The L-Rhamnose Biosynthetic Pathway in Trichomonas vaginalis: Identification and Characterization of UDP-D-Glucose 4,6-dehydratase
Abstract
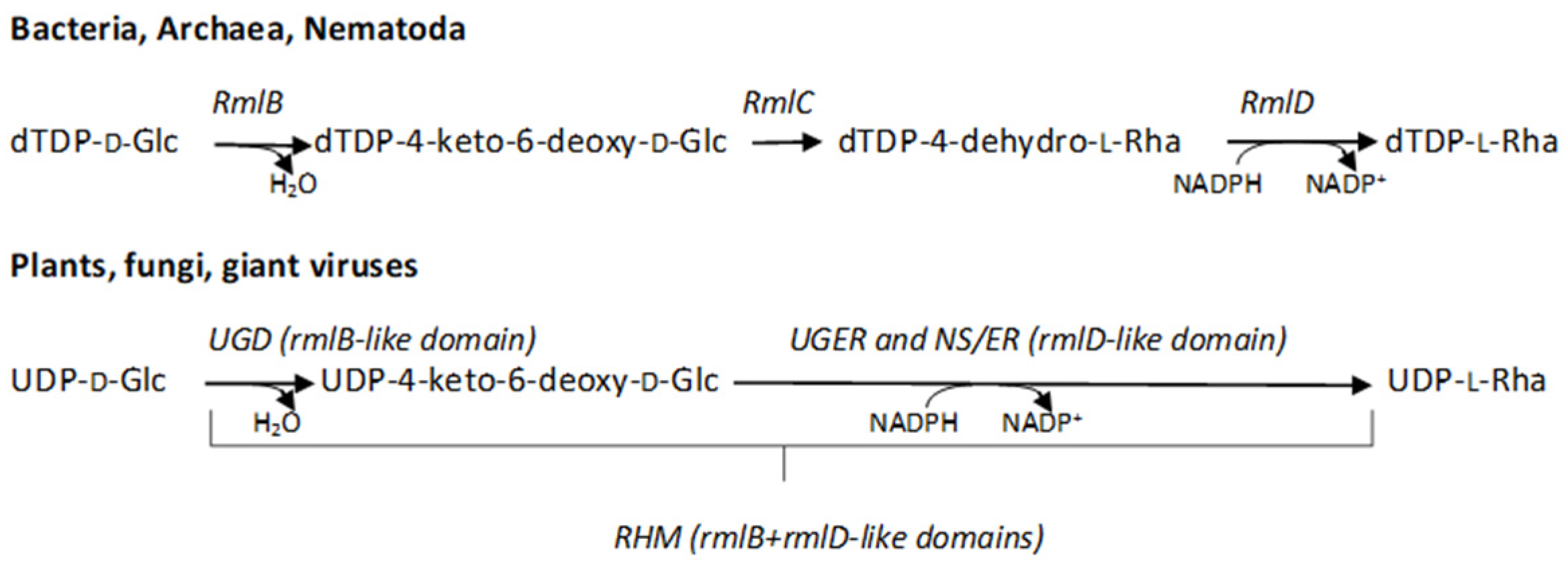
:1. Introduction
2. Results
2.1. Identification of the Genes Involved in the L-Rha Pathway in Tv
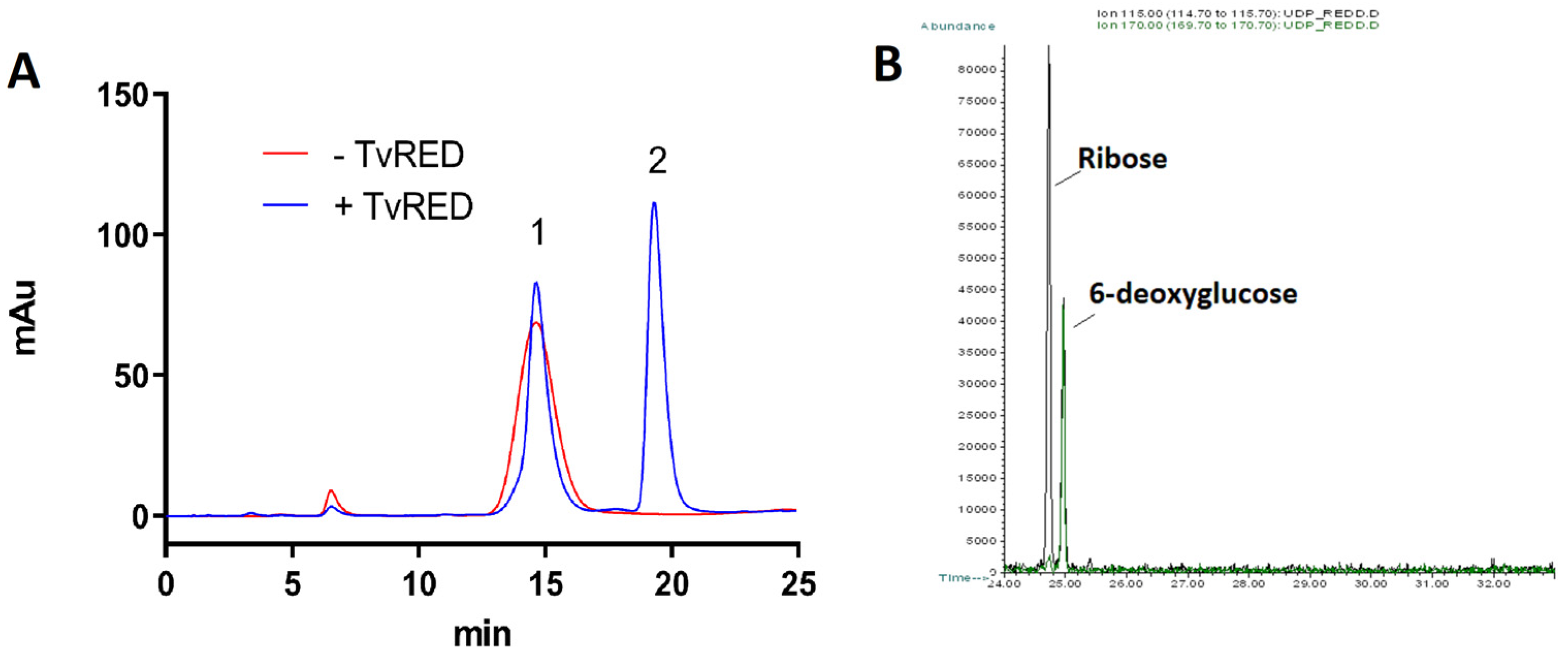
2.2. Production of the Recombinant Proteins and Identification of the Enzymatic Activities
2.3. TvUGD Kinetic Properties and Quaternary Structure Characterization
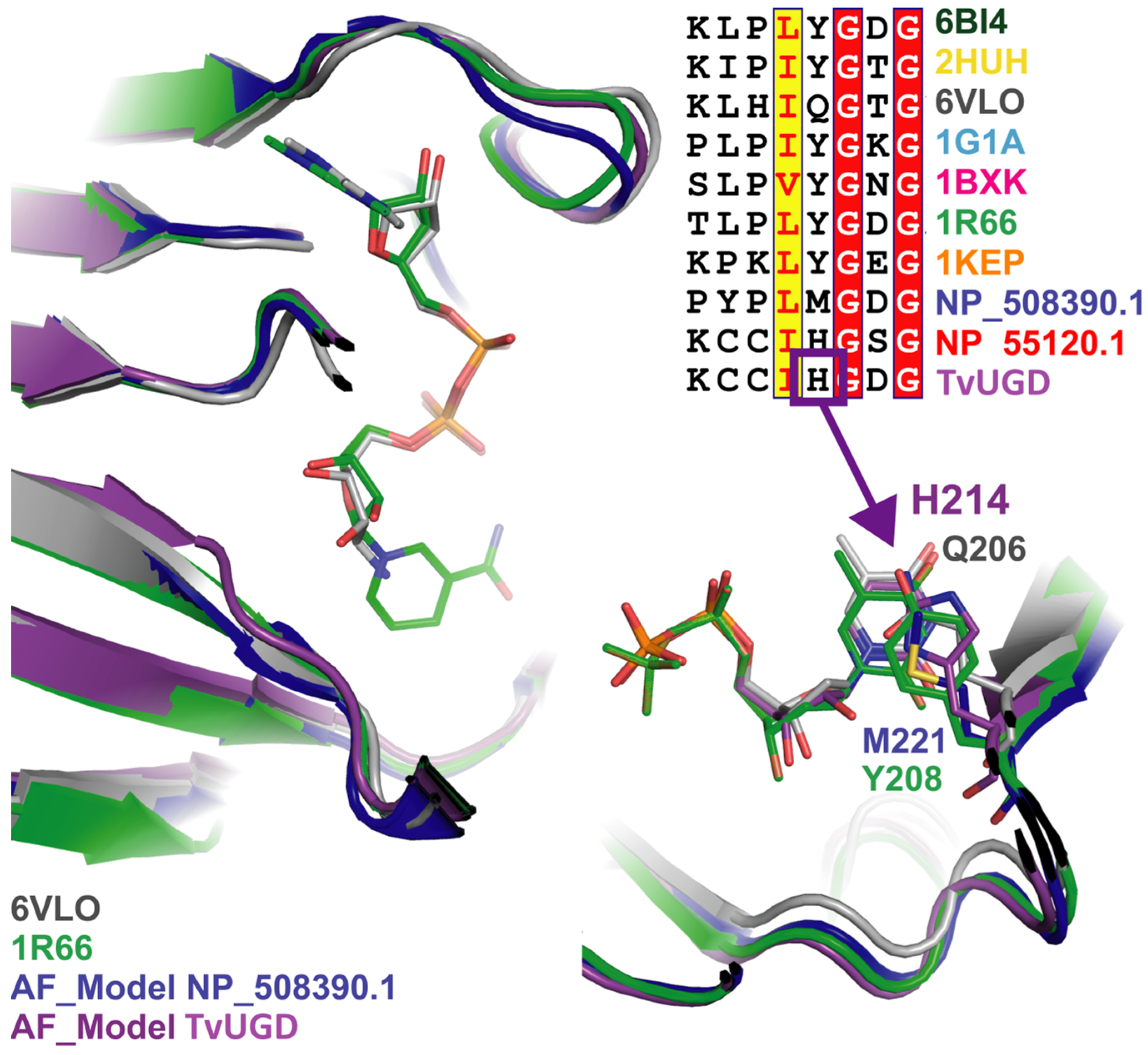
2.4. TvUGD Modeling
2.5. Phylogenetic Analysis of TvUGD
3. Discussion
4. Materials and Methods
4.1. Sequence Identification and Analysis
4.2. Cloning and Expression of the Recombinant Proteins in E. coli
4.3. Analysis of the Enzymatic Activities
4.4. Modeling of TvUGD in Complex with UDP
4.5. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Edwards, T.; Burke, P.; Smalley, H.; Hobbs, G. Trichomonas vaginalis: Clinical relevance, pathogenicity and diagnosis. Crit. Rev. Microbiol. 2016, 42, 406–417. [Google Scholar] [CrossRef]
- Kissinger, P. Trichomonas vaginalis: A review of epidemiologic, clinical and treatment issues. BMC Infect. Dis. 2015, 15, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Gerwen, O.T.; Craig-Kuhn, M.C.; Jones, A.T.; Schroeder, J.A.; Deaver, J.; Buekens, P.; Kissinger, P.J.; Muzny, C.A. Trichomoniasis and adverse birth outcomes: A systematic review and meta-analysis. BJOG 2021, 128, 1907–1915. [Google Scholar] [CrossRef] [PubMed]
- Rowley, J.; Vander Hoorn, S.; Korenromp, E.; Low, N.; Unemo, M.; Abu-Raddad, L.J.; Chico, R.M.; Smolak, A.; Newman, L.; Gottlieb, S.; et al. Chlamydia, gonorrhoea, trichomoniasis and syphilis: Global prevalence and incidence estimates, 2016. Bull. World Health Organ. 2019, 97, 548–562P. [Google Scholar] [CrossRef] [PubMed]
- Marques-Silva, M.; Lisboa, C.; Gomes, N.; Rodrigues, A.G. Trichomonas vaginalis and growing concern over drug resistance: A systematic review. J. Eur. Acad. Dermatol. Venereol. 2021, 35, 2007–2021. [Google Scholar] [CrossRef] [PubMed]
- Molgora, B.M.; Rai, A.K.; Sweredoski, M.J.; Moradian, A.; Hess, S.; Johnson, P.J. A Novel Trichomonas vaginalis Surface Protein Modulates Parasite Attachment via Protein: Host Cell Proteoglycan Interaction. mBio 2021, 12, e03374-20. [Google Scholar] [CrossRef]
- Bastida-Corcuera, F.D.; Okumura, C.Y.; Colocoussi, A.; Johnson, P.J. Trichomonas vaginalis lipophosphoglycan mutants have reduced adherence and cytotoxicity to human ectocervical cells. Eukaryot. Cell 2005, 4, 1951–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, B.N.; Hayes, G.R.; Lucas, J.J.; Sommer, U.; Viseux, N.; Mirgorodskaya, E.; Trifonova, R.T.; Sassi, R.R.; Costello, C.E.; Fichorova, R.N. Structural details and composition of Trichomonas vaginalis lipophosphoglycan in relevance to the epithelial immune function. Glycoconj. J. 2009, 26, 3–17. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, R.O.; Elia, G.; Beach, D.H.; Klaessig, S.; Singh, B.N. Cytopathogenic effect of Trichomonas vaginalis on human vaginal epithelial cells cultured in vitro. Infect. Immun. 2000, 68, 4200–4206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fichorova, R.N.; Trifonova, R.T.; Gilbert, R.O.; Costello, C.E.; Hayes, G.R.; Lucas, J.J.; Singh, B.N. Trichomonas vaginalis lipophosphoglycan triggers a selective upregulation of cytokines by human female reproductive tract epithelial cells. Infect. Immun. 2006, 74, 5773–5779. [Google Scholar] [CrossRef]
- Fichorova, R.N.; DeLong, A.K.; Cu-Uvin, S.; King, C.C.; Jamieson, D.J.; Klein, R.S.; Sobel, J.D.; Vlahov, D.; Yamamoto, H.S.; Mayer, K.H. Protozoan-Viral-Bacterial Co-Infections Alter Galectin Levels and Associated Immunity Mediators in the Female Genital Tract. Front. Cell Infect. Microbiol. 2021, 11, 649940. [Google Scholar] [CrossRef] [PubMed]
- Fichorova, R.N.; Yamamoto, H.S.; Fashemi, T.; Foley, E.; Ryan, S.; Beatty, N.; Dawood, H.; Hayes, G.R.; St-Pierre, G.; Sato, S.; et al. Trichomonas vaginalis Lipophosphoglycan Exploits Binding to Galectin-1 and -3 to Modulate Epithelial Immunity. J. Biol. Chem. 2016, 291, 998–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, C.M.; Mehlert, A.; Richardson, J.M.; Ferguson, M.A.; Johnson, P.J. Chemical structure of Trichomonas vaginalis surface lipoglycan: A role for short galactose (beta1-4/3) N-acetylglucosamine repeats in host cell interaction. J. Biol. Chem. 2011, 286, 40494–40508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allard, S.T.; Giraud, M.F.; Whitfield, C.; Graninger, M.; Messner, P.; Naismith, J.H. The crystal structure of dTDP-D-Glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium, the second enzyme in the dTDP-l-rhamnose pathway. J. Mol. Biol. 2001, 307, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Allard, S.T.; Cleland, W.W.; Holden, H.M. High resolution X-ray structure of dTDP-glucose 4,6-dehydratase from Streptomyces venezuelae. J. Biol. Chem. 2004, 279, 2211–2220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaminski, L.; Eichler, J. Haloferax volcanii N-glycosylation: Delineating the pathway of dTDP-rhamnose biosynthesis. PLoS ONE 2014, 9, e97441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, C.; Major, L.L.; Allen, A.; Blankenfeldt, W.; Maskell, D.; Naismith, J.H. High-resolution structures of RmlC from Streptococcus suis in complex with substrate analogs locate the active site of this class of enzyme. Structure 2003, 11, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Christendat, D.; Saridakis, V.; Dharamsi, A.; Bochkarev, A.; Pai, E.F.; Arrowsmith, C.H.; Edwards, A.M. Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP. J. Biol. Chem. 2000, 275, 24608–24612. [Google Scholar] [CrossRef] [Green Version]
- Giraud, M.F.; Leonard, G.A.; Field, R.A.; Berlind, C.; Naismith, J.H. RmlC, the third enzyme of dTDP-L-rhamnose pathway, is a new class of epimerase. Nature Struct. Biol. 2000, 7, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Blankenfeldt, W.; Kerr, I.D.; Giraud, M.F.; McMiken, H.J.; Leonard, G.; Whitfield, C.; Messner, P.; Graninger, M.; Naismith, J.H. Variation on a theme of SDR. dTDP-6-deoxy-L- lyxo-4-hexulose reductase (RmlD) shows a new Mg2+-dependent dimerization mode. Structure 2002, 10, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Shou, Q.; Butcher, R.A. Identification of a dTDP-rhamnose biosynthetic pathway that oscillates with the molting cycle in Caenorhabditis elegans. Biochem. J. 2016, 473, 1507–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, V.; Ingwers, M.; Smith, J.; Glushka, J.; Yang, T.; Bar-Peled, M. Biosynthesis of UDP-4-keto-6-deoxyglucose and UDP-rhamnose in pathogenic fungi Magnaporthe grisea and Botryotinia fuckeliana. J. Biol. Chem. 2012, 287, 879–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parakkottil Chothi, M.; Duncan, G.A.; Armirotti, A.; Abergel, C.; Gurnon, J.R.; Van Etten, J.L.; Bernardi, C.; Damonte, G.; Tonetti, M. Identification of an L-rhamnose synthetic pathway in two nucleocytoplasmic large DNA viruses. J. Virol. 2010, 84, 8829–8838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watt, G.; Leoff, C.; Harper, A.D.; Bar-Peled, M. A bifunctional 3,5-epimerase/4-keto reductase for nucleotide-rhamnose synthesis in Arabidopsis. Plant Physiol. 2004, 134, 1337–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oka, T.; Nemoto, T.; Jigami, Y. Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion. J. Biol. Chem. 2007, 282, 5389–5403. [Google Scholar] [CrossRef] [Green Version]
- Wagstaff, B.A.; Rejzek, M.; Kuhaudomlarp, S.; Hill, L.; Mascia, I.; Nepogodiev, S.A.; Dorfmueller, H.C.; Field, R.A. Discovery of an RmlC/D fusion protein in the microalga Prymnesium parvum and its implications for NDP-beta-l-rhamnose biosynthesis in microalgae. J. Biol. Chem. 2019, 294, 9172–9185. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.N.; BonDurant, R.H.; Campero, C.M.; Corbeil, L.B. Immunological and biochemical analysis of glycosylated surface antigens and lipophosphoglycan of Tritrichomonas foetus. J. Parasitol. 2001, 87, 770–777. [Google Scholar] [CrossRef]
- Singh, S.; Singh, G.; Singh, A.K.; Gautam, G.; Farmer, R.; Lodhi, S.S.; Wadhwa, G. Prediction and analysis of paralogous proteins in Trichomonas vaginalis genome. Bioinformation 2011, 6, 31–34. [Google Scholar] [CrossRef] [Green Version]
- Woehle, C.; Kusdian, G.; Radine, C.; Graur, D.; Landan, G.; Gould, S.B. The parasite Trichomonas vaginalis expresses thousands of pseudogenes and long non-coding RNAs independently from functional neighbouring genes. BMC Genom. 2014, 15, 906. [Google Scholar] [CrossRef] [Green Version]
- Carlton, J.M.; Hirt, R.P.; Silva, J.C.; Delcher, A.L.; Schatz, M.; Zhao, Q.; Wortman, J.R.; Bidwell, S.L.; Alsmark, U.C.; Besteiro, S.; et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 2007, 315, 207–212. [Google Scholar] [CrossRef]
- Huang, K.Y.; Chen, Y.Y.; Fang, Y.K.; Cheng, W.H.; Cheng, C.C.; Chen, Y.C.; Wu, T.E.; Ku, F.M.; Chen, S.C.; Lin, R.; et al. Adaptive responses to glucose restriction enhance cell survival, antioxidant capability, and autophagy of the protozoan parasite Trichomonas vaginalis. Biochim. Biophys. Acta 2014, 1840, 53–64. [Google Scholar] [CrossRef]
- Huang, K.Y.; Ku, F.M.; Cheng, W.H.; Lee, C.C.; Huang, P.J.; Chu, L.J.; Cheng, C.C.; Fang, Y.K.; Wu, H.H.; Tang, P. Novel insights into the molecular events linking to cell death induced by tetracycline in the amitochondriate protozoan Trichomonas vaginalis. Antimicrob. Agents Chemother. 2015, 59, 6891–6903. [Google Scholar] [CrossRef] [Green Version]
- Jornvall, H.; Hoog, J.O.; Persson, B. SDR and MDR: Completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett. 1999, 445, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Speciale, I.; Laugieri, M.E.; Noel, E.; Lin, S.; Lowary, T.L.; Molinaro, A.; Duncan, G.A.; Agarkova, I.V.; Garozzo, D.; Tonetti, M.G.; et al. Chlorovirus PBCV-1 protein A064R has three of the transferase activities necessary to synthesize its capsid protein N-linked glycans. Proc. Natl. Acad. Sci. USA 2020, 117, 28735–28742. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Beer, L.C.; Petrone-Garcia, V.M.; Graham, B.D.; Hargis, B.M.; Tellez-Isaias, G.; Vuong, C.N. Histomonosis in Poultry: A Comprehensive Review. Front. Vet. Sci. 2022, 9, 880738. [Google Scholar] [CrossRef]
- Falagas, M.E.; Siakavellas, E. Bacteroides, Prevotella, and Porphyromonas species: A review of antibiotic resistance and therapeutic options. Int. J. Antimicrob. Agents 2000, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Strese, A.; Backlund, A.; Alsmark, C. A recently transferred cluster of bacterial genes in Trichomonas vaginalis—Lateral gene transfer and the fate of acquired genes. BMC Evol. Biol. 2014, 14, 119. [Google Scholar] [CrossRef] [Green Version]
- Janssen, B.D.; Chen, Y.P.; Molgora, B.M.; Wang, S.E.; Simoes-Barbosa, A.; Johnson, P.J. CRISPR/Cas9-mediated gene modification and gene knock out in the human-infective parasite Trichomonas vaginalis. Sci. Rep. 2018, 8, 270. [Google Scholar] [CrossRef] [Green Version]
- Ferek, J.D.; Thoden, J.B.; Holden, H.M. Biochemical analysis of a sugar 4,6-dehydratase from Acanthamoeba polyphaga Mimivirus. Protein Sci. 2020, 29, 1148–1159. [Google Scholar] [CrossRef]
- Lal, P.; Sharma, D.; Pruthi, P.; Pruthi, V. Exopolysaccharide analysis of biofilm-forming Candida albicans. J. Appl. Microb. 2010, 109, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xin, Y.; McNeil, M.R.; Ma, Y. rmlB and rmlC genes are essential for growth of mycobacteria. Biochem. Biophys. Res. Commun. 2006, 342, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Sha, S.; Liu, L.; Li, X.; Ma, Y. A 96-well microtiter plate assay for high-throughput screening of Mycobacterium tuberculosis dTDP-d-glucose 4,6-dehydratase inhibitors. Anal. Biochem. 2016, 498, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Babaoglu, K.; Page, M.A.; Jones, V.C.; McNeil, M.R.; Dong, C.; Naismith, J.H.; Lee, R.E. Novel inhibitors of an emerging target in Mycobacterium tuberculosis; substituted thiazolidinones as inhibitors of dTDP-rhamnose synthesis. Bioorg. Med. Chem. Lett. 2003, 13, 3227–3230. [Google Scholar] [CrossRef] [PubMed]
- Van der Beek, S.L.; Zorzoli, A.; Canak, E.; Chapman, R.N.; Lucas, K.; Meyer, B.H.; Evangelopoulos, D.; de Carvalho, L.P.S.; Boons, G.J.; Dorfmueller, H.C.; et al. Streptococcal dTDP-L-rhamnose biosynthesis enzymes: Functional characterization and lead compound identification. Mol. Microbiol. 2019, 111, 951–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehmke, N.; Caliebe, A.; Koenig, R.; Kant, S.G.; Stark, Z.; Cormier-Daire, V.; Wieczorek, D.; Gillessen-Kaesbach, G.; Hoff, K.; Kawalia, A.; et al. Homozygous and compound-heterozygous mutations in TGDS cause Catel-Manzke syndrome. Am. J. Hum. Genet. 2014, 95, 763–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.H.C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomic Protocol Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Zuccotti, S.; Zanardi, D.; Rosano, C.; Sturla, L.; Tonetti, M.; Bolognesi, M. Kinetic and crystallographic analyses support a sequential-ordered bi bi catalytic mechanism for Escherichia coli glucose-1-phosphate thymidylyltransferase. J. Mol. Biol. 2001, 313, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Piacente, F.; Marin, M.; Molinaro, A.; De Castro, C.; Seltzer, V.; Salis, A.; Damonte, G.; Bernardi, C.; Claverie, J.M.; Abergel, C.; et al. Giant DNA virus mimivirus encodes pathway for biosynthesis of unusual sugar 4-amino-4,6-dideoxy-D-glucose (Viosamine). J. Biol. Chem. 2012, 287, 3009–3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosano, C.; Bisso, A.; Izzo, G.; Tonetti, M.; Sturla, L.; De Flora, A.; Bolognesi, M. Probing the catalytic mechanism of GDP-4-keto-6-deoxy-d-mannose Epimerase/Reductase by kinetic and crystallographic characterization of site-specific mutants. J. Mol. Biol. 2000, 303, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Zidek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Cryst. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997, 53, 240–255. [Google Scholar] [CrossRef]
- Holm, L. Using Dali for Protein Structure Comparison. Meth. Mol. Biol. 2020, 2112, 29–42. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. CABIOS 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]



| Gene Name | Length | Domain(s) | Pfam acc. n. | Predicted Activity | Ortholog in T. Foetus (% id.) |
|---|---|---|---|---|---|
| TVAG_414560 | 348 aa |  | PF16363 | 4,6-dehydratase | TRFO_03667 (77%) |
| TVAG_357160 | 285 aa |  | PF04321 | 4-keto-reductase | ND |
| TVAG_463120 | 450 aa |  | PF00908/PF04321 | 3,5-epi,4-keto-red | TRFO_30760 (59%) |
| TVAG_016600 | 449 aa |  | PF00908/PF04321 | 3,5-epi,4-keto-red | TRFO_30760 (59%) |
| TVAG_401850 | 448 aa |  | PF00908/PF04321 | 3,5-epi,4-keto-red | TRFO_30760 (51%) |
| TVAG_340730 | 347 aa |  | PF00908 | 3,5-epimerase | TRFO_42104 (37%) |
| TVAG_313960 | 420 aa |  | PF00908 | 3,5-epimerase | TRFO_42104 (42%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaglianone, M.; Laugieri, M.E.; Rojas, A.L.; Coppola, M.R.; Piacente, F.; Fiori, P.L.; Tonetti, M.G. The L-Rhamnose Biosynthetic Pathway in Trichomonas vaginalis: Identification and Characterization of UDP-D-Glucose 4,6-dehydratase. Int. J. Mol. Sci. 2022, 23, 14587. https://doi.org/10.3390/ijms232314587
Gaglianone M, Laugieri ME, Rojas AL, Coppola MR, Piacente F, Fiori PL, Tonetti MG. The L-Rhamnose Biosynthetic Pathway in Trichomonas vaginalis: Identification and Characterization of UDP-D-Glucose 4,6-dehydratase. International Journal of Molecular Sciences. 2022; 23(23):14587. https://doi.org/10.3390/ijms232314587
Chicago/Turabian StyleGaglianone, Matteo, Maria Elena Laugieri, Adriana Lucely Rojas, Maria Rosaria Coppola, Francesco Piacente, Pier Luigi Fiori, and Michela Giulia Tonetti. 2022. "The L-Rhamnose Biosynthetic Pathway in Trichomonas vaginalis: Identification and Characterization of UDP-D-Glucose 4,6-dehydratase" International Journal of Molecular Sciences 23, no. 23: 14587. https://doi.org/10.3390/ijms232314587
APA StyleGaglianone, M., Laugieri, M. E., Rojas, A. L., Coppola, M. R., Piacente, F., Fiori, P. L., & Tonetti, M. G. (2022). The L-Rhamnose Biosynthetic Pathway in Trichomonas vaginalis: Identification and Characterization of UDP-D-Glucose 4,6-dehydratase. International Journal of Molecular Sciences, 23(23), 14587. https://doi.org/10.3390/ijms232314587






