Promoter Methylation of Cancer Stem Cell Surface Markers as an Epigenetic Biomarker for Prognosis of Oral Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Results
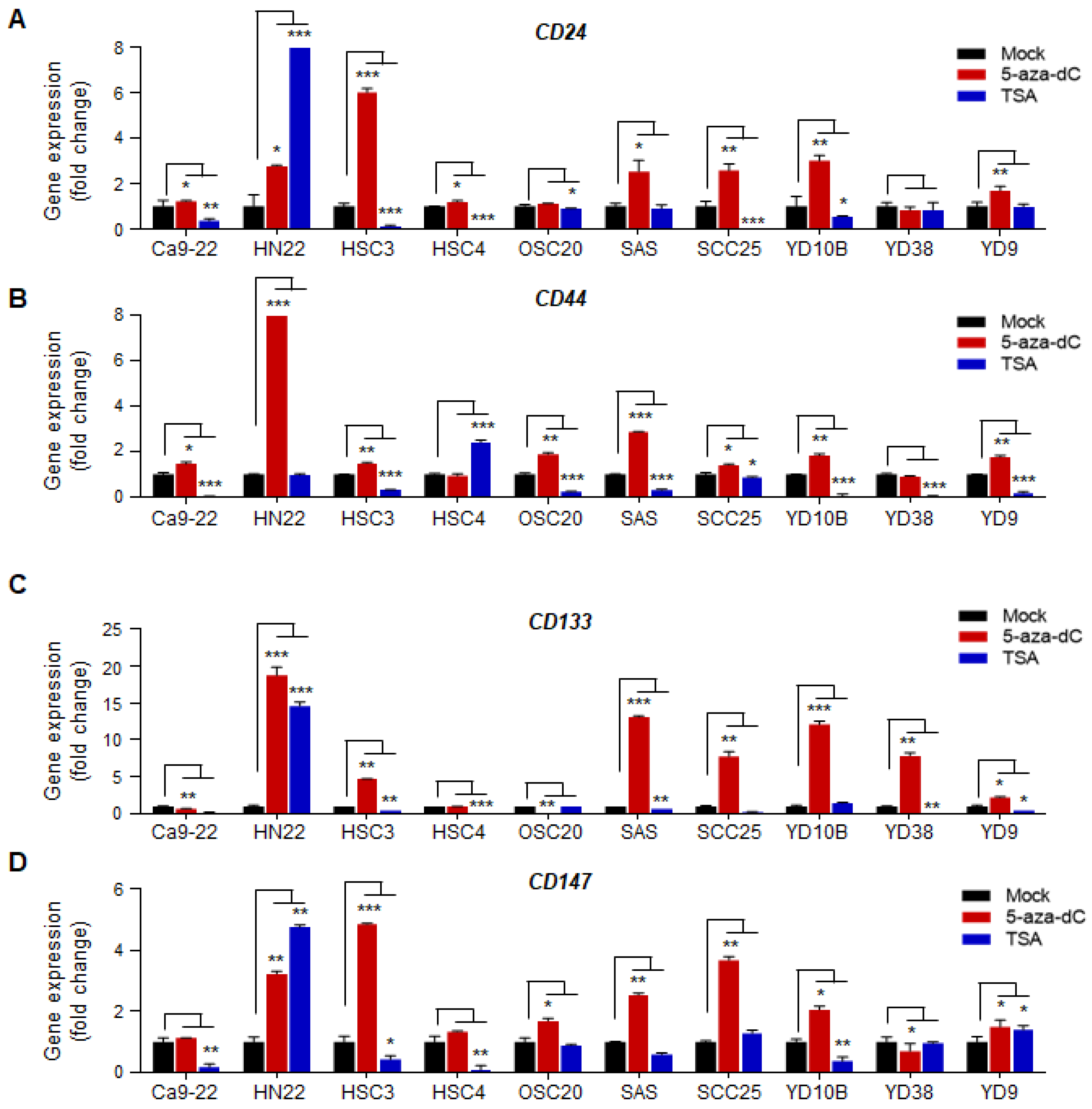
2.1. Transcriptional Expression of CSC Surface Markers Is Regulated by Promoter DNA Hypermethylation in OSCC Cell Lines
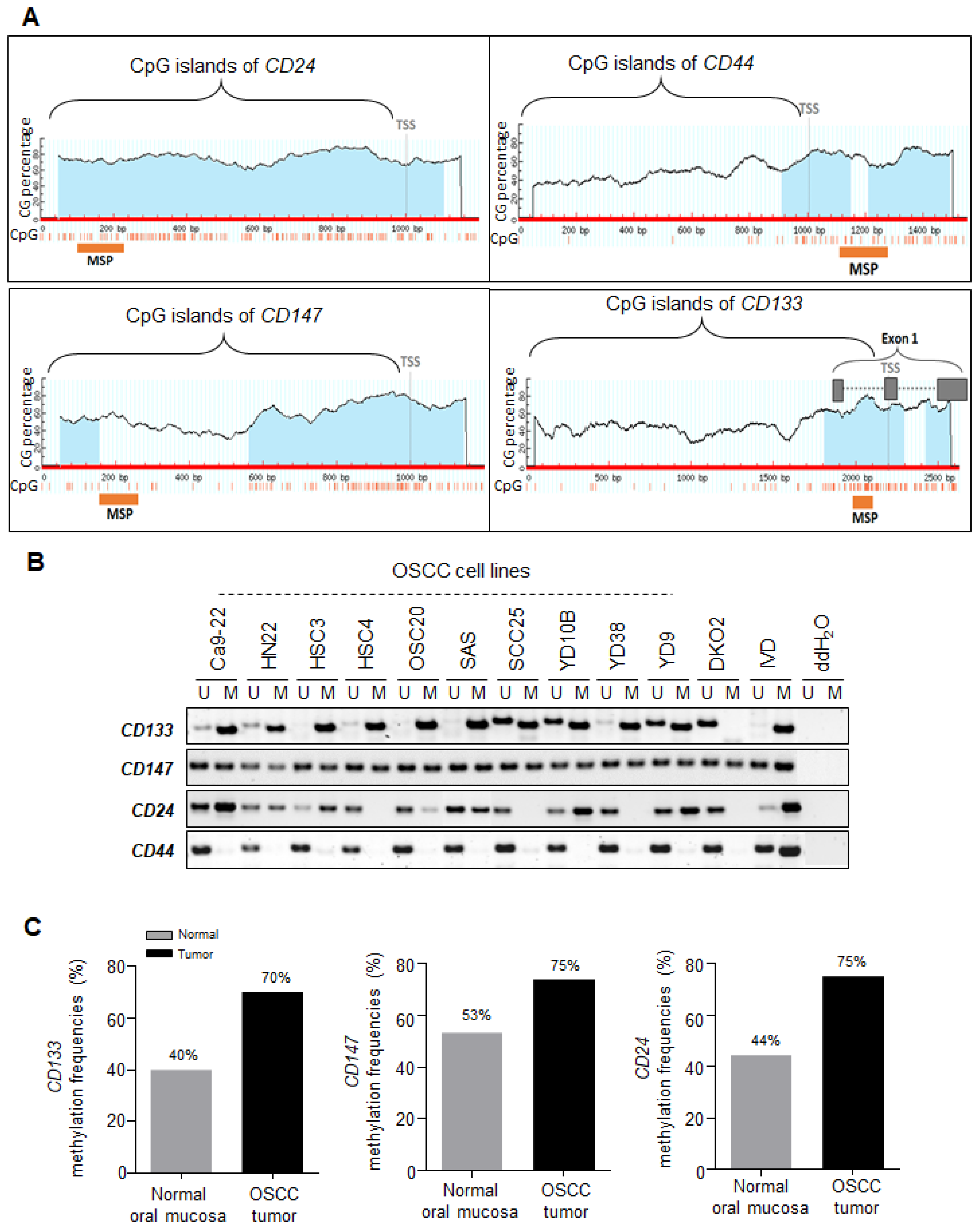
2.2. Correlation between the Transcriptional Expression of CD133 and CD147 and Promoter Hypermethylation in OSCC Cell Lines
2.3. Promoter Hypermethylation of CSC Surface Markers Frequently Occurred in Primary OSCC Tumor Tissues
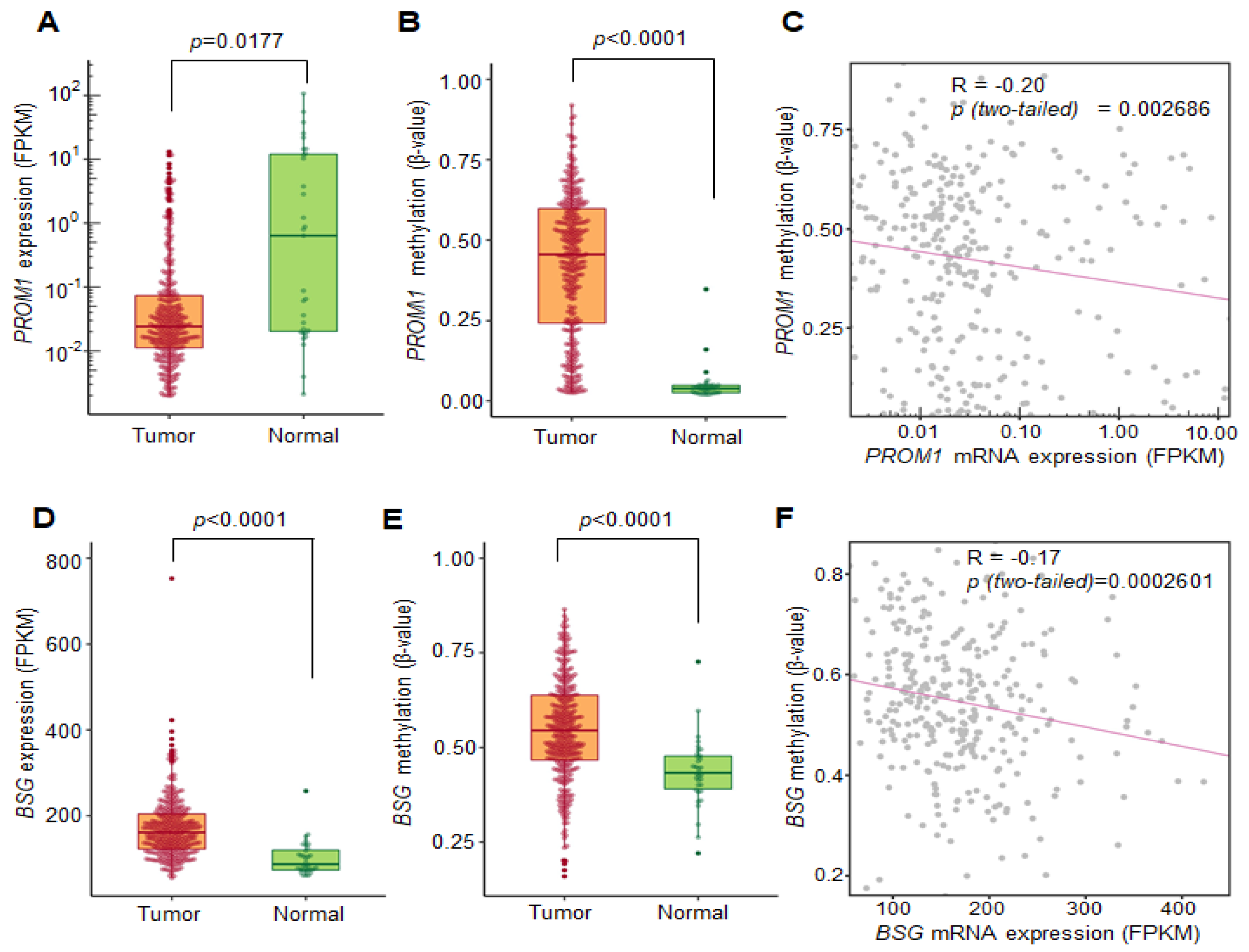
2.4. Correlation between DNA Methylation and the Transcriptional Expression of CD133 and CD147 Using the TCGA Database
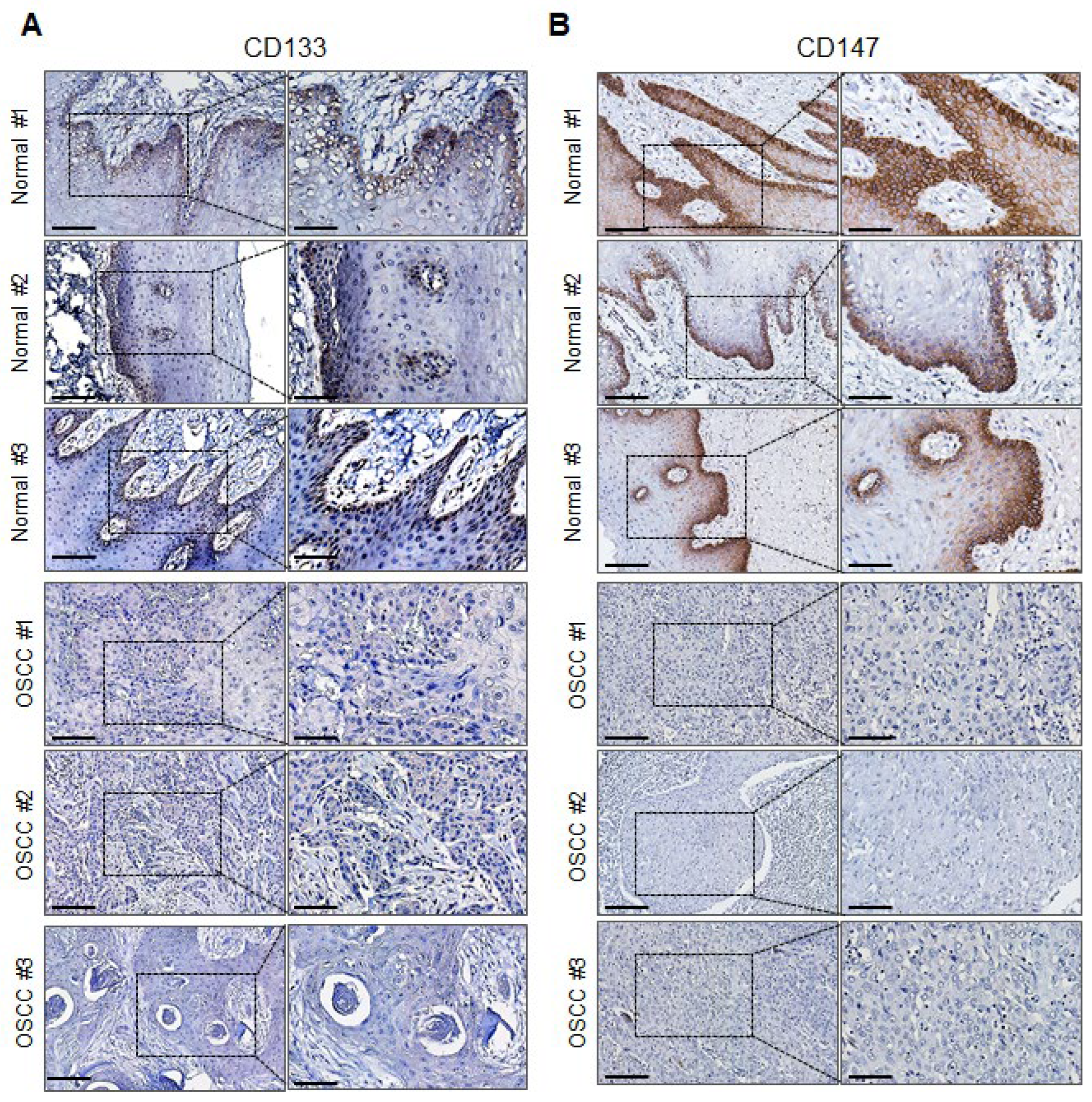
2.5. CD147 and CD133 Protein Expression Is Downregulated in Primary OSCC Tumor Tissues
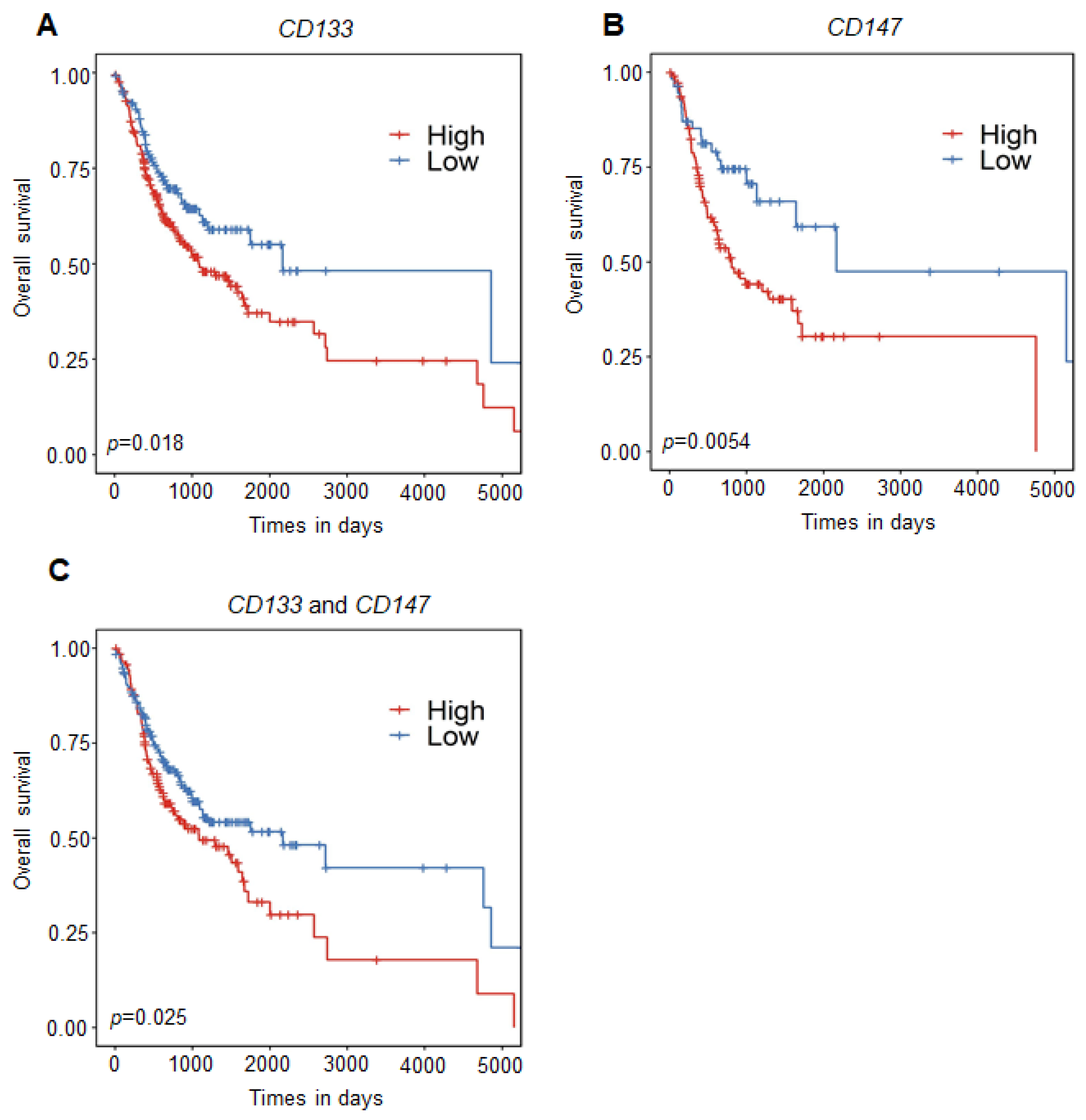
2.6. Promoter DNA Hypermethylation of CD133 and CD147 Is Associated with Poor Prognosis in OSCC Patients
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cell Culture and Treatment
5.2. Primer Design
5.3. Quantitative Real-Time RT-PCR
5.4. Methylation Analyses
5.5. Bisulfite Sequencing Analysis
5.6. Tissue Samples
5.7. Validation Analysis from the TCGA Database
5.8. Immunohistochemistry (IHC)
5.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tang, Q.; Xie, M.; Yu, S.; Zhou, X.; Xie, Y.; Chen, G.; Guo, F.; Chen, L. Periodic Oxaliplatin Administration in Synergy with PER2-Mediated PCNA Transcription Repression Promotes Chronochemotherapeutic Efficacy of OSCC. Adv. Sci. 2019, 6, 1900667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Ausoni, S.; Boscolo-Rizzo, P.; Singh, B.; Da Mosto, M.C.; Spinato, G.; Tirelli, G.; Spinato, R.; Azzarello, G. Targeting cellular and molecular drivers of head and neck squamous cell carcinoma: Current options and emerging perspectives. Cancer Metastasis Rev. 2016, 35, 413–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxi, S.; Fury, M.; Ganly, I.; Rao, S.; Pfister, D.G. Ten Years of Progress in Head and Neck Cancers. J. Natl. Compr. Cancer Netw. 2012, 10, 806–810. [Google Scholar] [CrossRef] [Green Version]
- Esteller, M. Epigenetic gene silencing in cancer: The DNA hypermethylome. Hum. Mol. Genet. 2007, 16, R50–R59. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef]
- Yi, J.M.; Tsai, H.-C.; Glöckner, S.C.; Lin, S.; Ohm, J.E.; Easwaran, H.; James, C.D.; Costello, J.F.; Riggins, G.; Eberhart, C.G.; et al. Abnormal DNA Methylation of CD133 in Colorectal and Glioblastoma Tumors. Cancer Res. 2008, 68, 8094–8103. [Google Scholar] [CrossRef] [Green Version]
- Kito, H.; Suzuki, H.; Ichikawa, T.; Sekita, N.; Kamiya, N.; Akakura, K.; Igarashi, T.; Nakayama, T.; Watanabe, M.; Harigaya, K.; et al. Hypermethylation of the CD44 gene is associated with progression and metastasis of human prostate cancer. Prostate 2001, 49, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Verkaik, N.S.; Trapman, J.; Romijn, J.C.; Van der Kwast, T.H.; Van Steenbrugge, G.J. Down-regulation of CD44 expression in human prostatic carcinoma cell lines is correlated with DNA hypermethylation. Int. J. Cancer 1999, 80, 439–443. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Hartman, Y.E.; Warram, J.M.; Knowles, J.A.; Sweeny, L.; Zhou, T.; Rosenthal, E.L. Fibroblast Growth Factor Receptor Mediates Fibroblast-Dependent Growth in EMMPRIN-Depleted Head and Neck Cancer Tumor Cells. Mol. Cancer Res. 2011, 9, 1008–1017. [Google Scholar] [CrossRef] [Green Version]
- Lescaille, G.; Menashi, S.; Cavelier-Balloy, B.; Khayati, F.; Quemener, C.; Podgorniak, M.P.; Naïmi, B.; Calvo, F.; Lebbe, C.; Mourah, S. EMMPRIN/CD147 up-regulates urokinase-type plasminogen activator: Implications in oral tumor progression. BMC Cancer 2012, 12, 115. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas, N.; Lawrence, M.S.; Sougnez, C.; Lichtenstein, L.; Cibulskis, K.; Lander, E.; Gabriel, S.B.; Getz, G.; Ally, A.; Balasundaram, M.; et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576. [Google Scholar] [CrossRef] [Green Version]
- Baylin, S.B.; Jones, P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019505. [Google Scholar] [CrossRef] [Green Version]
- Ehrlich, M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J. Nutr. 2002, 132, 2424S–2429S. [Google Scholar] [CrossRef] [Green Version]
- Schuebel, K.E.; Chen, W.; Cope, L.; Glöckner, S.C.; Suzuki, H.; Yi, J.-M.; Chan, T.A.; Neste, L.V.; Criekinge, W.V.; Bosch, S.v.d.; et al. Comparing the DNA Hypermethylome with Gene Mutations in Human Colorectal Cancer. PLoS Genet. 2007, 3, e157. [Google Scholar] [CrossRef]
- Yi, J.M. DNA methylation of stem cell surface markers in colon cancers. Genes Genom. 2012, 34, 299–304. [Google Scholar] [CrossRef]
- Saluja, T.S.; Ali, M.; Mishra, P.; Kumar, V.; Singh, S.K. Prognostic Value of Cancer Stem Cell Markers in Potentially Malignant Disorders of Oral Mucosa: A Meta-analysis. Cancer Epidemiol. Biomark. Prev. 2019, 28, 144–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohanta, S.; Siddappa, G.; Valiyaveedan, S.G.; Dodda Thimmasandra Ramanjanappa, R.; Das, D.; Pandian, R.; Khora, S.S.; Kuriakose, M.A.; Suresh, A. Cancer stem cell markers in patterning differentiation and in prognosis of oral squamous cell carcinoma. Tumor Biol. 2017, 39, 1010428317703656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.K.; Kim, S.H.; Choi, S.H.; Kim, K.H.; Yoo, B.C.; Ku, J.L.; Park, J.G. Promoter hypermethylation and loss of CD133 gene expression in colorectal cancers. World J. Gastroenterol. 2010, 16, 3153–3160. [Google Scholar] [CrossRef] [PubMed]
- Richard, V.; Pillai, M.R. The stem cell code in oral epithelial tumorigenesis: ‘The cancer stem cell shift hypothesis’. Biochim. Biophys. Acta Rev. Cancer 2010, 1806, 146–162. [Google Scholar] [CrossRef]
- Higashi, K.; Yagi, M.; Arakawa, T.; Asano, K.; Kobayashi, K.; Tachibana, T.; Saito, K. A Novel Marker for Undifferentiated Human Embryonic Stem Cells. Monoclon. Antibodies Immunodiagn. Immunother. 2015, 34, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cui, P.; Chen, J.; Li, W. Isolation and phenotypic characterization of side population cells in oral squamous cell carcinoma. Mol. Med. Rep. 2015, 11, 3642–3646. [Google Scholar] [CrossRef] [Green Version]
- Sidhu, S.S.; Nawroth, R.; Retz, M.; Lemjabbar-Alaoui, H.; Dasari, V.; Basbaum, C. EMMPRIN regulates the canonical Wnt/β-catenin signaling pathway, a potential role in accelerating lung tumorigenesis. Oncogene 2010, 29, 4145–4156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurt, E.M.; Kawasaki, B.T.; Klarmann, G.J.; Thomas, S.B.; Farrar, W.L. CD44+CD24− prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br. J. Cancer 2008, 98, 756–765. [Google Scholar] [CrossRef]
- Chiu, C.C.; Lee, L.Y.; Li, Y.C.; Chen, Y.J.; Lu, Y.C.; Li, Y.L.; Wang, H.M.; Chang, J.T.; Cheng, A.J. Grp78 as a therapeutic target for refractory head–neck cancer with CD24−CD44+ stemness phenotype. Cancer Gene Ther. 2013, 20, 606–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herman, J.G.; Graff, J.R.; Myohanen, S.; Nelkin, B.D.; Baylin, S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 1996, 93, 9821–9826. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | N | ||
|---|---|---|---|
| Total Number of Patients | 102 | ||
| Age (years) | |||
| Mean (St. dev) | 63.8 (13.7) | ||
| Median (range) | 63.5 (30-90) | ||
| Gender, n (%) | |||
| Female | 39 (38.2%) | ||
| Male | 63 (61.8%) | ||
| Tumor origin, n (%) | |||
| Primary | 95 (93.18%) | ||
| Recurrent | 7 (6.9%) | ||
| Differentiation, n (%) | |||
| Moderate | 38 (37.3%) | ||
| Poor | 4 (3.9%) | ||
| Well | 60 (58.8%) | ||
| Grade, n (%) | |||
| 1 | 33 (32.4%) | ||
| 2 | 41 (40.2%) | ||
| 3 | 24 (23.5%) | ||
| 4 | 3 (2.9%) | ||
| Unknown | 1 (1.0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.K.; Park, H.Y.; Park, S.-G.; Hwang, J.J.; Park, H.R.; Yi, J.M. Promoter Methylation of Cancer Stem Cell Surface Markers as an Epigenetic Biomarker for Prognosis of Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 14624. https://doi.org/10.3390/ijms232314624
Han YK, Park HY, Park S-G, Hwang JJ, Park HR, Yi JM. Promoter Methylation of Cancer Stem Cell Surface Markers as an Epigenetic Biomarker for Prognosis of Oral Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2022; 23(23):14624. https://doi.org/10.3390/ijms232314624
Chicago/Turabian StyleHan, Yu Kyeong, Ha Young Park, Sae-Gwang Park, Jae Joon Hwang, Hae Ryoun Park, and Joo Mi Yi. 2022. "Promoter Methylation of Cancer Stem Cell Surface Markers as an Epigenetic Biomarker for Prognosis of Oral Squamous Cell Carcinoma" International Journal of Molecular Sciences 23, no. 23: 14624. https://doi.org/10.3390/ijms232314624
APA StyleHan, Y. K., Park, H. Y., Park, S.-G., Hwang, J. J., Park, H. R., & Yi, J. M. (2022). Promoter Methylation of Cancer Stem Cell Surface Markers as an Epigenetic Biomarker for Prognosis of Oral Squamous Cell Carcinoma. International Journal of Molecular Sciences, 23(23), 14624. https://doi.org/10.3390/ijms232314624







