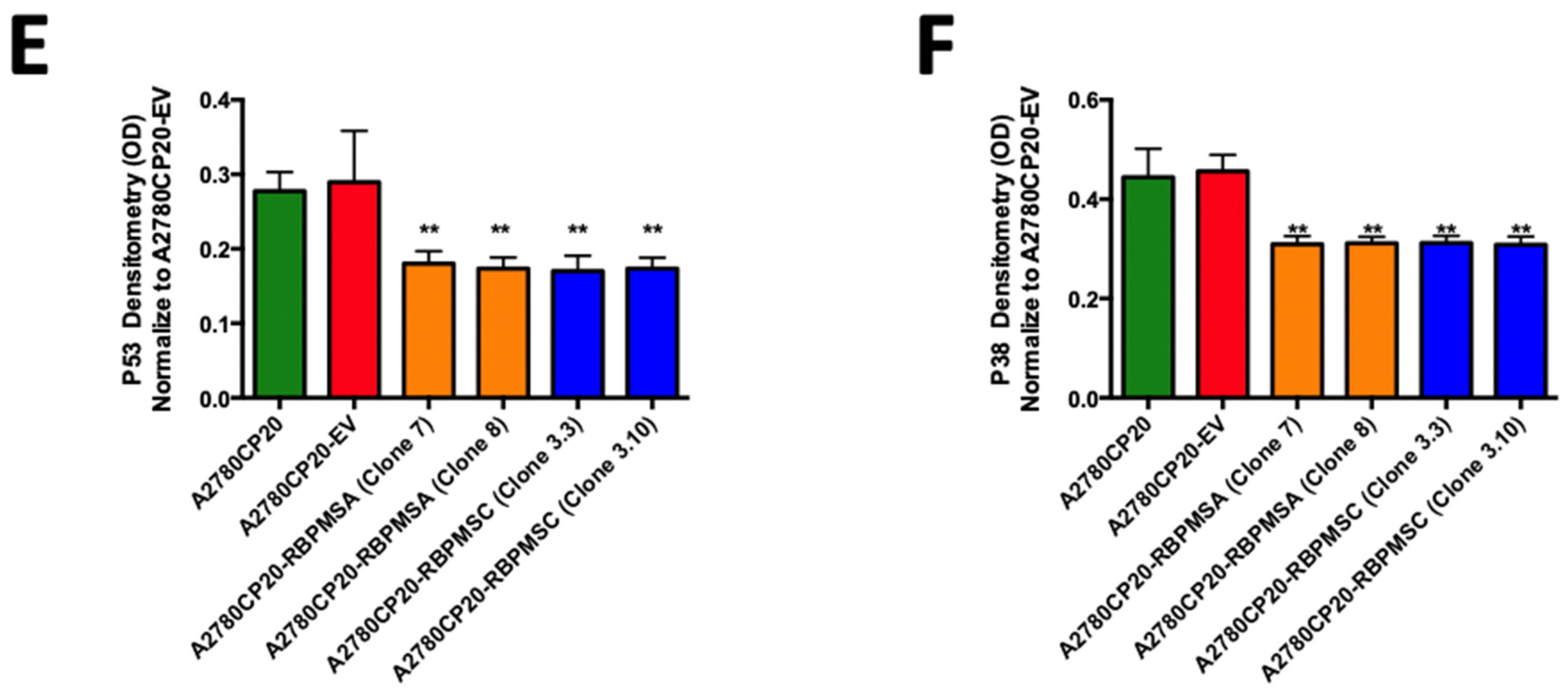
2.5. RBPMSA and RBPMSC Overexpression Decreased the Senescence-Associated β-Galactosidase Levels
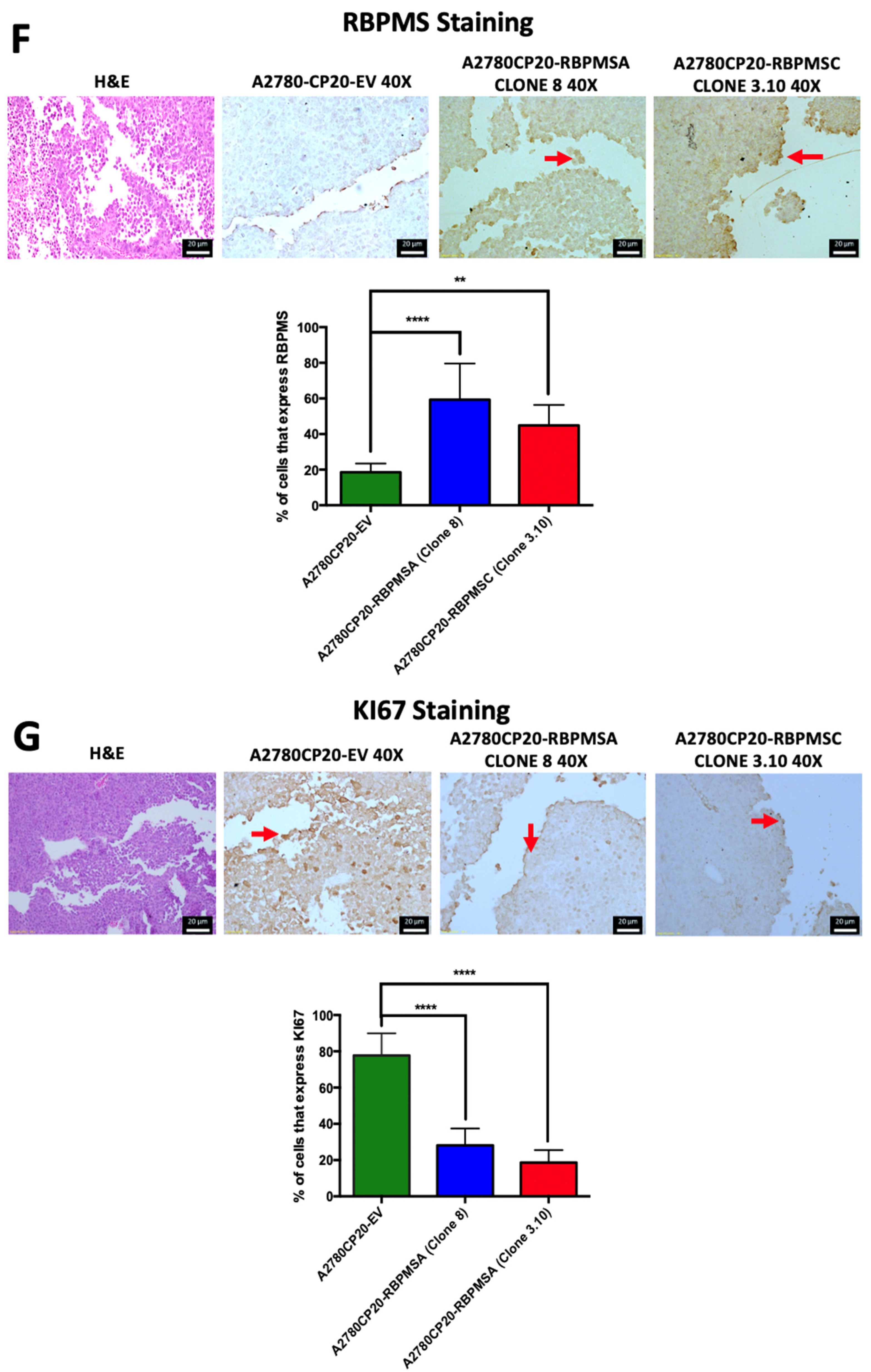
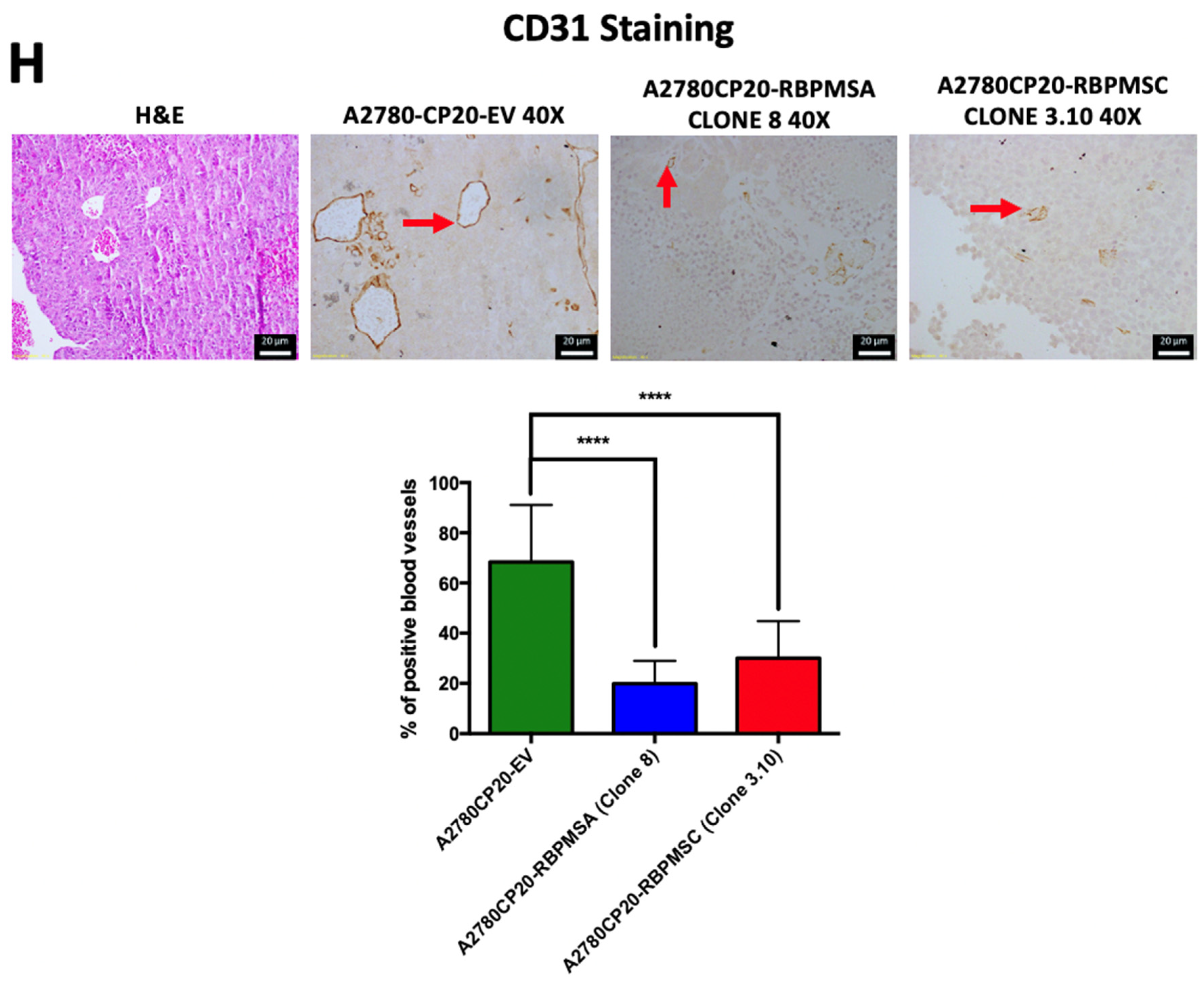
2.6. Effects of Subcutaneous Implantation of RBPMSA and RBPMSC Overexpressing Cells in an Ovarian Cancer Mouse Model
2.7. Identification of RBPMSA and RBPMSC Downstream Signaling Pathways by RNAseq
2.8. Prognostic Value of RBPMSA and RBPMSC Downstream Effectors
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture Conditions
4.2. Western Blot Analysis
4.3. RNA Isolation, cDNA Synthesis, and RT-PCR
4.4. Stable Transfection for RBPMS exp ression
4.5. Transient Transfection of RBPMSA and RBPMSC in OVCAR3CIS Cells
4.6. Colony Formation, Cell Growth, and Cell Viability Assays
4.7. Migration and Invasion Assays
4.8. Mice Experiments
4.9. Immunohistochemistry
4.10. Senescence Associated β-Galactosidase Activity Assays
4.11. RNA Sequencing Library Preparation, Data Processing, and Statistics
4.12. Validation of the RNAseq Results by SYBR-GREEN Based qRT-PCR
4.13. Clustering and Network Analysis
4.14. Kaplan-Meyer (KM) Survival Analysis
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.W.; Ruiz, B.; Killeen, J.L.; Coté, T.R.; Wu, X.C.; Correa, C.N.; Howe, H.L. Pathology and classification of ovarian tumors. Cancer 2003, 97, 2631–2642. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.R.; Shih Ie, M. Ovarian cancer. Annu. Rev. Pathol. 2009, 4, 287–313. [Google Scholar] [CrossRef] [PubMed]
- Nezhat, F.R.; Apostol, R.; Nezhat, C.; Pejovic, T. New insights in the pathophysiology of ovarian cancer and implications for screening and prevention. Am. J. Obstet. Gynecol. 2015, 213, 262–267. [Google Scholar] [CrossRef]
- Mok, S.C.; Kwong, J.; Welch, W.R.; Samimi, G.; Ozbun, L.; Bonome, T.; Birrer, M.J.; Berkowitz, R.S. Etiology and pathogenesis of epithelial ovarian cancer. Dis. Markers 2007, 23, 367–376. [Google Scholar] [CrossRef]
- Prat, J. New insights into ovarian cancer pathology. Ann. Oncol. 2012, 23 (Suppl. 10), x111–x117. [Google Scholar] [CrossRef]
- Matulonis, U.A.; Sood, A.K.; Fallowfield, L.; Howitt, B.E.; Sehouli, J.; Karlan, B.Y. Ovarian cancer. Nat. Rev. Dis. Primers 2016, 2, 16061. [Google Scholar] [CrossRef]
- Shen, D.-W.; Pouliot, L.M.; Hall, M.D.; Gottesman, M.M. Cisplatin resistance: A cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol. Rev. 2012, 64, 706–721. [Google Scholar] [CrossRef]
- Reyes-González, J.M.; Quiñones-Díaz, B.I.; Santana, Y.; Báez-Vega, P.M.; Soto, D.; Valiyeva, F.; Marcos-Martínez, M.J.; Thomas, R.J.F.-D.; Vivas-Mejía, P.E. Downstream Effectors of ILK in Cisplatin-Resistant Ovarian Cancer. Cancers 2020, 12, 880. [Google Scholar] [CrossRef]
- Xu, J.-H.; Hu, S.-L.; Shen, G.-D.; Shen, G. Tumor suppressor genes and their underlying interactions in paclitaxel resistance in cancer therapy. Cancer Cell Int. 2016, 16, 13. [Google Scholar] [CrossRef]
- Wang, F.; Liu, M.; Li, X.; Tang, H. MiR-214 reduces cell survival and enhances cisplatin-induced cytotoxicity via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett. 2013, 587, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Michels, J.; Brenner, C.; Szabadkai, G.; Harel-Bellan, A.; Castedo, M.; Kroemer, G. Systems biology of cisplatin resistance: Past, present and future. Cell Death Dis. 2014, 5, e1257. [Google Scholar] [CrossRef] [PubMed]
- Gerber, W.V.; Yatskievych, T.A.; Antin, P.; Correia, K.M.; Conlon, R.A.; Krieg, P.A. The RNA-binding protein gene, hermes, is expressed at high levels in the developing heart. Mech. Dev. 1999, 80, 77–86. [Google Scholar] [CrossRef]
- Kwong, J.M.K.; Caprioli, J.; Piri, N. RNA binding protein with multiple splicing: A new marker for retinal ganglion cells. Investig. Opthalmol. Vis. Sci. 2010, 51, 1052–1058. [Google Scholar] [CrossRef] [PubMed]
- Farazi, T.A.; Leonhardt, C.S.; Mukherjee, N.; Mihailovic, A.; Li, S.; Max, K.E.; Meyer, C.; Yamaji, M.; Cekan, P.; Jacobs, N.C.; et al. Identification of the RNA recognition element of the RBPMS family of RNA-binding proteins and their transcriptome-wide mRNA targets. RNA 2014, 20, 1090–1102. [Google Scholar] [CrossRef]
- Shimamoto, A.; Kitao, S.; Ichikawa, K.; Suzuki, N.; Yamabe, Y.; Imamura, O.; Tokutake, Y.; Satoh, M.; Matsumoto, T.; Kuromitsu, J.; et al. A unique human gene that spans over 230 kb in the human chromosome 8p11-12 and codes multiple family proteins sharing RNA-binding motifs. Proc. Natl. Acad. Sci. USA 1996, 93, 10913–10917. [Google Scholar] [CrossRef]
- Rabelo-Fernández, R.J.; Santiago-Sánchez, G.S.; Sharma, R.K.; Roche-Lima, A.; Carrion, K.C.; Rivera, R.A.N.; Quiñones-Díaz, B.I.; Rajasekaran, S.; Siddiqui, J.; Miles, W.; et al. Reduced RBPMS Levels Promote Cell Proliferation and Decrease Cisplatin Sensitivity in Ovarian Cancer Cells. Int. J. Mol. Sci. 2022, 23, 535. [Google Scholar] [CrossRef]
- Fu, J.; Cheng, L.; Wang, Y.; Yuan, P.; Xu, X.; Ding, L.; Zhang, H.; Jiang, K.; Song, H.; Chen, Z.; et al. The RNA-binding protein RBPMS1 represses AP-1 signaling and regulates breast cancer cell proliferation and migration. Biochim. Biophys. Acta 2015, 1853, 1–13. [Google Scholar] [CrossRef]
- Sun, Y.; Ding, L.; Zhang, H.; Han, J.; Yang, X.; Yan, J.; Zhu, Y.; Li, J.; Song, H.; Ye, Q. Potentiation of Smad-mediated transcriptional activation by the RNA-binding protein RBPMS. Nucleic Acids Res. 2006, 34, 6314–6326. [Google Scholar] [CrossRef] [PubMed]
- Santana-Rivera, Y.; Rabelo-Fernández, R.J.; Quiñones-Díaz, B.I.; Grafals-Ruíz, N.; Santiago-Sánchez, G.; Lozada-Delgado, E.L.; Echevarría-Vargas, I.M.; Apiz, J.; Soto, D.; Rosado, A.; et al. Reduced expression of enolase-1 correlates with high intracellular glucose levels and increased senescence in cisplatin-resistant ovarian cancer cells. Am. J. Transl. Res. 2020, 12, 1275–1292. [Google Scholar] [PubMed]
- Huang, J.; Yao, W.-Y.; Zhu, Q.; Tu, S.-P.; Yuan, F.; Wang, H.-F.; Zhang, Y.-P.; Yuan, Y.-Z. XAF1 as a prognostic biomarker and therapeutic target in pancreatic cancer. Cancer Sci. 2010, 101, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, U.R.; Samant, R.S.; Fodstad, O.; Shevde, L.A. Emerging role of nuclear protein 1 (NUPR1) in cancer biology. Cancer Metastasis Rev. 2009, 28, 225–232. [Google Scholar] [PubMed]
- Flores, E.R. The roles of p63 in cancer. Cell Cycle 2007, 6, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Hallen, L.; Burki, Y.; Ebeling, M.; Broger, C.; Siegrist, F.; Oroszlan-Szovik, K.; Bohrmann, B.; Certa, U.; Foser, S. Antiproliferative activity of the human IFN-alpha-inducible protein IFI44. J. Interf. Cytokine Res. 2007, 27, 675–680. [Google Scholar] [CrossRef]
- Pan, H.; Wang, X.; Huang, W.; Dai, Y.; Yang, M.; Liang, H.; Wu, X.; Zhang, L.; Huang, W.; Yuan, L.; et al. Interferon-Induced Protein 44 Correlated With Immune Infiltration Serves as a Potential Prognostic Indicator in Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 557157. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Langevin, S.M.; Kuhnell, D.; Niu, L.; Biesiada, J.; Leung, Y.K.; Deka, R.; Chen, A.; Medvedovic, M.; Kelsey, K.T.; Kasper, S.; et al. Comprehensive mapping of the methylation landscape of 16 CpG-dense regions in oral and pharyngeal squamous cell carcinoma. Epigenomics 2019, 11, 987–1002. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, N.; Yao, W.; Li, S.; Ren, Z. RAD51 is a potential marker for prognosis and regulates cell proliferation in pancreatic cancer. Cancer Cell Int. 2019, 19, 356. [Google Scholar] [CrossRef]
- Huang, W.-C.; Tung, S.-L.; Chen, Y.-L.; Chen, P.-M.; Chu, P.-Y. IFI44L is a novel tumor suppressor in human hepatocellular carcinoma affecting cancer stemness, metastasis, and drug resistance via regulating met/Src signaling pathway. BMC Cancer 2018, 18, 609. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, X.; Liang, Q.; Wang, S.; Xiwen, L.; Pan, F.; Chen, H.; Li, D. Distinct prognostic value of mRNA expression of guanylate-binding protein genes in skin cutaneous melanoma. Oncol. Lett. 2018, 15, 7914–7922. [Google Scholar] [CrossRef]
- Song, F.; Yi, Y.; Li, C.; Hu, Y.; Wang, J.; Smith, D.E.; Jiang, H. Regulation and biological role of the peptide/histidine transporter SLC15A3 in Toll-like receptor-mediated inflammatory responses in macrophage. Cell Death Dis. 2018, 9, 770. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Wu, J.; Lu, C.; Yan, T.; Qian, Y.; Shen, H.; Zhao, Y.; Wang, J.; Kong, P.; Zhang, X. Systematic Transcriptome Analysis Reveals the Inhibitory Function of Cinnamaldehyde in Non-Small Cell Lung Cancer. Front. Pharmacol. 2020, 11, 611060. [Google Scholar] [CrossRef] [PubMed]
- Mukai, S.; Oue, N.; Oshima, T.; Mukai, R.; Tatsumoto, Y.; Sakamoto, N.; Sentani, K.; Tanabe, K.; Egi, H.; Hinoi, T.; et al. Overexpression of Transmembrane Protein BST2 is Associated with Poor Survival of Patients with Esophageal, Gastric, or Colorectal Cancer. Ann. Surg. Oncol. 2017, 24, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Hu, W.; Zhang, L.; Jiang, F.; Mao, X.; Yang, G.; Li, L. FGF21 facilitates autophagy in prostate cancer cells by inhibiting the PI3K-Akt-mTOR signaling pathway. Cell Death Dis. 2021, 12, 303. [Google Scholar] [CrossRef] [PubMed]
- Herrin, B.; Groeger, A.L.; Justement, L. The adaptor protein HSH2 attenuates apoptosis in response to ligation of the B cell antigen receptor complex on the B lymphoma cell line, WEHI-231. J. Biol. Chem. 2005, 280, 3507–3515. [Google Scholar] [CrossRef]
- Masuda, T.; Ishikawa, T.; Mogushi, K.; Okazaki, S.; Ishiguro, M.; Iida, S.; Mizushima, H.; Tanaka, H.; Uetake, H.; Sugihara, K. Overexpression of the S100A2 protein as a prognostic marker for patients with stage II and III colorectal cancer. Int. J. Oncol. 2016, 48, 975–982. [Google Scholar] [CrossRef]
- Zou, A.; Lin, Z.; Humble, M.; Creech, C.D.; Wagoner, P.K.; Krafte, D.; Jegla, T.J.; Wickenden, A.D. Distribution and functional properties of human KCNH8 (Elk1) potassium channels. Am. J. Physiol. Cell Physiol. 2003, 285, C1356–C1366. [Google Scholar] [CrossRef]
- Dieci, G.; Preti, M.; Montanini, B. Eukaryotic snoRNAs: A paradigm for gene expression flexibility. Genomics 2009, 94, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Voss, F.K.; Ullrich, F.; Münch, J.; Lazarow, K.; Lutter, D.; Mah, N.; Andrade-Navarro, M.A.; von Kries, J.P.; Stauber, T.; Jentsch, T.J. Identification of LRRC8 heteromers as an essential component of the volume-regulated anion channel VRAC. Science 2014, 344, 634–638. [Google Scholar] [CrossRef]
- Suzuki, N.; Nara, K.; Suzuki, T. Skewed Th1 responses caused by excessive expression of Txk, a member of the Tec family of tyrosine kinases, in patients with Behcet’s disease. Clin. Med. Res. 2006, 4, 147–151. [Google Scholar] [CrossRef][Green Version]
- Piovani, G.; Savio, G.; Traversa, M.; Pilotta, A.; De Petro, G.; Barlati, S.; Magri, C. De novo 1Mb interstitial deletion of 8p22 in a patient with slight mental retardation and speech delay. Mol. Cytogenet. 2014, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tian, R.; Gao, H.; Yang, Y.; Williams, B.R.G.; Gantier, M.P.; McMillan, N.A.J.; Xu, D.; Hu, Y.; Gao, Y. Identification of a histone family gene signature for predicting the prognosis of cervical cancer patients. Sci. Rep. 2017, 7, 16495. [Google Scholar] [CrossRef]
- Li, Y.; Su, Z.; Wei, B.; Qin, M.; Liang, Z. Bioinformatics analysis identified MMP14 and COL12A1 as immune-related biomarkers associated with pancreatic adenocarcinoma prognosis. Math. Biosci. Eng. 2021, 18, 5921–5942. [Google Scholar] [CrossRef]
- Yang, M.-H.; Yen, C.-H.; Chen, Y.-F.; Fang, C.-C.; Li, C.-H.; Lee, K.-J.; Lin, Y.-H.; Weng, C.-H.; Liu, T.-T.; Huang, S.-F.; et al. Somatic mutations of PREX2 gene in patients with hepatocellular carcinoma. Sci. Rep. 2019, 9, 2552. [Google Scholar] [CrossRef] [PubMed]
- Craig, M.J.; Loberg, R.D. CCL2 (Monocyte Chemoattractant Protein-1) in cancer bone metastases. Cancer Metastasis Rev. 2006, 25, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhang, Y.; Jiang, L.; Zhang, M.; Chen, Z.; Liu, D.; Huang, Q. Disabled homolog 2 is required for migration and invasion of prostate cancer cells. Front. Med. 2015, 9, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, L.; Allen, W.L.; Proutski, I.; Stewart, G.; Johnston, L.; McCloskey, K.; Wilson, P.M.; Longley, D.B.; Johnston, P.G. Calbindin 2 (CALB2) regulates 5-fluorouracil sensitivity in colorectal cancer by modulating the intrinsic apoptotic pathway. PLoS ONE 2011, 6, e20276. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chen, Y.H.; Hong, H.; Zeng, Y.; Zhang, J.; Lu, J.P.; Jeansonne, B.; Lu, Q. Increased nucleotide polymorphic changes in the 5′-untranslated region of delta-catenin (CTNND2) gene in prostate cancer. Oncogene 2009, 28, 555–564. [Google Scholar] [CrossRef]
- Sakaki, T.; Yasuda, K.; Kittaka, A.; Yamamoto, K.; Chen, T.C. CYP24A1 as a potential target for cancer therapy. Anti-Cancer Agents Med. Chem. 2014, 14, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Brandenberger, R.; Wei, H.; Zhang, S.; Lei, S.; Murage, J.; Fisk, G.J.; Li, Y.; Xu, C.; Fang, R.; Guegler, K.; et al. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat. Biotechnol. 2004, 22, 707–716. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, X.; Nan, C.; Zhao, Z.; Ma, S.; Li, W.; Hu, H.; Liang, Z. MicroRNA-182 targets protein phosphatase 1 regulatory inhibitor subunit 1C in glioblastoma. Oncotarget 2017, 8, 114677–114684. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luan, J.; Gao, X.; Hu, F.; Zhang, Y.; Gou, X. SLFN11 is a general target for enhancing the sensitivity of cancer to chemotherapy (DNA-damaging agents). J. Drug Target. 2020, 28, 33–40. [Google Scholar] [CrossRef]
- Murn, J.; Alibert, O.; Wu, N.; Tendil, S.; Gidrol, X. Prostaglandin E2 regulates B cell proliferation through a candidate tumor suppressor, Ptger4. J. Exp. Med. 2008, 205, 3091–3103. [Google Scholar] [CrossRef]
- Wan, N.; Yang, W.; Cheng, H.; Wang, J. FOXD3-AS1 Contributes to the Progression of Melanoma Via miR-127-3p/FJX1 Axis. Cancer Biother. Radiopharm. 2020, 35, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Requena, T.; Cabrera, S.; Martín-Sierra, C.; Price, S.D.; Lysakowski, A.; Lopez-Escamez, J.A. Identification of two novel mutations in FAM136A and DTNA genes in autosomal-dominant familial Meniere’s disease. Hum. Mol. Genet. 2015, 24, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Tan, N.-N.; Tang, H.-L.; Lin, G.-W.; Chen, Y.-H.; Lu, P.; Li, H.-J.; Gao, M.-M.; Zhao, Q.-H.; Yi, Y.-H.; Liao, W.-P.; et al. Epigenetic Downregulation of Scn3a Expression by Valproate: A Possible Role in Its Anticonvulsant Activity. Mol. Neurobiol. 2017, 54, 2831–2842. [Google Scholar] [CrossRef]
- Sanuki, R.; Omori, Y.; Koike, C.; Sato, S.; Furukawa, T. Panky, a novel photoreceptor-specific ankyrin repeat protein, is a transcriptional cofactor that suppresses CRX-regulated photoreceptor genes. FEBS Lett. 2010, 584, 753–758. [Google Scholar] [CrossRef]
- Bhushan, A.; Singh, A.; Kapur, S.; Borthakar, B.B.; Sharma, J.; Rai, A.K.; Kataki, A.C.; Saxena, S. Identification and Validation of Fibroblast Growth Factor 12 Gene as a Novel Potential Biomarker in Esophageal Cancer Using Cancer Genomic Datasets. OMICS J. Integr. Biol. 2017, 21, 616–631. [Google Scholar] [CrossRef]
- Yan, L.; Gong, Y.Z.; Shao, M.N.; Ruan, G.T.; Xie, H.L.; Liao, X.W.; Wang, X.K.; Han, Q.F.; Zhou, X.; Zhu, L.C.; et al. Distinct diagnostic and prognostic values of gamma-aminobutyric acid type A receptor family genes in patients with colon adenocarcinoma. Oncol. Lett. 2020, 20, 275–291. [Google Scholar] [CrossRef]
- Guan, Y.; Bhandari, A.; Xia, E.; Yang, F.; Xiang, J.; Wang, O. lncRNA FOXD3-AS1 is associated with clinical progression and regulates cell migration and invasion in breast cancer. Cell Biochem. Funct. 2019, 37, 239–244. [Google Scholar] [CrossRef]
- Northrop, J.P.; Ho, S.N.; Chen, L.; Thomas, D.J.; Timmerman, L.A.; Nolan, G.P.; Admon, A.; Crabtree, G.R. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature 1994, 369, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, R.E.; Myers, M.; Duncan, W.C. Novel regulated expression of the SLIT/ROBO pathway in the ovary: Possible role during luteolysis in women. Endocrinology 2008, 149, 5024–5034. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shimoyama, Y.; Gotoh, M.; Terasaki, T.; Kitajima, M.; Hirohashi, S. Isolation and sequence analysis of human cadherin-6 complementary DNA for the full coding sequence and its expression in human carcinoma cells. Cancer Res. 1995, 55, 2206–2211. [Google Scholar] [PubMed]
- Zhang, Y.; Yu, Y.; Su, X.; Lu, Y. HOXD8 inhibits the proliferation and migration of triple-negative breast cancer cells and induces apoptosis in them through regulation of AKT/mTOR pathway. Reprod. Biol. 2021, 21, 100544. [Google Scholar] [CrossRef] [PubMed]
- Rudnicki, M.A.; Jackowski, G.; Saggin, L.; McBurney, M.W. Actin and myosin expression during development of cardiac muscle from cultured embryonal carcinoma cells. Dev. Biol. 1990, 138, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Niwa, R.; Nagata-Ohashi, K.; Takeichi, M.; Mizuno, K.; Uemura, T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 2002, 108, 233–246. [Google Scholar] [CrossRef]
- Liu, D.-B.; Gu, Z.-D.; Cao, X.-Z.; Liu, H.; Li, J.-Y. Immunocytochemical detection of HoxD9 and Pbx1 homeodomain protein expression in Chinese esophageal squamous cell carcinomas. World J. Gastroenterol. 2005, 11, 1562–1566. [Google Scholar] [CrossRef]
- Lee, Y.-R.; Yuan, W.-C.; Ho, H.-C.; Chen, C.-H.; Shih, H.-M.; Chen, R.-H. The Cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J. 2010, 29, 1748–1761. [Google Scholar] [CrossRef]
- Hogan, A.; Shepherd, L.; Chabot, J.; Quenneville, S.; Prescott, S.M.; Topham, M.K.; Gee, S.H. Interaction of gamma 1-syntrophin with diacylglycerol kinase-zeta. Regulation of nuclear localization by PDZ interactions. J. Biol. Chem. 2001, 276, 26526–26533. [Google Scholar] [CrossRef]
- Gagnon, M.L.; Bielenberg, D.R.; Gechtman, Z.; Miao, H.Q.; Takashima, S.; Soker, S.; Klagsbrun, M. Identification of a natural soluble neuropilin-1 that binds vascular endothelial growth factor: In vivo expression and antitumor activity. Proc. Natl. Acad. Sci. USA 2000, 97, 2573–2578. [Google Scholar] [CrossRef]
- Liu, D.; Zhuang, Y.; Zhang, L.; Gao, H.; Neavin, D.; Carrillo-Roa, T.; Wang, Y.; Yu, J.; Qin, S.; Kim, D.C.; et al. ERICH3: Vesicular association and antidepressant treatment response. Mol. Psychiatry 2021, 26, 2415–2428. [Google Scholar] [CrossRef]
- Liu, Z.; Zhu, Y.; Li, F.; Xie, Y. GATA1-regulated JAG1 promotes ovarian cancer progression by activating Notch signal pathway. Protoplasma 2020, 257, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Rossjohn, J.; Gras, S.; Miles, J.J.; Turner, S.J.; Godfrey, D.I.; McCluskey, J. T Cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 2015, 33, 169–200. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef] [PubMed]
- Nakagaki-Silva, E.E.; Gooding, C.; Llorian, M.; Jacob, A.G.; Richards, F.; Buckroyd, A.; Sinha, S.; Smith, C.W. Identification of RBPMS as a mammalian smooth muscle master splicing regulator via proximity of its gene with super-enhancers. eLife 2019, 8, e46327. [Google Scholar] [CrossRef]
- Rastgoo, N.; Pourabdollah, M.; Abdi, J.; Reece, D.; Chang, H. Dysregulation of EZH2/miR-138 axis contributes to drug resistance in multiple myeloma by downregulating RBPMS. Leukemia 2018, 32, 2471–2482. [Google Scholar] [CrossRef]
- Gordon, R.R.; Nelson, P.S. Cellular senescence and cancer chemotherapy resistance. Drug Resist. Updat. 2012, 15, 123–131. [Google Scholar] [CrossRef]
- Guo, X.; Ma, N.; Wang, J.; Song, J.; Bu, X.; Cheng, Y.; Sun, K.; Xiong, H.; Jiang, G.; Zhang, B.; et al. Increased p38-MAPK is responsible for chemotherapy resistance in human gastric cancer cells. BMC Cancer 2008, 8, 375. [Google Scholar] [CrossRef]
- Guillon, J.; Petit, C.; Toutain, B.; Guette, C.; Lelievre, E.; Coqueret, O. Chemotherapy-induced senescence, an adaptive mechanism driving resistance and tumor heterogeneity. Cell Cycle 2019, 18, 2385–2397. [Google Scholar] [CrossRef]
- Bavik, C.; Coleman, I.; Dean, J.P.; Knudsen, B.; Plymate, S.; Nelson, P.S. The gene expression program of prostate fibroblast senescence modulates neoplastic epithelial cell proliferation through paracrine mechanisms. Cancer Res. 2006, 66, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Krtolica, A.; Parrinello, S.; Lockett, S.; Desprez, P.-Y.; Campisi, J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: A link between cancer and aging. Proc. Natl. Acad. Sci. USA 2001, 98, 12072–12077. [Google Scholar] [CrossRef] [PubMed]
- Coppé, J.-P.; Patil, C.K.; Rodier, F.; Sun, Y.; Muñoz, D.P.; Goldstein, J.; Nelson, P.S.; Desprez, P.-Y.; Campisi, J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008, 6, e301. [Google Scholar] [CrossRef] [PubMed]
- McConkey, D.J.; Choi, W.; Marquis, L.; Martin, F.; Williams, M.B.; Shah, J.; Svatek, R.; Das, A.; Adam, L.; Kamat, A.; et al. Role of epithelial-to-mesenchymal transition (EMT) in drug sensitivity and metastasis in bladder cancer. Cancer Metastasis Rev. 2009, 28, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Mou, Z.; Zhang, Z.; Wu, S.; Zhou, Q.; Chen, Y.; Gong, J.; Xu, C.; Ou, Y.; Chen, X.; et al. Circular RNA RBPMS inhibits bladder cancer progression via miR-330-3p/RAI2 regulation. Mol. Ther. Nucleic Acids 2021, 23, 872–886. [Google Scholar] [CrossRef]
- Shanmugaapriya, S.; van Caam, A.; de Kroon, L.; Vitters, E.L.; Walgreen, B.; van Beuningen, H.; Davidson, E.B.; van der Kraan, P.M. Expression of TGF-beta Signaling Regulator RBPMS (RNA-Binding Protein With Multiple Splicing) Is Regulated by IL-1beta and TGF-beta Superfamily Members, and Decreased in Aged and Osteoarthritic Cartilage. Cartilage 2016, 7, 333–345. [Google Scholar] [CrossRef]
- Jiang, W.G.; Davies, G.; Martin, T.A.; Kynaston, H.; Mason, M.D.; Fodstad, O. Com-1/p8 acts as a putative tumour suppressor in prostate cancer. Int. J. Mol. Med. 2006, 18, 981–986. [Google Scholar] [CrossRef]
- Zhou, H.-H.; Chen, L.; Liang, H.-F.; Li, G.-Z.; Zhang, B.-X.; Chen, X.-P. Smad3 Sensitizes Hepatocelluar Carcinoma Cells to Cisplatin by Repressing Phosphorylation of AKT. Int. J. Mol. Sci. 2016, 17, 610. [Google Scholar] [CrossRef]
- Clark, D.; Mitra, A.; Fillmore, R.; Jiang, W.; Samant, R.; Fodstad, O.; Shevde, L. NUPR1 interacts with p53, transcriptionally regulates p21 and rescues breast epithelial cells from doxorubicin-induced genotoxic stress. Curr. Cancer Drug Targets 2008, 8, 421–430. [Google Scholar] [CrossRef]
- Jiang, W.G.; Watkins, G.; Douglas-Jones, A.; Mokbel, K.; Mansel, R.E.; Fodstad, O. Expression of Com-1/P8 in human breast cancer and its relevance to clinical outcome and ER status. Int. J. Cancer 2005, 117, 730–737. [Google Scholar] [CrossRef]
- Huang, T.-C.; Chung, Y.-L.; Wu, M.-L.; Chuang, S.-M. Cinnamaldehyde enhances Nrf2 nuclear translocation to upregulate phase II detoxifying enzyme expression in HepG2 cells. J. Agric. Food Chem. 2011, 59, 5164–5171. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Ma, W.; Hong, Z.; Wu, L.; Chen, X.; Yuan, Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, 11. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Ye, L.; Ji, K.; Toms, A.-M.; Davies, M.L.; Ruge, F.; Ji, J.; Hargest, R.; Jiang, W.G. Death associated protein 1 is correlated with the clinical outcome of patients with colorectal cancer and has a role in the regulation of cell death. Oncol. Rep. 2014, 31, 175–182. [Google Scholar] [CrossRef]
- Winkler, C.; Armenia, J.; Jones, G.N.; Tobalina, L.; Sale, M.J.; Petreus, T.; Baird, T.; Serra, V.; Wang, A.T.; Lau, A.; et al. SLFN11 informs on standard of care and novel treatments in a wide range of cancer models. Br. J. Cancer 2021, 124, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.P.; Ely, B.D.; Pong, R.C.; Wang, Z.; Zhou, J.; Hsieh, J.T. The role of DOC-2/DAB2 protein phosphorylation in the inhibition of AP-1 activity. An underlying mechanism of its tumor-suppressive function in prostate cancer. J. Biol. Chem. 1999, 274, 31981–31986. [Google Scholar] [CrossRef] [PubMed]
- Melino, G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011, 18, 1487–1499. [Google Scholar] [CrossRef]
- Sundqvist, A.; Vasilaki, E.; Voytyuk, O.; Bai, Y.; Morikawa, M.; Moustakas, A.; Miyazono, K.; Heldin, C.-H.; Dijke, P.T.; van Dam, H. TGFβ and EGF signaling orchestrates the AP-1- and p63 transcriptional regulation of breast cancer invasiveness. Oncogene 2020, 39, 4436–4449. [Google Scholar] [CrossRef]
- Lau, C.P.Y.; Ng, P.K.S.; Li, M.S.; Tsui, S.K.W.; Huang, L.; Kumta, S.M. p63 regulates cell proliferation and cell cycle progression-associated genes in stromal cells of giant cell tumor of the bone. Int. J. Oncol. 2013, 42, 437–443. [Google Scholar] [CrossRef]
- Senoo, M.; Pinto, F.; Crum, C.P.; McKeon, F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 2007, 129, 523–536. [Google Scholar] [CrossRef]
- He, Z.; Tessier-Lavigne, M. Neuropilin is a receptor for the axonal chemorepellent Semaphorin III. Cell 1997, 90, 739–751. [Google Scholar] [CrossRef]
- Soker, S.; Takashima, S.; Miao, H.Q.; Neufeld, G.; Klagsbrun, M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell 1998, 92, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; Huang, C.; Wang, H.; Wang, C.; Dong, L.; Gu, X.; Feng, X.; Chen, B. Association study between CYP24A1 gene polymorphisms and cancer risk. Pathol. Res. Pract. 2020, 216, 152735. [Google Scholar] [CrossRef] [PubMed]
- Reyes-González, J.M.; Vivas-Mejía, P.E. c-MYC and Epithelial Ovarian Cancer. Front. Oncol. 2021, 11, 601512. [Google Scholar] [CrossRef]
- Echevarria-Vargas, I.M.; Valiyeva, F.; Vivas-Mejia, P.E. Upregulation of miR-21 in cisplatin resistant ovarian cancer via JNK-1/c-Jun pathway. PLoS ONE 2014, 9, e97094. [Google Scholar] [CrossRef]
- Vivas-Mejia, P.E.; Rodriguez-Aguayo, C.; Han, H.-D.; Shahzad, M.M.; Valiyeva, F.; Shibayama, M.; Chavez-Reyes, A.; Sood, A.K.; Lopez-Berestein, G. Silencing survivin splice variant 2B leads to antitumor activity in taxane-resistant ovarian cancer. Clin. Cancer Res. 2011, 17, 3716–3726. [Google Scholar] [CrossRef] [PubMed]
- Quiñones-Díaz, B.I.; Reyes-González, J.M.; Sánchez-Guzmán, V.; Moral, I.C.-D.; Valiyeva, F.; Santiago-Sánchez, G.S.; Vivas-Mejía, P.E. MicroRNA-18a-5p Suppresses Tumor Growth via Targeting Matrix Metalloproteinase-3 in Cisplatin-Resistant Ovarian Cancer. Front. Oncol. 2020, 10, 602670. [Google Scholar] [CrossRef] [PubMed]
- Bahar, E.; Yoon, H. Modeling and Predicting the Cell Migration Properties from Scratch Wound Healing Assay on Cisplatin-Resistant Ovarian Cancer Cell Lines Using Artificial Neural Network. Healthcare 2021, 9, 911. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016, 44, D184–D189. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Gerace, E.; Moazed, D. Affinity Pull-Down of Proteins Using Anti-FLAG M2 Agarose Beads. Methods Enzymol. 2015, 559, 99–110. [Google Scholar] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef]
- Győrffy, B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput. Struct. Biotechnol. J. 2021, 19, 4101–4109. [Google Scholar] [CrossRef] [PubMed]




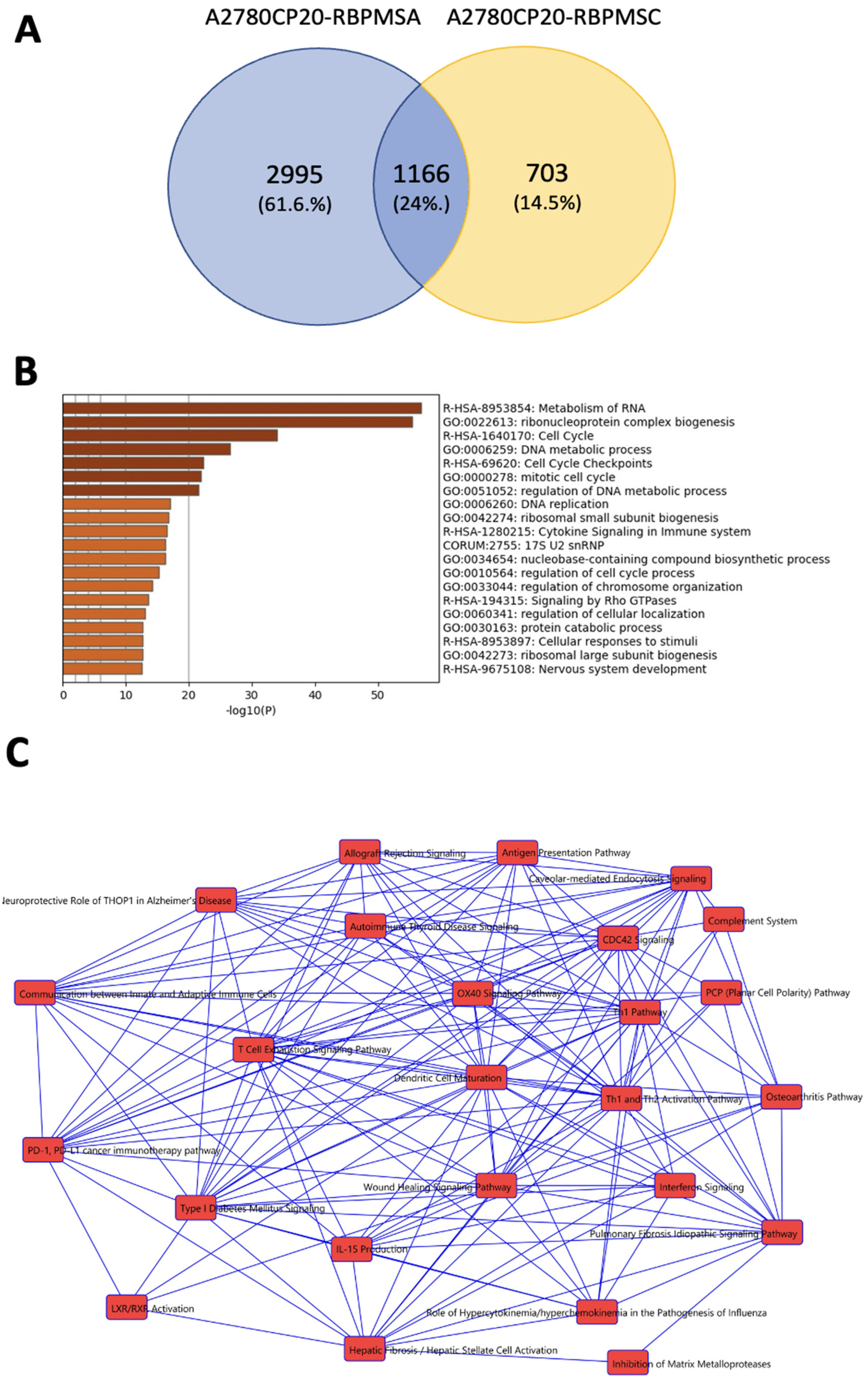

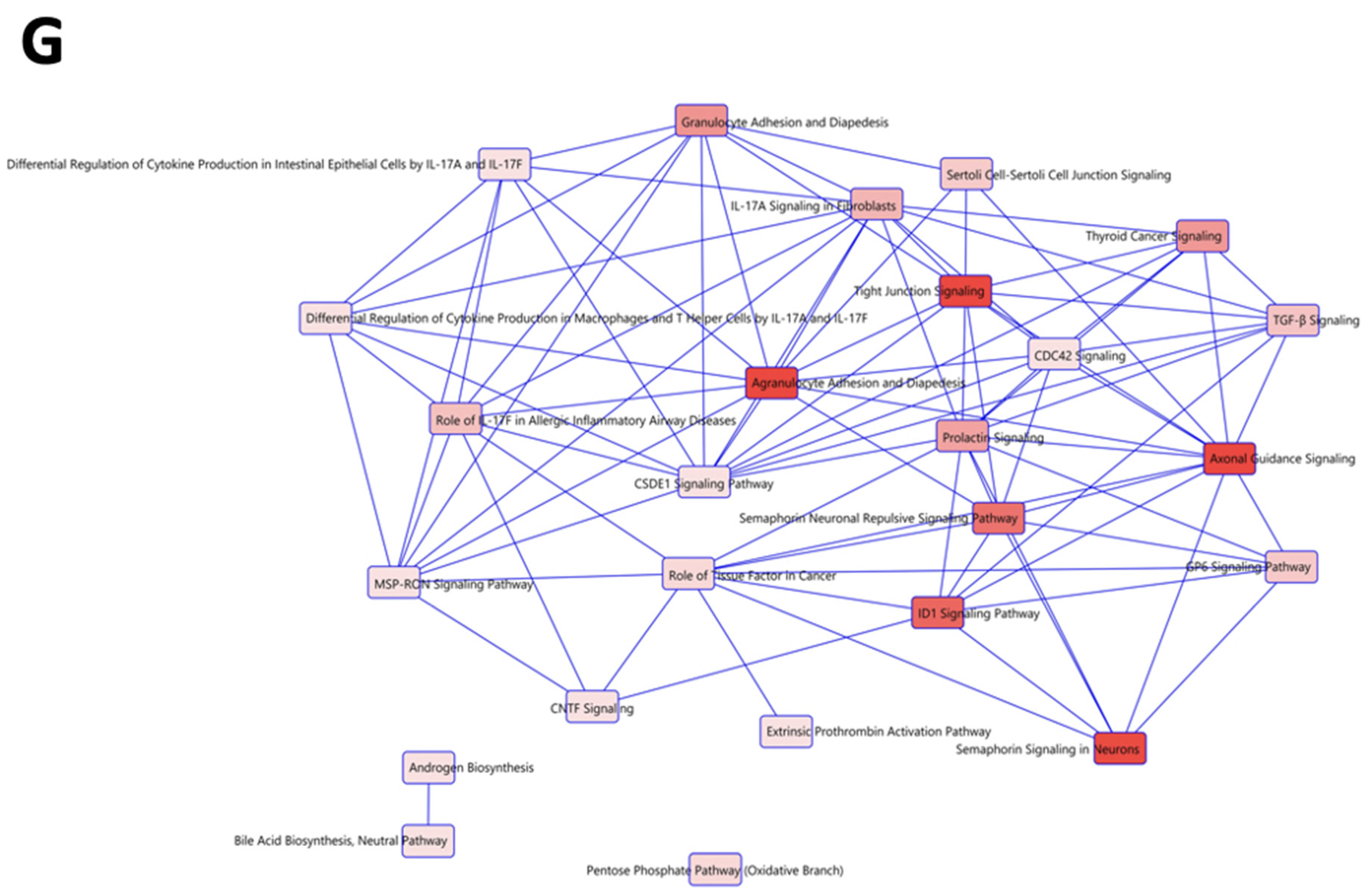
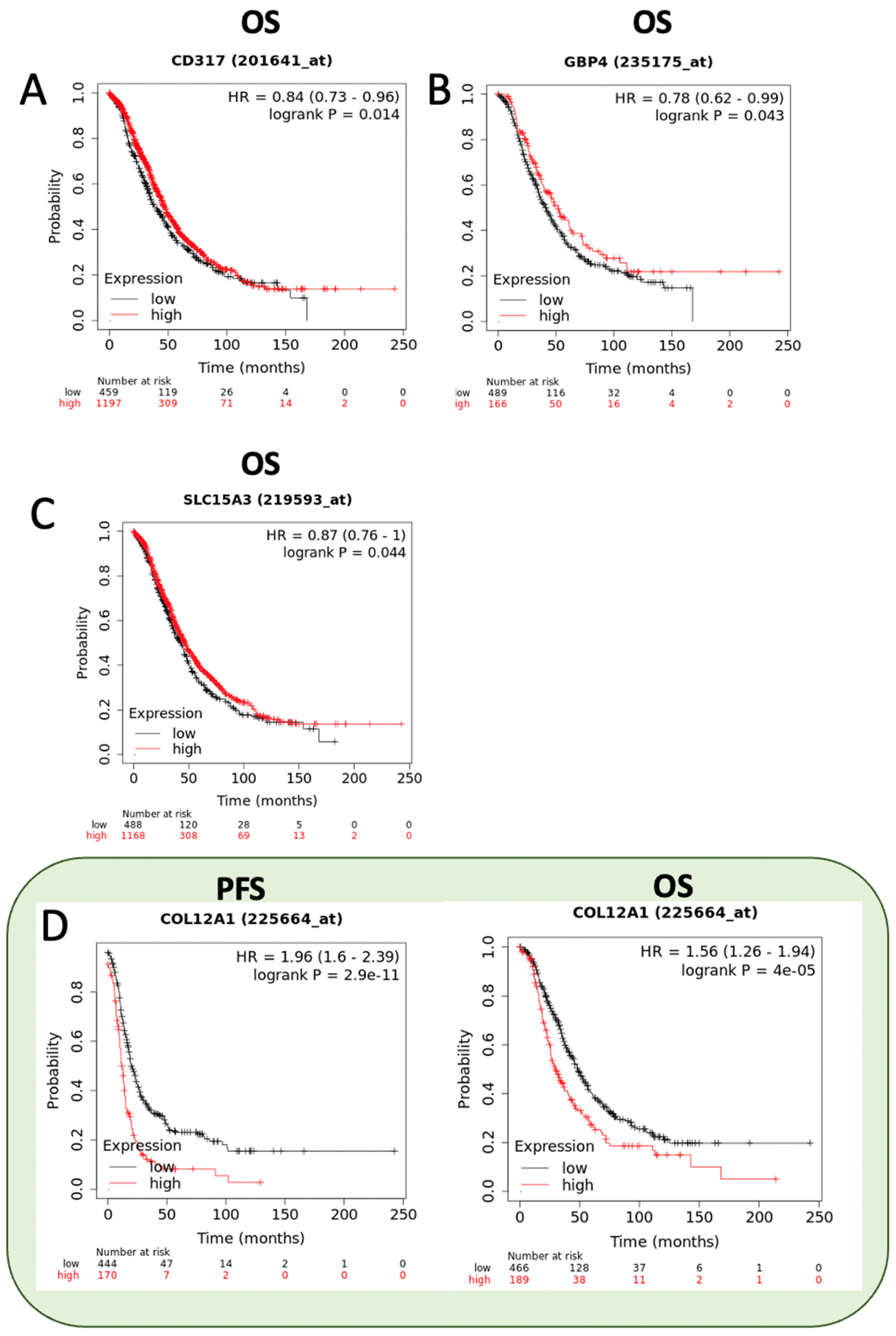
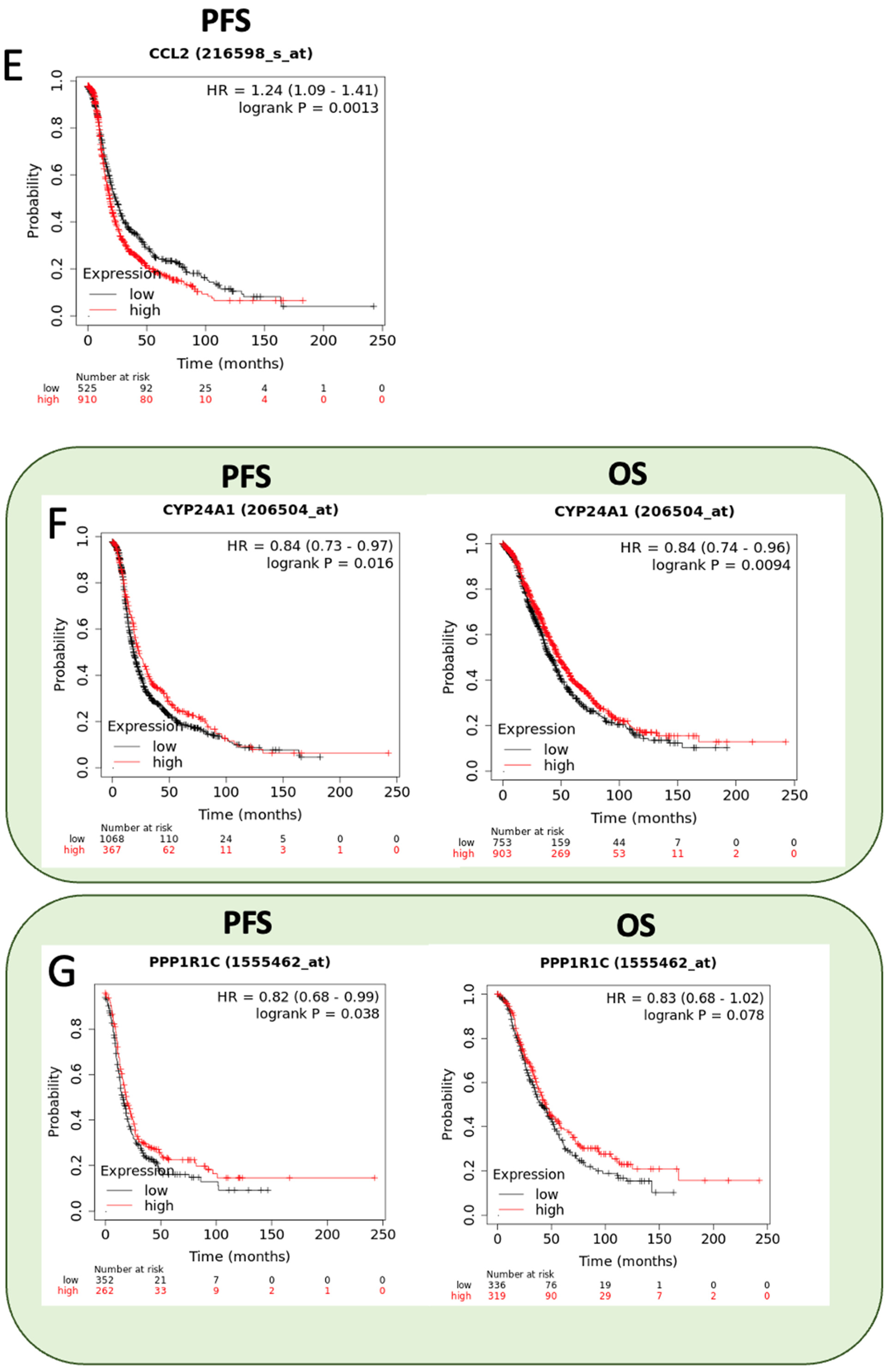

| Symbol | Gene Name | Fold Change | Biological Role | Reference |
|---|---|---|---|---|
| IFI44 | Interferon induced protein 44 | 9.66541828 | Plays a role in the immune response during autoimmune diseases. | [29] |
| XAF1 | XIAP Associated Factor 1 | 8.297767889 | A putative tumor suppressor candidate that junction to several pathways leading to apoptosis. | [21] |
| GBP4 | Guanylate Binding Protein 4 | 6.931543382 | Involved in the host-defense mechanisms response against cellular pathogens and tumorigenesis. | [30] |
| SLC15A3 | Solute Carrier Family 15 Member 3 | 6.865730827 | Transporting histidine, peptides and peptidomimetics from inside the lysosome to cytosol. | [31] |
| RBPMS | RNA Binding Protein | 6.758908087 | Regulate the RNA transport, stability and localization. | [17] |
| LINC01504 | Long Intergenic Non-Protein Coding RNA 1504 | 6.554246988 | A lncRNA which has a role on the suppression of malignant phenotypes of lung cancer. | [32] |
| NUPR1 | Nuclear Protein 1, Transcriptional Regulator | 6.087442834 | Upregulation of this protein is associated with malignant characteristics of cancer as well as with chemoresistance. | [22] |
| BST2 | Bone Marrow Stromal Cell Antigen 2 | 5.971957997 | Lipid raft-associated type II transmembrane glycoprotein which mediates various facets of cancer progression and metastasis | [33] |
| FGF21 | Fibroblast Growth Factor 21 | 5.930365363 | Member of the FGF family which possess broad mitogenic and cell survival activities. | [34] |
| HSH2D | Hematopoietic SH2 Domain Containing | 5.864666169 | Play a role in various cellular functions such as apoptosis, membrane-associated intracellular trafficking and the biogenesis of lipid and collagen remodeling. | [35] |
| S100A2 | S100 Calcium Binding Protein A2 | −2.477696881 | Plays a role in metastasis process by transforming growth factor-β (TGF-β) mediated cancer cell invasion and migration. | [36] |
| KCNH4 | Potassium Voltage-Gated Channel Subfamily H Member 4 | −2.510279699 | Transport positively charged potassium atoms between neighboring cells. KCNH4 plays a key role in the ability of cells to generate and transmit electrical signals. | [37] |
| SNORD99 | Small Nucleolar RNA, C/D Box 99 | −2.521724113 | Related with diverse cellular functions such as regulation of T cell proliferation and death balance to promoting cancer cell plasticity. | [38] |
| LRRC8D-DT | LRRC8D Divergent Transcript | −3.051305443 | Plays important pharmacological and physiological roles in supporting the transport of anti-cancer drugs and of the organic osmolyte taurine. | [39] |
| TXK | TXK Tyrosine Kinase | −3.120303742 | Play important roles in the immune response and pathway signaling mediator | [40] |
| SGCZ | Sarcoglycan Zeta | −4.110780038 | Part of the sarcoglycan complex which have a structural role in connecting cytoskeletal proteins with the extracellular matrix. | [41] |
| HIST1H2BH | H2B Clustered Histone 9 | −4.323395136 | Responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Low levels of HIST1H2BEH caused decreased proliferation in breast cancer cell lines. | [42] |
| COL12A1 | Collagen Type XII Alpha 1 Chain | −4.332051747 | Found in several cancer types and could be involved in tumor progression. | [43] |
| PREX2 | Phosphatidylinositol-3,4,5-Trisphosphate Dependent Rac Exchange Factor 2 | −4.381347741 | Implicated in the inhibition of phosphatase and tensin homolog (PTEN). Overexpression significantly increases the proliferation, invasion, and migration of pancreatic cancer. | [44] |
| CCL2 | C-C Motif Chemokine Ligand 2 | −4.644149886 | Strongest chemoattractant synthesized and secreted mainly by monocytic cells. | [45] |
| Symbol | Gene Name | Fold Change | Biological Role | Reference |
|---|---|---|---|---|
| DAB2 | DAB Adaptor Protein 2 | 7.15380118 | Multi-function signaling molecule which catalytic enzyme activity suggest that it is an adaptor molecule involved in multiple receptor-mediated signalling pathways that plays a pivotal role in the cellular homeostasis. | [46] |
| CALB2 | Calbindin 2 | 6.574845254 | Important mediator of 5-FU-induced cell death and specific marker for the diagnosis of malignant mesothelioma. | [47] |
| CTNND2 | Catenin Delta 2 | 6.484328261 | Recognized to be a biomarker for cancers, overexpressed in various types of cancers, including prostate, breast, lung and ovarian cancer. | [48] |
| CYP24A1 | Cytochrome P450 Family 24 Subfamily A Member 1 | 6.041287981 | Member of the cytochrome P450 superfamily of enzymes which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. | [49] |
| FAR2P2 | Fatty Acyl-CoA Reductase 2 Pseudogene 2 | 5.29742507 | Catalyzes the reduction in saturated but not unsaturated C16 or C18 fatty acyl-CoA to fatty alcohols. | [50] |
| RBPMS | RNA Binding Protein | 4.920050075 | Regulate the RNA transport, stability and localization. | [17] |
| PPP1R1C | Protein Phosphatase 1 Regulatory Inhibitor Subunit 1C | 4.253043369 | Major serine/threonine phosphatase that regulates a variety of cellular functions and themselves regulated by phosphorylation. | [51] |
| SLFN11 | Schlafen Family Member 11 | 3.827804248 | DNA/RNA helicase that is recruited during stressed replication fork and irreversibly triggers replication block and cell death. | [52] |
| PTGER4 | Prostaglandin E Receptor 4 | 3.770525307 | Member of the G-protein coupled receptor family which bind and mediate cellular responses to PGE2 and other prostanoids. | [53] |
| FOXD3-AS1 | FOXD3 Antisense RNA 1 | 3.654548595 | Is abnormally expressed in many disease types. Reports suggest that FOXD3-AS1 is highly expressed in different cancer types promoting migration and invasion capacity. | [54] |
| TP63 | Tumor Protein P63 | −2.226163472 | Functions as a transcription factor interacting with other proteins to turn different genes on and off at different times. | [23] |
| DTNA | Dystrobrevin Alpha | −2.582128781 | Belongs to the dystrobrevin subfamily of the dystrophin family. Reports suggest that DTNA binds and activates STAT3 to induce TGFβ1 expression and repress P53 expression. | [55] |
| SCN3A | Sodium Voltage-Gated Channel Alpha Subunit 3 | −4.437260362 | Is a transmembrane glycoprotein responsible for the generation and propagation of action potentials in neurons and muscle. | [56] |
| Symbol | Gene Name | Fold Change | Biological Role | Reference |
|---|---|---|---|---|
| FAR2P2 | Fatty Acyl-CoA Reductase 2 Pseudogene 2 | 5.29742507 | Acts as guanine nucleotide exchange factor that activates RAC1. Also, plays a role in the response to class 3 semaphorins and remodeling of the actin cytoskeleton. | [50] |
| RBPMS | RNA Binding Protein | 4.920050075 | Regulate the RNA transport, stability and localization. | [17] |
| ANKRD33B | Ankyrin Repeat Domain 33B | 4.556503793 | Involved in negative regulation of transcription by RNA polymerase II and negative regulation of transcription regulatory region DNA binding activity. | [57] |
| PPP1R1C | Protein Phosphatase 1 Regulatory Inhibitor Subunit 1C | 4.253043369 | Major serine/threonine phosphatase that regulates a variety of cellular functions and themselves regulated by phosphorylation. | [51] |
| FGF12 | Fibroblast Growth Factor 12 | 3.920423579 | Involved in a broad mitogenic and cell survival activities, including embryonic development, cell growth, morphogenesis, tissue repair, tumor growth, and invasion. | [58] |
| GABRA2 | Gamma-Aminobutyric Acid Type A Receptor Subunit Alpha2 | 3.844344607 | Component of the heteropentameric receptor for GABA, the major inhibitory neurotransmitter in the brain. | [59] |
| FOXD3-AS1 | FOXD3 Antisense RNA 1 | 3.654548595 | Is abnormally expressed in many disease types. Reports suggest that FOXD3-AS1 is highly expressed in different cancer types promoting migration and invasion capacity. | [60] |
| NFATC1 | Nuclear Factor of Activated T Cells 1 | 3.620469318 | Family of proteins that play a central role in inducible gene transcription during immune response. | [61] |
| ROBO2 | Roundabout Guidance Receptor 2 | 3.448549593 | Transmembrane receptor for the slit homolog 2 protein that play a function in axon guidance and cell migration. | [62] |
| CDH6 | Cadherin 6 | 3.421265843 | Membrane glycoprotein that mediates homophilic cell-cell adhesion and play critical roles in cell differentiation and morphogenesis. | [63] |
| HOXD8 | Homeobox D8 | −2.593778164 | Gene belongs to the homeobox family of genes which play an important role in morphogenesis in all multicellular organisms. | [64] |
| MYL7 | Myosin Light Chain 7 | −2.677248207 | Part of the family motor proteins that have ATPase enzyme activity, actin binding and potential for kinetic energy transduction. | [65] |
| SSUH2 | Ssu-2 Homolog | −2.71336991 | Gene that encodes a protein tyrosine phosphatase that plays a key role in the regulation of actin filaments. | [66] |
| HOXD9 | Homeobox D9 | −2.800133712 | Transcription factor which is part of a developmental regulatory system providing cells the specific positional identities on the anterior-posterior axis. | [67] |
| DAPK1 | Death-Associated Protein Kinase 1 | −3.221475672 | Mediator of gamma-interferon involved in multiple cellular signaling pathways that trigger cell survival, apoptosis, and autophagy. | [68] |
| SNTG1 | Syntrophin Gamma 1 | −3.228723507 | Cytoplasmic peripheral membrane proteins that contain 2 pleckstrin domains. | [69] |
| NRP1 | Neuropilin 1 | −3.454159744 | Cell membrane receptor involved in the development of cardiovascular system, angiogenesis, certain neuronal circuits and organogenesis in nervous system. | [70] |
| ERICH3 | Glutamate Rich 3 | −3.951576843 | Interacts with proteins function in vesicle biogenesis and may play a significant role in vesicular function in serotonergic and other neuronal cell types. | [71] |
| JAG1 | Jagged Canonical Notch Ligand 1 | −6.91254142 | Ligand for multiple Notch receptors involved in the mediation of Notch signaling, cell-fate decisions during and cardiovascular development. | [72] |
| TRBV12-4 | T Cell Receptor Beta Variable 12-4 | −6.91254142 | Antigen specific receptor which are essential to the immune response and are present on the cell surface of T lymphocytes | [73] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rabelo-Fernández, R.J.; Noriega Rivera, R.A.; Rivera, Y.S.; Tous-Beveraggi, J.; Valiyeva, F.; Vivas-Mejia, P.E. Increased Expression of the RBPMS Splice Variants Inhibits Cell Proliferation in Ovarian Cancer Cells. Int. J. Mol. Sci. 2022, 23, 14742. https://doi.org/10.3390/ijms232314742
Rabelo-Fernández RJ, Noriega Rivera RA, Rivera YS, Tous-Beveraggi J, Valiyeva F, Vivas-Mejia PE. Increased Expression of the RBPMS Splice Variants Inhibits Cell Proliferation in Ovarian Cancer Cells. International Journal of Molecular Sciences. 2022; 23(23):14742. https://doi.org/10.3390/ijms232314742
Chicago/Turabian StyleRabelo-Fernández, Robert J., Ricardo A. Noriega Rivera, Yasmarie Santana Rivera, José Tous-Beveraggi, Fatima Valiyeva, and Pablo E. Vivas-Mejia. 2022. "Increased Expression of the RBPMS Splice Variants Inhibits Cell Proliferation in Ovarian Cancer Cells" International Journal of Molecular Sciences 23, no. 23: 14742. https://doi.org/10.3390/ijms232314742
APA StyleRabelo-Fernández, R. J., Noriega Rivera, R. A., Rivera, Y. S., Tous-Beveraggi, J., Valiyeva, F., & Vivas-Mejia, P. E. (2022). Increased Expression of the RBPMS Splice Variants Inhibits Cell Proliferation in Ovarian Cancer Cells. International Journal of Molecular Sciences, 23(23), 14742. https://doi.org/10.3390/ijms232314742








