DNA Sequence-Dependent Properties of Nucleosome Positioning in Regions of Distinct Chromatin States in Mouse Embryonic Stem Cells
Abstract
1. Introduction
2. Results and Discussion
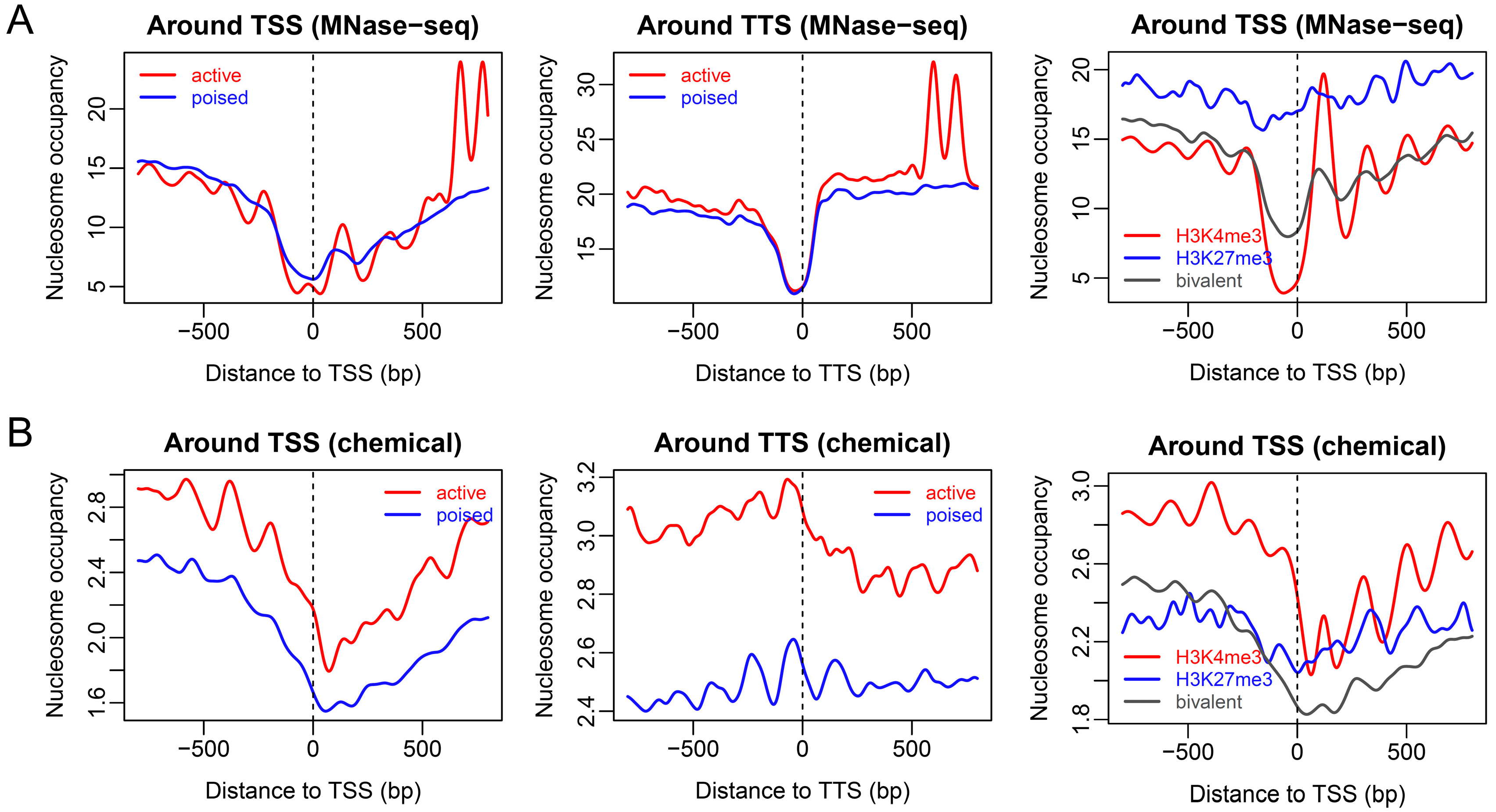
2.1. Nucleosome Organization around Promoters with Distinct Chromatin States in ESCs
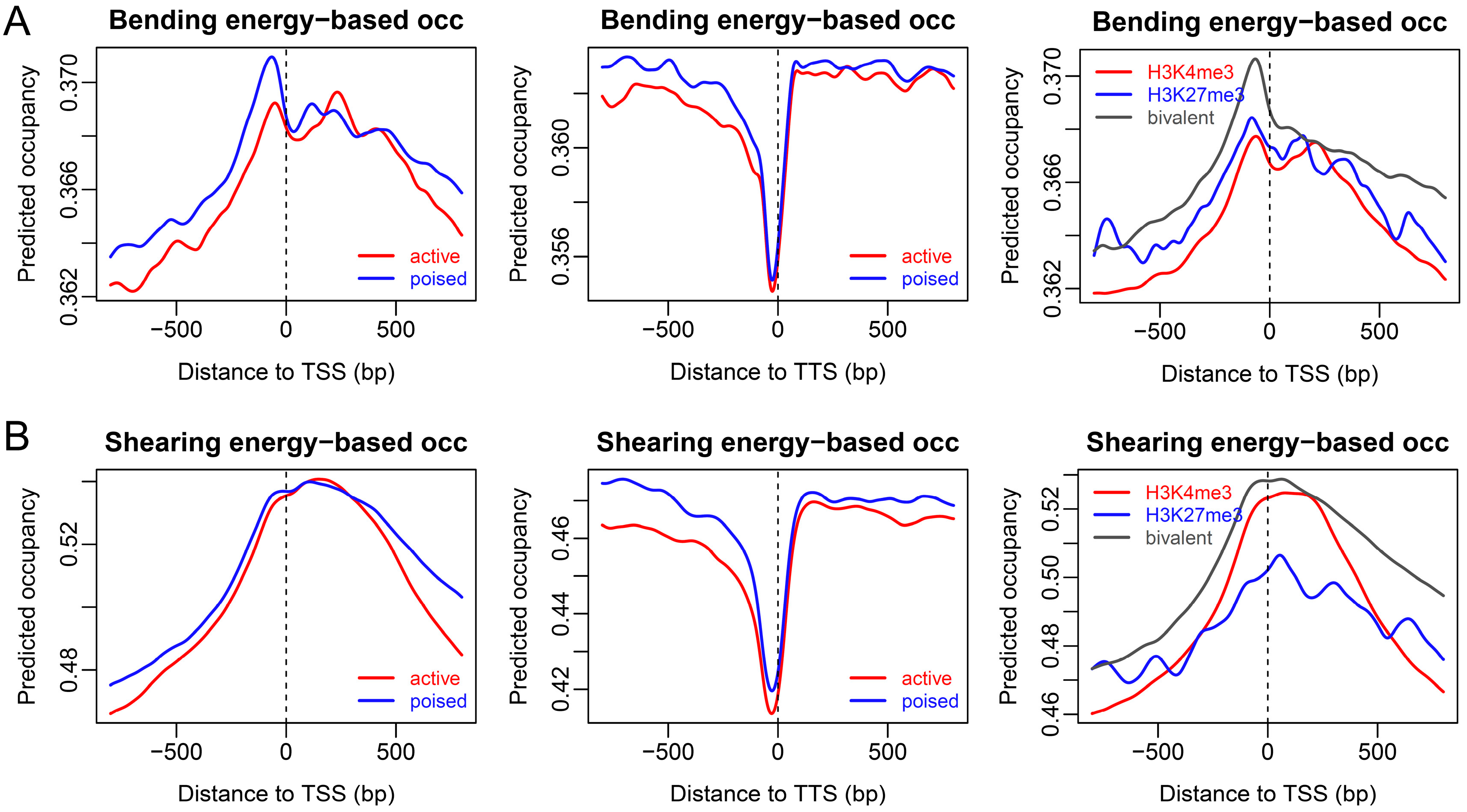
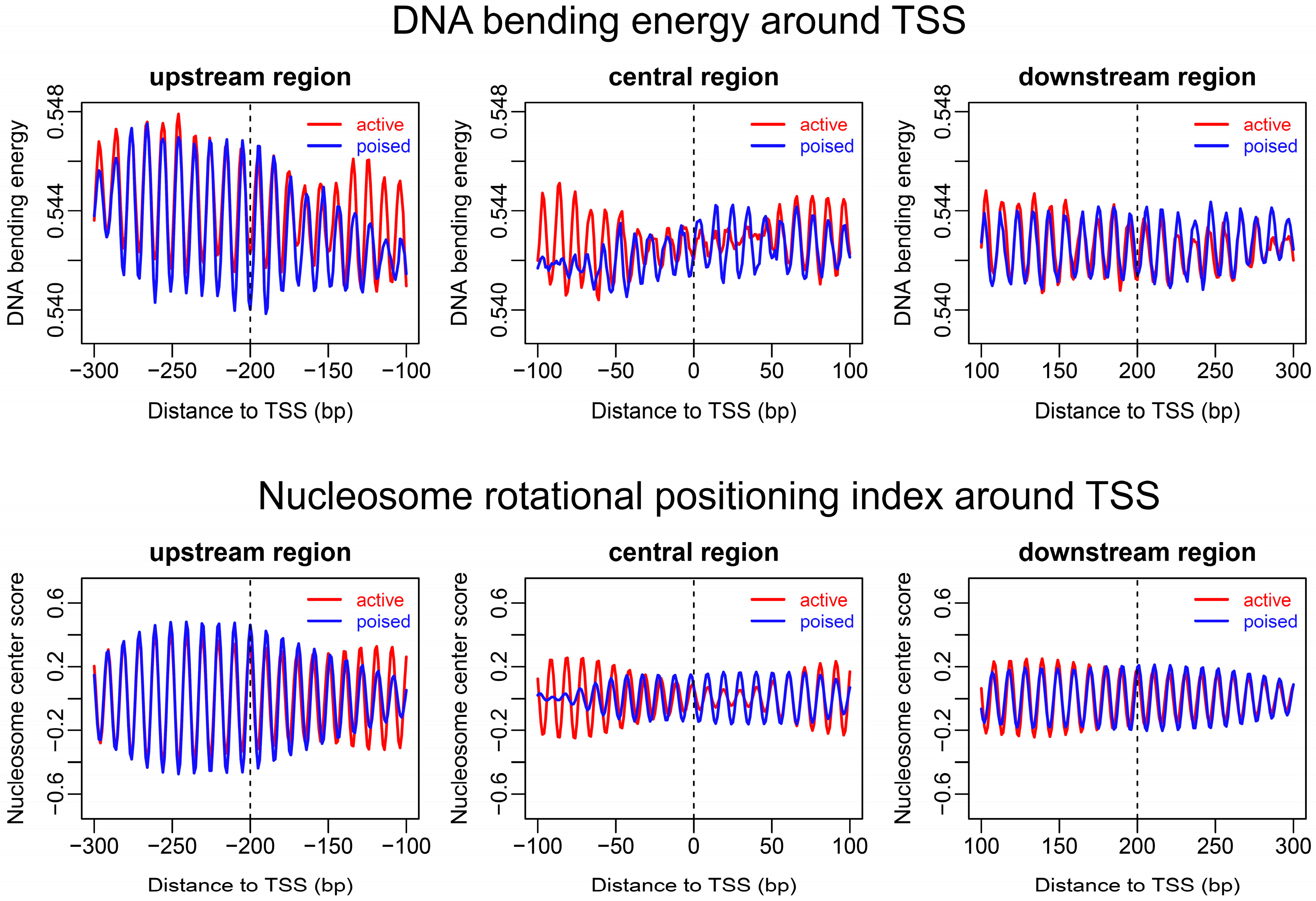
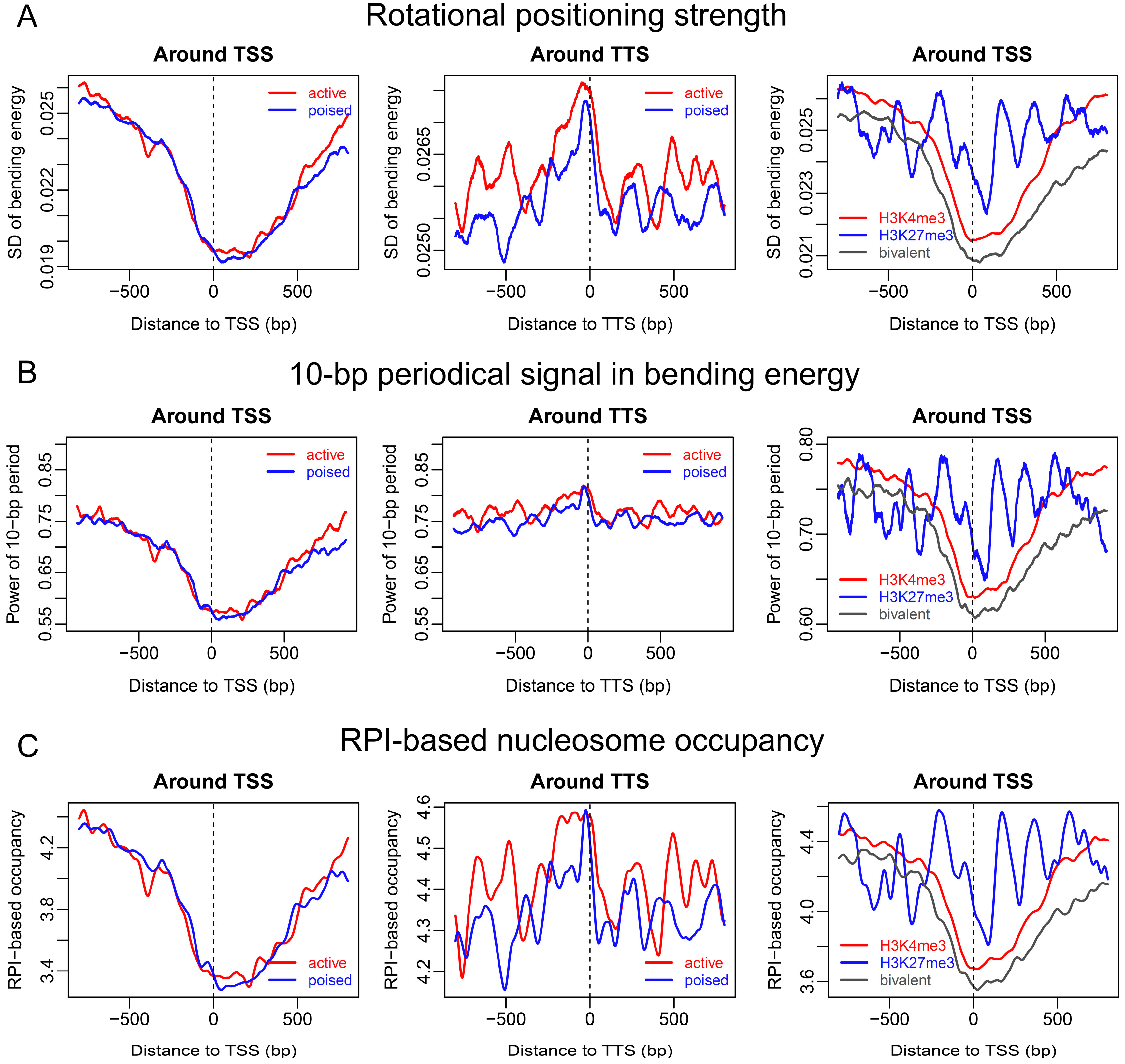
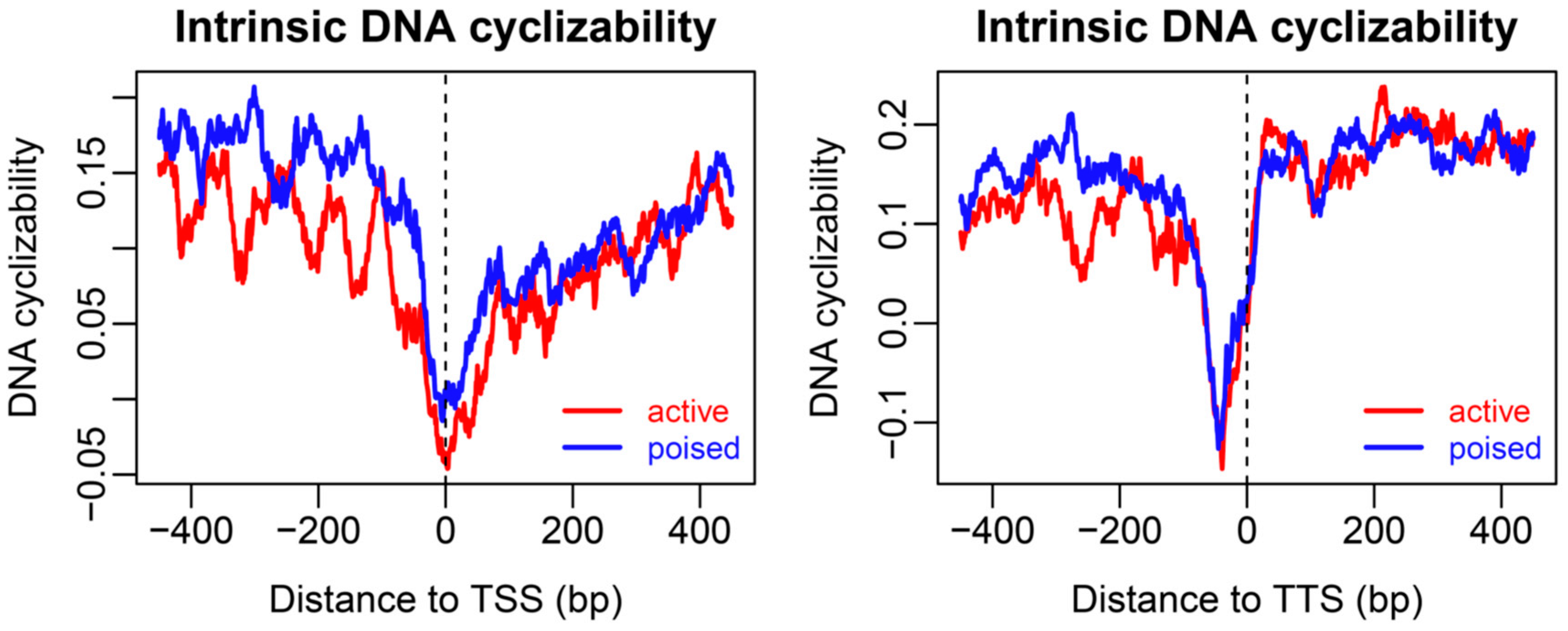
2.2. Sequence-Dependent Nucleosome Positioning around Promoters with Distinct Chromatin States in ESCs
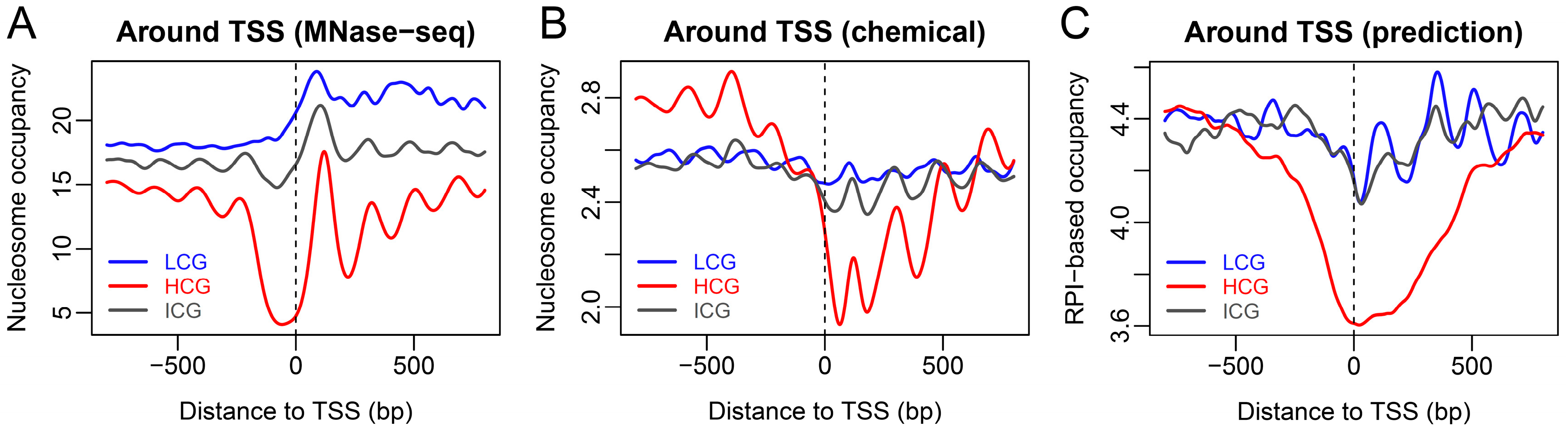
2.3. Nucleosome Organization around Promoters with Different CpG Composition in ESCs
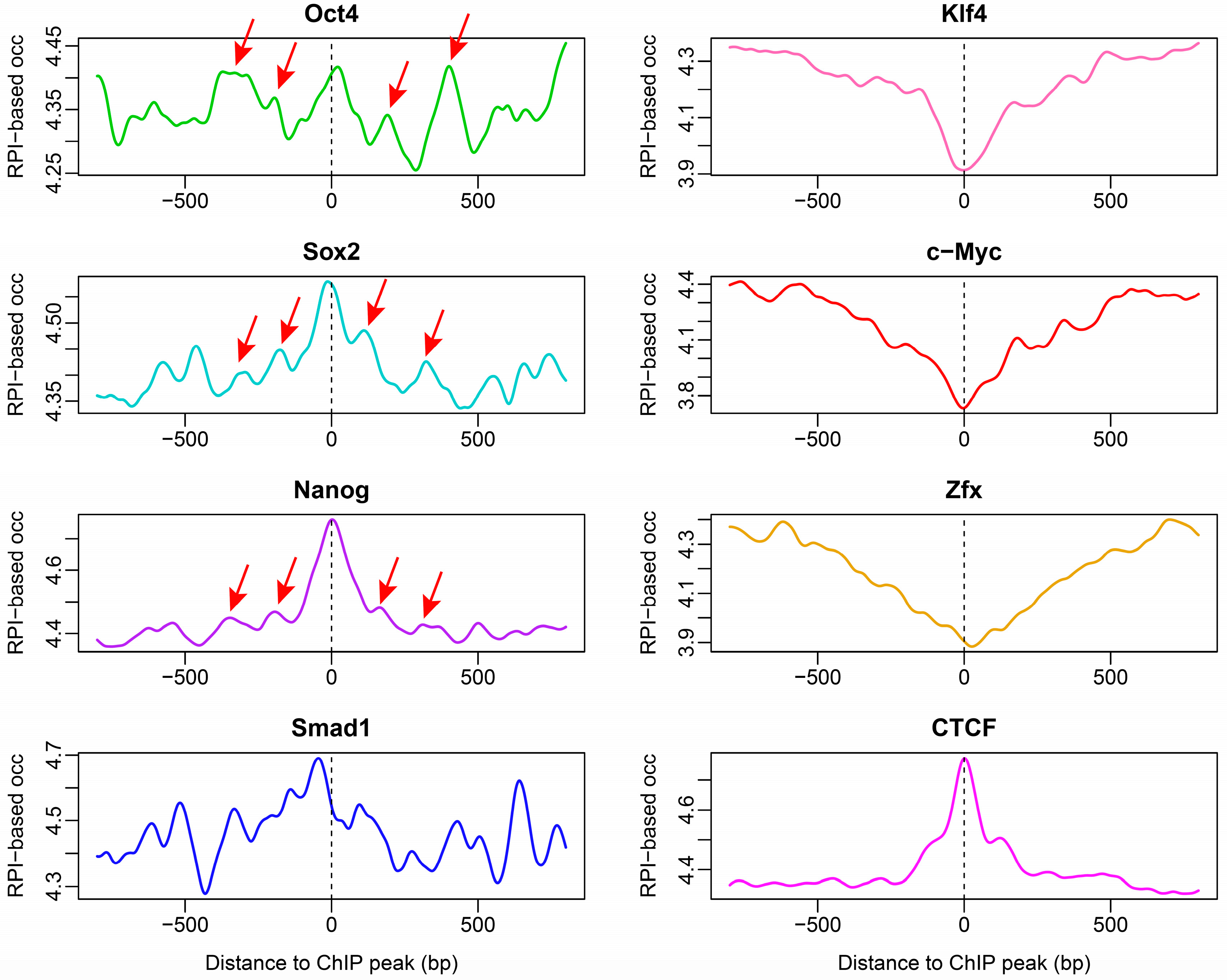
2.4. Nucleosome Organization around Transcription Factor-Binding Sites in ESCs
2.5. Conclusions
3. Materials and Methods
3.1. Genes with Distinct Chromatin States
3.2. Experimental Nucleosome Maps of Mouse ESCs
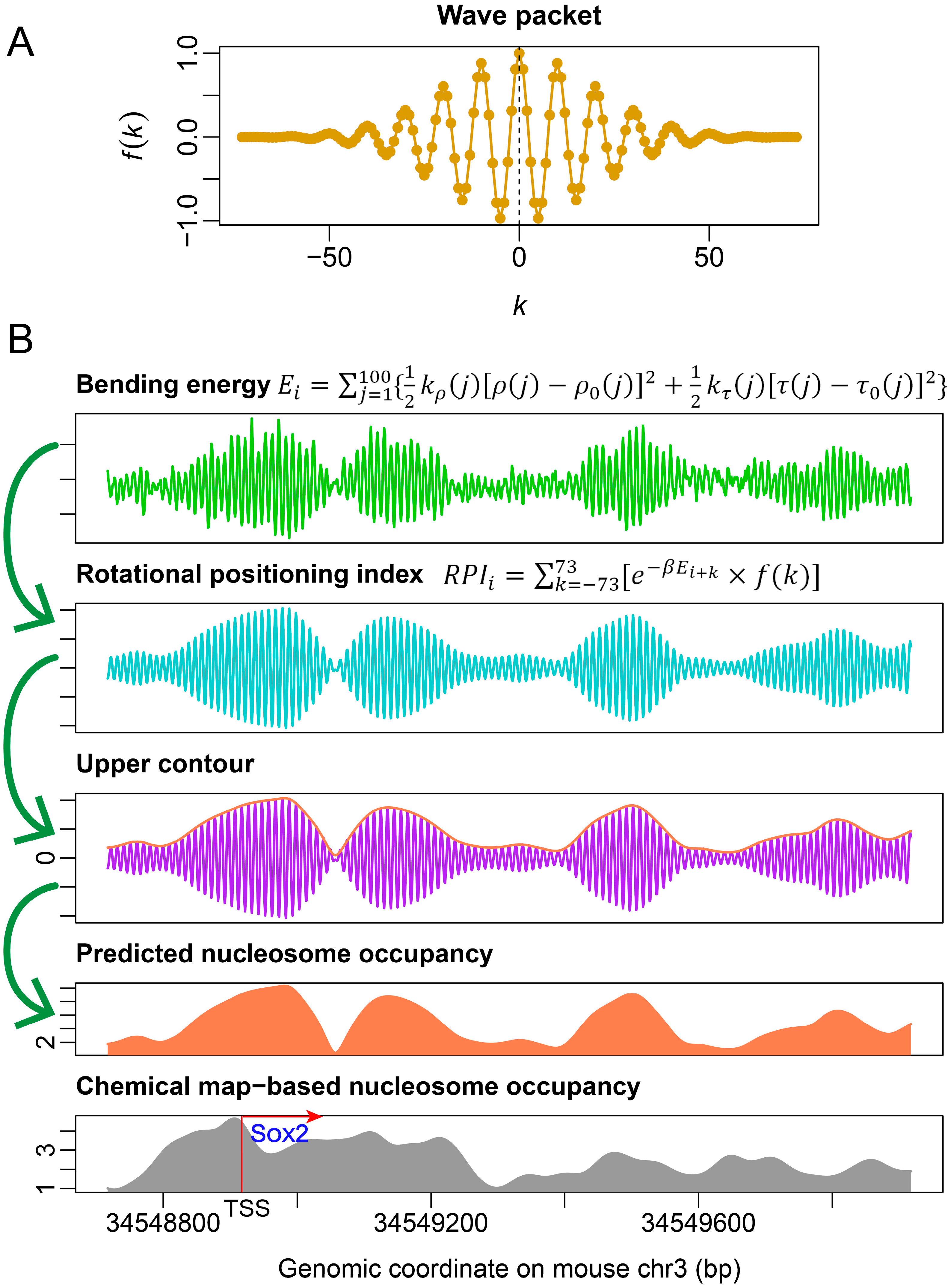
3.3. DNA Deformation-Energy-Based Descriptors
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ekundayo, B.; Richmond, T.J.; Schalch, T. Capturing structural heterogeneity in chromatin fibers. J. Mol. Biol. 2017, 429, 3031–3042. [Google Scholar] [CrossRef] [PubMed]
- Grigoryev, S.A. Chromatin Higher-Order Folding: A Perspective with Linker DNA Angles. Biophys. J. 2018, 114, 2290–2297. [Google Scholar] [CrossRef] [PubMed]
- Collepardo-Guevara, R.; Schlick, T. Chromatin fiber polymorphism triggered by variations of DNA linker lengths. Proc. Natl. Acad. Sci. USA 2014, 111, 8061–8066. [Google Scholar] [CrossRef] [PubMed]
- Mcginty, R.K.; Tan, S. Nucleosome structure and function. Chem. Rev. 2015, 115, 2255–2273. [Google Scholar] [CrossRef]
- Willcockson, M.A.; Healton, S.E.; Weiss, C.N.; Bartholdy, B.A.; Botbol, Y.; Mishra, L.N.; Sidhwani, D.S.; Wilson, T.J.; Pinto, H.B.; Maron, M.I.; et al. H1 histones control the epigenetic landscape by local chromatin compaction. Nature 2021, 589, 293–298. [Google Scholar] [CrossRef]
- Ohno, M.; Ando, T.; Priest, D.G.; Taniguchi, Y. Hi-CO: 3D genome structure analysis with nucleosome resolution. Nat. Protoc. 2021, 16, 3439–3469. [Google Scholar] [CrossRef]
- Ohno, M.; Priest, D.G.; Taniguchi, Y. Nucleosome-level 3D organization of the genome. Biochem. Soc. Trans. 2018, 46, 491–501. [Google Scholar] [CrossRef]
- Hsieh, T.H.; Weiner, A.; Lajoie, B.; Dekker, J.; Friedman, N.; Rando, O.J. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell 2015, 162, 108–119. [Google Scholar] [CrossRef]
- Core, L.; Adelman, K. Promoter-proximal pausing of RNA polymerase II: A nexus of gene regulation. Genes Dev. 2019, 33, 960–982. [Google Scholar] [CrossRef] [PubMed]
- Voong, L.N.; Xi, L.; Sebeson, A.C.; Xiong, B.; Wang, J.P.; Wang, X. Insights into nucleosome organization in mouse embryonic stem cells through chemical mapping. Cell 2016, 167, 1555–1570. [Google Scholar] [CrossRef]
- Dingwall, C.; Lomonossoff, G.P.; Laskey, R.A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981, 9, 2659–2673. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.R.; Dunkel, I.; Heise, F.; Linke, C.; Krobitsch, S.; Ehrenhofer-Murray, A.E.; Sperling, S.R.; Vingron, M. The effect of micrococcal nuclease digestion on nucleosome positioning data. PLoS ONE 2010, 5, e15754. [Google Scholar] [CrossRef] [PubMed]
- Mieczkowski, J.; Cook, A.; Bowman, S.K.; Mueller, B.; Alver, B.H.; Kundu, S.; Deaton, A.M.; Urban, J.A.; Larschan, E.; Park, P.J.; et al. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 2016, 7, 11485. [Google Scholar] [CrossRef] [PubMed]
- Chereji, R.V.; Ramachandran, S.; Bryson, T.D.; Henikoff, S. Precise genome-wide mapping of single nucleosomes and linkers in vivo. Genome Biol. 2018, 19, 19. [Google Scholar] [CrossRef] [PubMed]
- Brogaard, K.; Xi, L.; Wang, J.P.; Widom, J. A map of nucleosome positions in yeast at base-pair resolution. Nature 2012, 486, 496–501. [Google Scholar] [CrossRef]
- Segal, E.; Fondufe-Mittendorf, Y.; Chen, L.; Thåström, A.; Field, Y.; Moore, I.K.; Wang, J.P.; Widom, J. A genomic code for nucleosome positioning. Nature 2006, 442, 772–778. [Google Scholar] [CrossRef]
- Cui, F.; Chen, L.; LoVerso, P.R.; Zhurkin, V.B. Prediction of nucleosome rotational positioning in yeast and human genomes based on sequence-dependent DNA anisotropy. BMC Bioinform. 2014, 15, 313. [Google Scholar] [CrossRef]
- Tolstorukov, M.Y.; Colasanti, A.V.; McCandlish, D.M.; Olson, W.K.; Zhurkin, V.B. A novel roll-and-slide mechanism of DNA folding in chromatin: Implications for nucleosome positioning. J. Mol. Biol. 2007, 371, 725–738. [Google Scholar] [CrossRef]
- Bishop, T.C. Geometry of the nucleosomal DNA superhelix. Biophys. J. 2008, 95, 1007–1017. [Google Scholar] [CrossRef][Green Version]
- Morozov, A.V.; Fortney, K.; Gaykalova, D.A.; Studitsky, V.M.; Widom, J.; Siggia, E.D. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 2009, 37, 4707–4722. [Google Scholar] [CrossRef]
- Liu, G.; Xing, Y.; Zhao, H.; Wang, J.; Shang, Y.; Cai, L. A deformation energy-based model for predicting nucleosome dyads and occupancy. Sci. Rep. 2016, 6, 24133. [Google Scholar] [CrossRef]
- Liu, G.; Zhao, H.; Meng, H.; Xing, Y.; Cai, L. A deformation energy model reveals sequence-dependent property of nucleosome positioning. Chromosoma 2021, 130, 27–40. [Google Scholar] [CrossRef]
- Wang, J.Y.; Wang, J.; Liu, G. Calculation of nucleosomal DNA deformation energy: Its implication for nucleosome positioning. Chromosome Res. 2012, 20, 889–902. [Google Scholar] [CrossRef]
- Liu, G.; Zhao, H.; Meng, H.; Xing, Y.; Yang, H.; Lin, H. Deform-nu: A DNA Deformation Energy-Based Predictor for Nucleosome Positioning. Front. Cell Dev. Biol. 2020, 8, 596341. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef]
- Mikkelsen, T.S.; Ku, M.; Jaffe, D.B.; Issac, B.; Lieberman, E.; Giannoukos, G.; Alvarez, P.; Brockman, W.; Kim, T.K.; Koche, R.P.; et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007, 448, 553–560. [Google Scholar] [CrossRef]
- Teif, V.B.; Vainshtein, Y.; Caudron-Herger, M.; Mallm, J.P.; Marth, C.; Höfer, T.; Rippe, K. Genome-wide nucleosome positioning during embryonic stem cell development. Nat. Struct. Mol. Biol. 2012, 19, 1185–1192. [Google Scholar] [CrossRef]
- Yu, J.; Xiong, C.; Zhuo, B.; Wen, Z.; Shen, J.; Liu, C.; Chang, L.; Wang, K.; Wang, M.; Wu, C.; et al. Analysis of local chromatin states reveals gene transcription potential during mouse neural progenitor cell differentiation. Cell Rep. 2020, 32, 107953. [Google Scholar] [CrossRef]
- Soufi, A.; Garcia, M.F.; Jaroszewicz, A.; Osman, N.; Pellegrini, M.; Zaret, K.S. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 2015, 161, 555–568. [Google Scholar] [CrossRef]
- Tang, Z.; Luo, O.J.; Li, X.; Zheng, M.; Zhu, J.J.; Szalaj, P.; Trzaskoma, P.; Magalska, A.; Wlodarczyk, J.; Ruszczycki, B.; et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell 2015, 163, 1611–1627. [Google Scholar] [CrossRef]
- Li, K.; Carroll, M.; Vafabakhsh, R.; Wang, X.A.; Wang, J.P. DNAcycP: A deep learning tool for DNA cyclizability prediction. Nucleic Acids Res. 2022, 50, 3142–3154. [Google Scholar] [CrossRef] [PubMed]
- Dreos, R.; Ambrosini, G.; Bucher, P. Influence of Rotational Nucleosome Positioning on Transcription Start Site Selection in Animal Promoters. PLoS Comput. Biol. 2016, 12, e1005144. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Farnung, L.; Kaasinen, E.; Sahu, B.; Yin, Y.; Wei, B.; Dodonova, S.O.; Nitta, K.R.; Morgunova, E.; Taipale, M.; et al. The interaction landscape between transcription factors and the nucleosome. Nature 2018, 562, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, H.; Yuan, P.; Fang, F.; Huss, M.; Vega, V.B.; Wong, E.; Orlov, Y.L.; Zhang, W.; Jiang, J.; et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008, 133, 1106–1117. [Google Scholar] [CrossRef]
- Xi, L.; Brogaard, K.; Zhang, Q.; Lindsay, B.; Widom, J.; Wang, J.P. A locally convoluted cluster model for nucleosome positioning signals in chemical map. J. Am. Stat. Assoc. 2014, 109, 48–62. [Google Scholar] [CrossRef]
- Olson, W.K.; Bansal, M.; Burley, S.K.; Dickerson, R.E.; Gerstein, M.; Harvey, S.C.; Heinemann, U.; Lu, X.J.; Neidle, S.; Shakked, Z.; et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001, 313, 229–237. [Google Scholar] [CrossRef]
- Richmond, T.J.; Davey, C.A. The structure of DNA in the nucleosome core. Nature 2003, 423, 145–150. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Zhang, Z.; Dong, B.; Liu, J. DNA Sequence-Dependent Properties of Nucleosome Positioning in Regions of Distinct Chromatin States in Mouse Embryonic Stem Cells. Int. J. Mol. Sci. 2022, 23, 14488. https://doi.org/10.3390/ijms232214488
Liu G, Zhang Z, Dong B, Liu J. DNA Sequence-Dependent Properties of Nucleosome Positioning in Regions of Distinct Chromatin States in Mouse Embryonic Stem Cells. International Journal of Molecular Sciences. 2022; 23(22):14488. https://doi.org/10.3390/ijms232214488
Chicago/Turabian StyleLiu, Guoqing, Zhi Zhang, Biyu Dong, and Jia Liu. 2022. "DNA Sequence-Dependent Properties of Nucleosome Positioning in Regions of Distinct Chromatin States in Mouse Embryonic Stem Cells" International Journal of Molecular Sciences 23, no. 22: 14488. https://doi.org/10.3390/ijms232214488
APA StyleLiu, G., Zhang, Z., Dong, B., & Liu, J. (2022). DNA Sequence-Dependent Properties of Nucleosome Positioning in Regions of Distinct Chromatin States in Mouse Embryonic Stem Cells. International Journal of Molecular Sciences, 23(22), 14488. https://doi.org/10.3390/ijms232214488




