Mutation of Leaf Senescence 1 Encoding a C2H2 Zinc Finger Protein Induces ROS Accumulation and Accelerates Leaf Senescence in Rice
Abstract
1. Introduction
2. Results
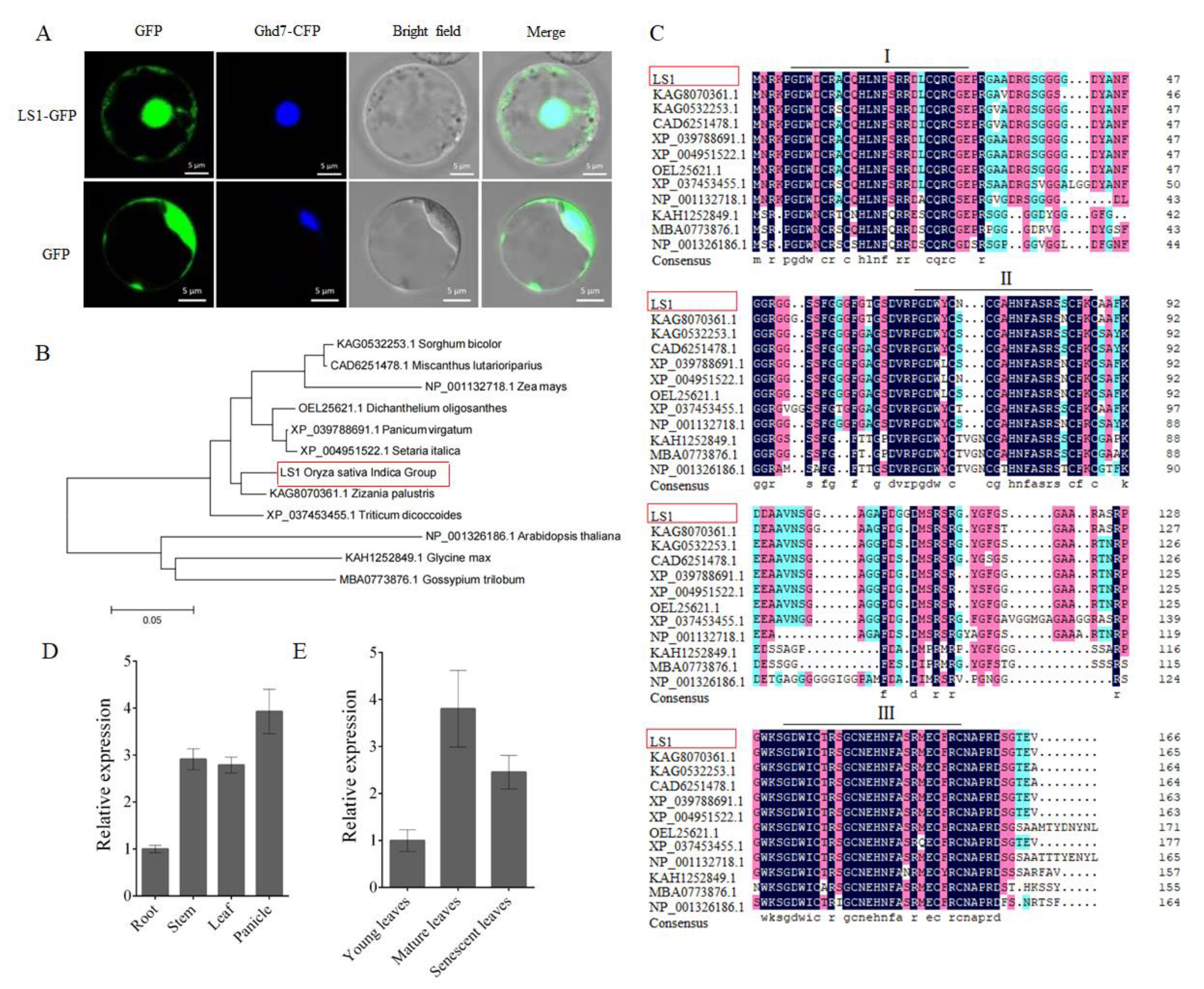
2.1. Phylogenetic Tree and Expression Pattern Analysis
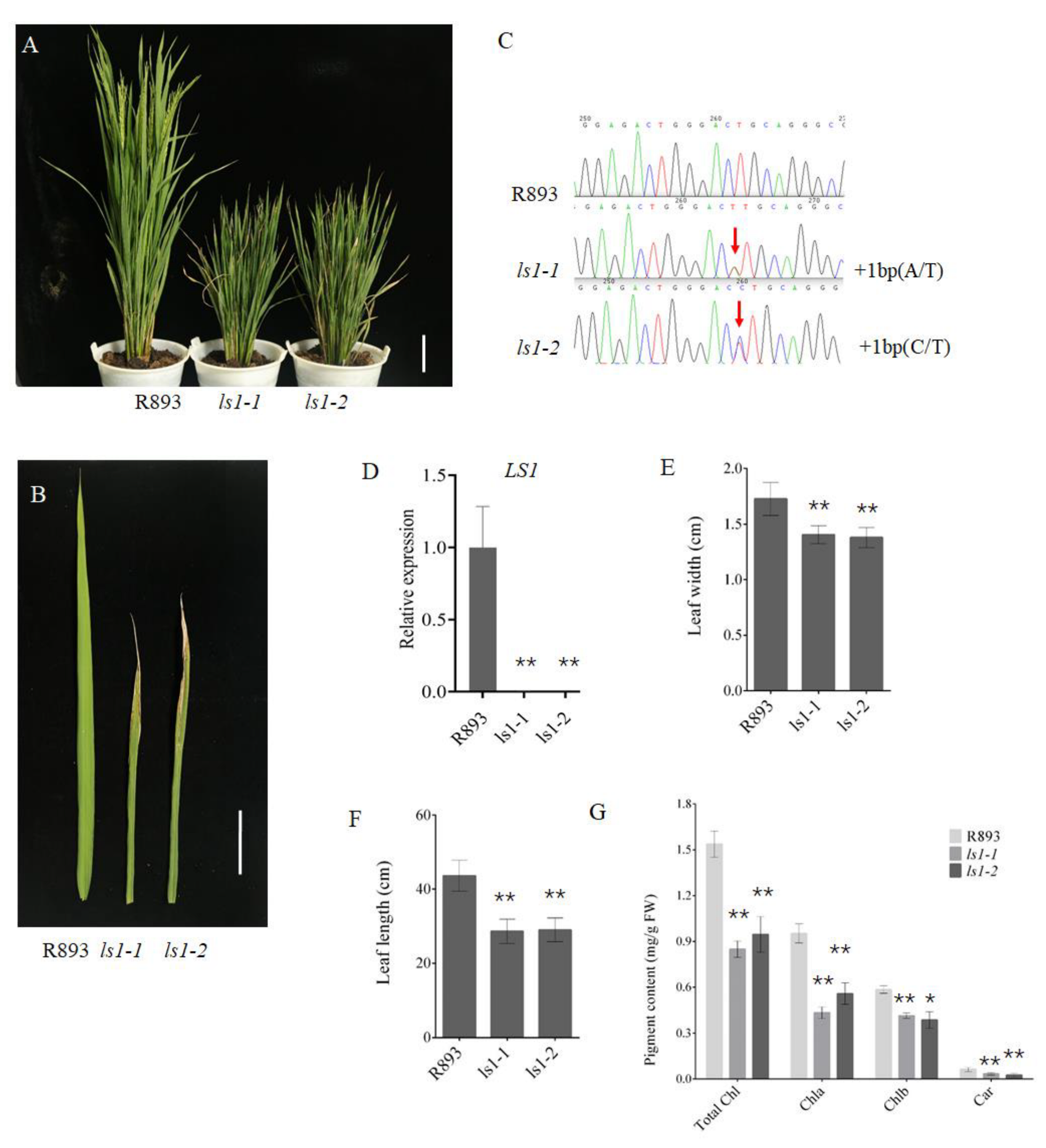
2.2. Loss of LS1 Function Accelerates Leaf Senescence
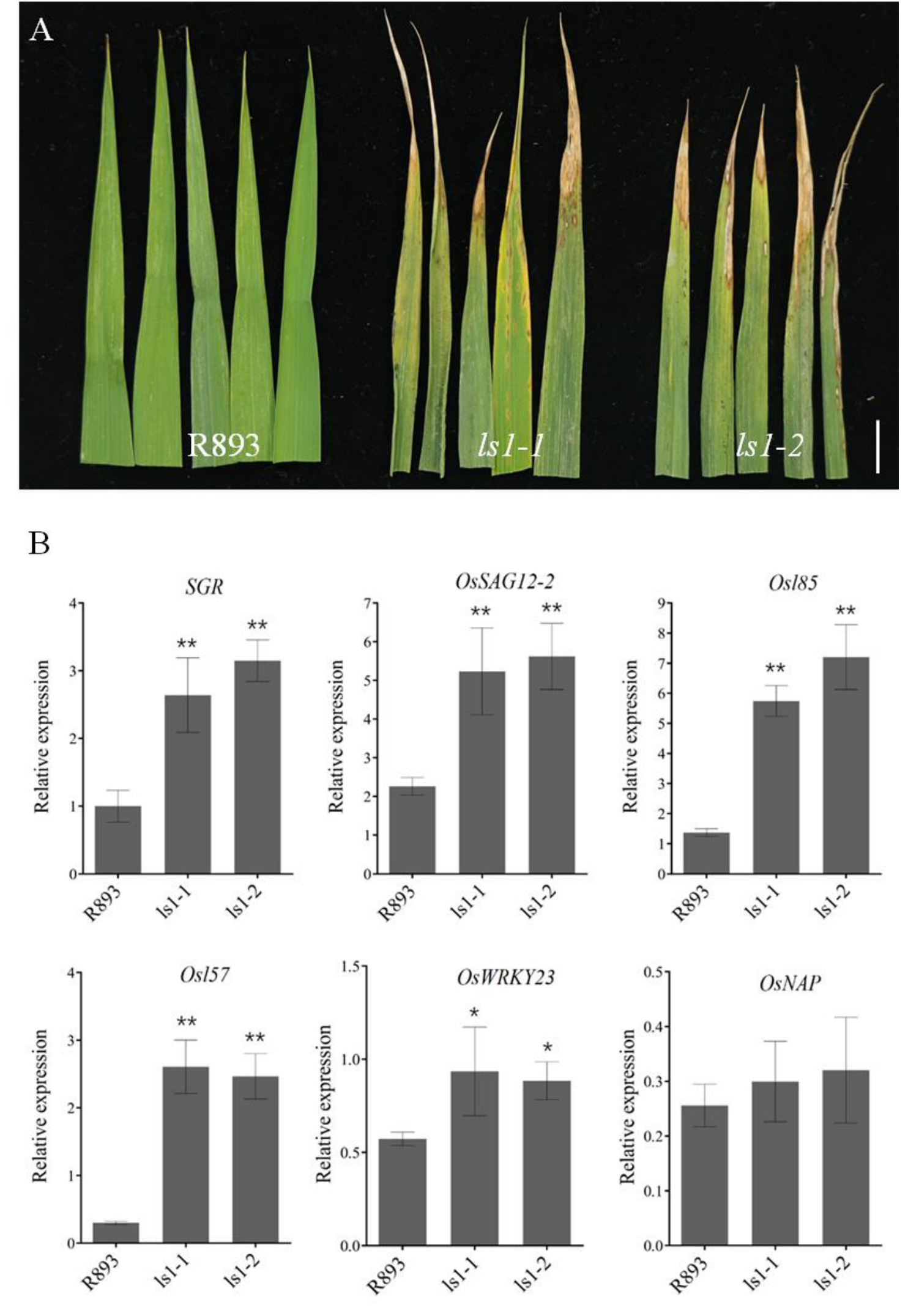
2.3. Mutations in LS1 Lead to Increased Expression of Senescence-Related Genes
2.4. The LS1 Mutants Exhibited More ROS Accumulation and Cell Death
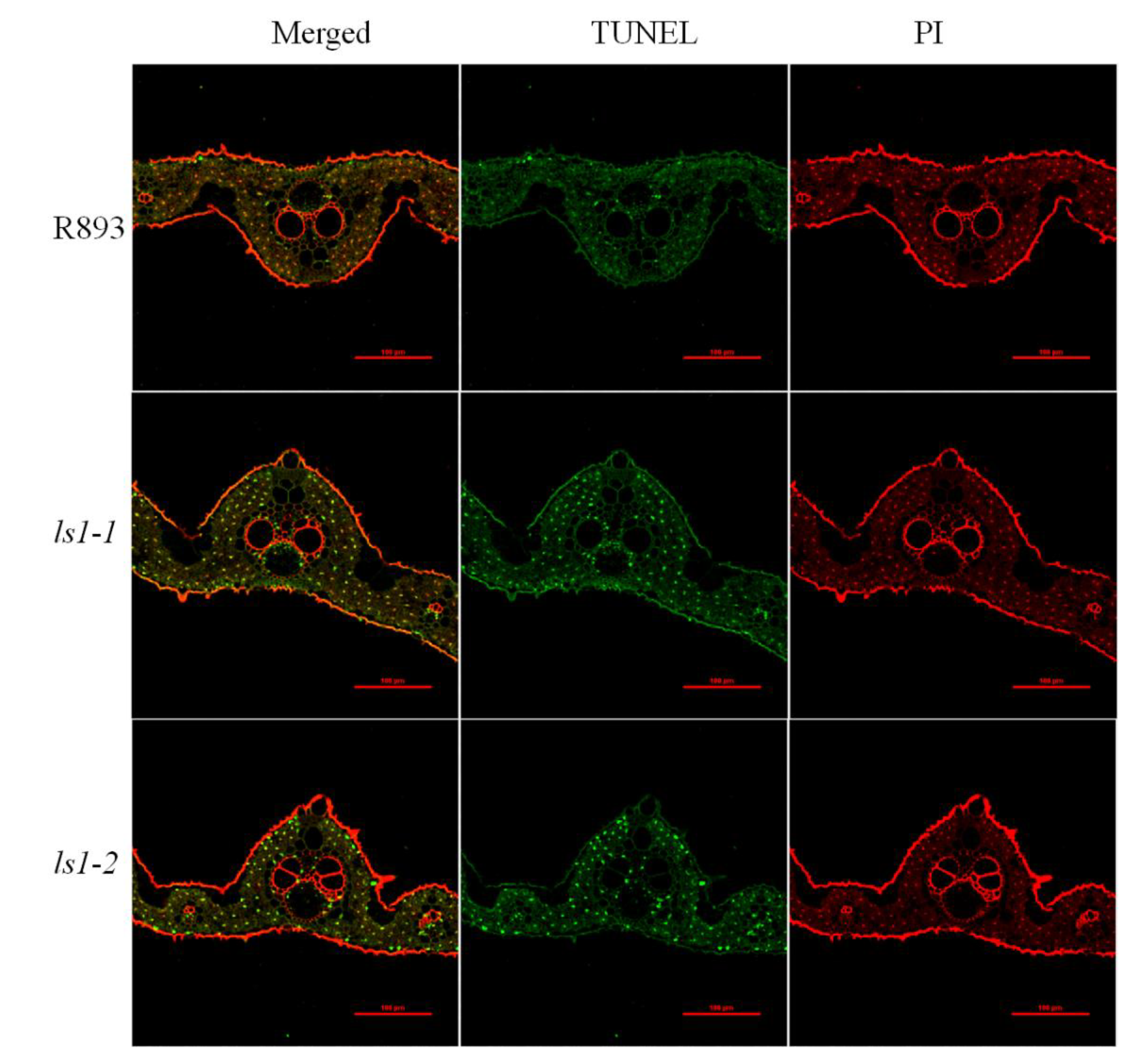
2.5. Loss of Function of LS1 Causes DNA Damage and Triggers PCD
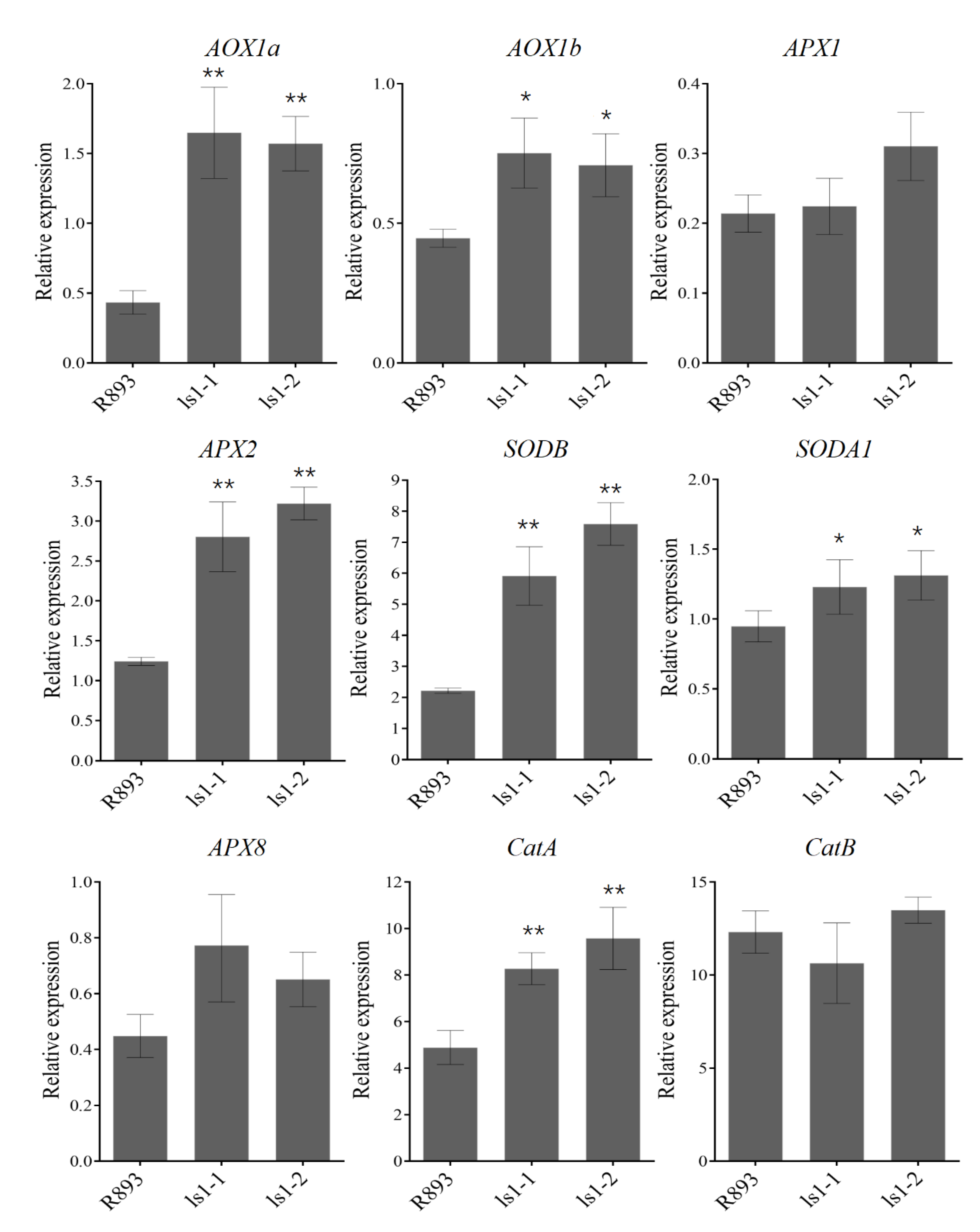
2.6. Alteration in mRNA Levels of ROS-Related Genes in LS1 Plants
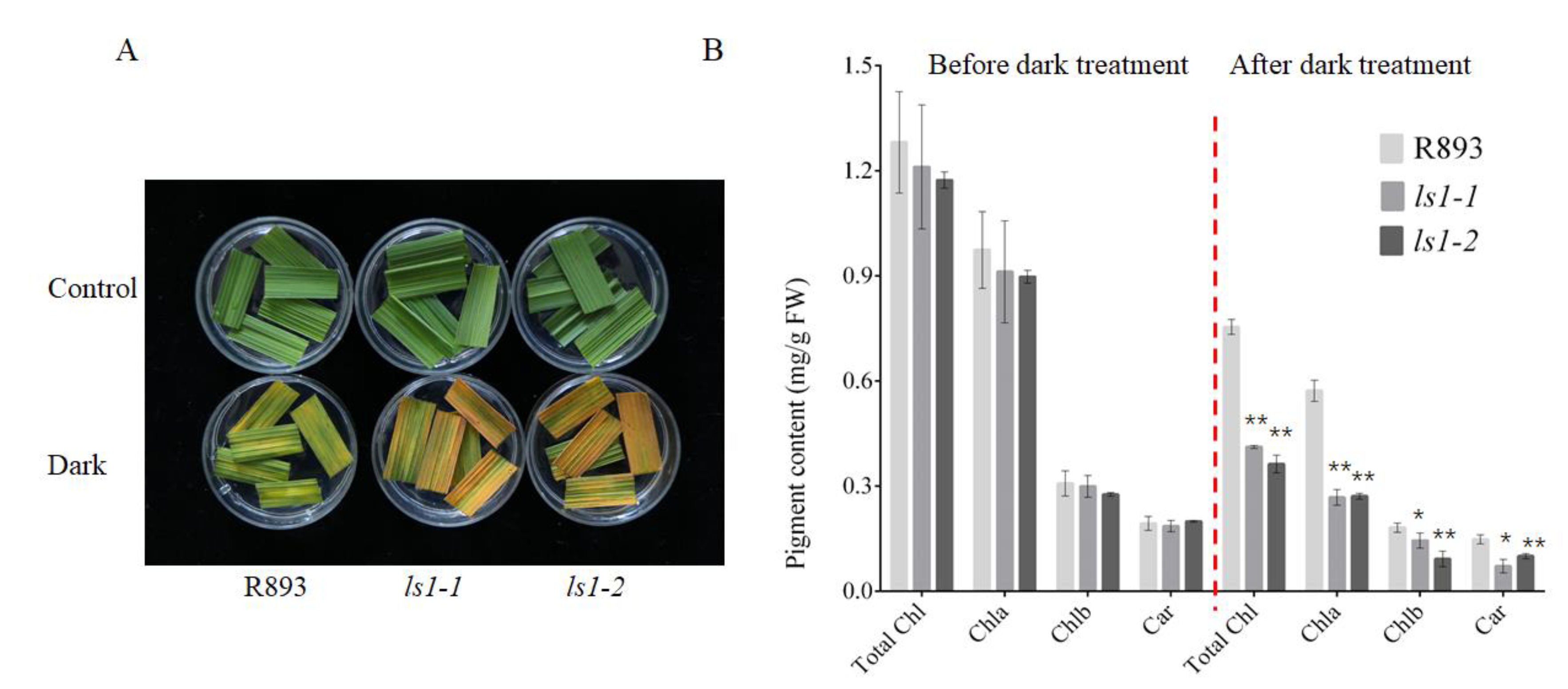
2.7. Acceleration of Dark-Induced Leaf Senescence in LS1 Plants
3. Discussion
3.1. Mutation of LS1 Leads to Leaf Senescence in Rice
3.2. Disruption of LS1 Function Leads to ROS Accumulation and Cell Death in Rice
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. CRISPR/Cas9 Vector Construction, Rice Transformations, and Generation of Cloned Lines
4.3. Histochemical Marker Staining Assay
4.4. Dark Treatment
4.5. Analysis of H2O2, MDA Content, and Enzyme Activity
4.6. Determination of Photosynthetic Pigment Content
4.7. TUNEL Assay
4.8. Subcellular Localization
4.9. Phylogenetic Analysis and Sequence Alignment
4.10. RNA Extraction and Quantitative Real-Time PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, N.; Long, T.; Yao, W.; Xiong, L.; Zhang, Q.; Wu, C. Mutant resources for the functional analysis of the rice genome. Mol. Plant 2013, 6, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, P.L.; Culetic, A.; Boschian, L.; Krupinska, K. Plant senescence and crop productivity. Plant Mol. Biol. 2013, 82, 603–622. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Schippers, J.H. Transcriptional networks in leaf senescence. Curr. Opin. Plant Biol. 2015, 27, 77–83. [Google Scholar] [CrossRef]
- Zhang, W.; Peng, K.; Cui, F.; Wang, D.; Zhao, J.; Zhang, Y.; Yu, N.; Wang, Y.; Zeng, D.; Wang, Y.; et al. Cytokinin oxidase/dehydrogenase OsCKX11 coordinates source and sink relationship in rice by simultaneous regulation of leaf senescence and grain number. Plant Biotechnol. J. 2021, 19, 335–350. [Google Scholar] [CrossRef]
- Wu, X.-Y.; Kuai, B.-K.; Jia, J.-Z.; Jing, H.-C. Regulation of leaf senescence and crop genetic improvement. J. Integr. Plant Biol. 2012, 54, 936–952. [Google Scholar] [CrossRef]
- Thompson, B.G.; Lake, B.H. The effect of radiation on the long term productivity of a plant based CELSS. Adv. Space Res. 1987, 7, 133–140. [Google Scholar] [CrossRef][Green Version]
- Lee, R.H.; Hsu, J.H.; Huang, H.J.; Lo, S.F.; Chen, S.C. Alkaline alpha-galactosidase degrades thylakoid membranes in the chloroplast during leaf senescence in rice. New Phytol. 2009, 184, 596–606. [Google Scholar] [CrossRef]
- Cui, Y.; Peng, Y.; Zhang, Q.; Xia, S.; Ruan, B.; Xu, Q.; Yu, X.; Zhou, T.; Liu, H.; Zeng, D.; et al. Disruption of EARLY LESION LEAF 1, encoding a cytochrome P450 monooxygenase, induces ROS accumulation and cell death in rice. Plant J. 2021, 105, 942–956. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Kim, H.J.; Nam, H.G.; Lim, P.O. Plant leaf senescence and death-regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 2013, 126, 4823–4833. [Google Scholar] [CrossRef] [PubMed]
- Agüera, E.; Cabello, P.; de la Haba, P. Induction of leaf senescence by low nitrogen nutrition in sunflower (Helianthus annuus) plants. Physiol. Plant 2010, 138, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Masclaux-Daubresse, C. Current Understanding of Leaf Senescence in Rice. Int. J. Mol. Sci. 2021, 22, 4515. [Google Scholar] [CrossRef] [PubMed]
- Durian, G.; Jeschke, V.; Rahikainen, M.; Vuorinen, K.; Gollan, P.J.; Brosché, M.; Salojärvi, J.; Glawischnig, E.; Winter, Z.; Li, S.; et al. PROTEIN PHOSPHATASE 2A-B′γ Controls Botrytis cinerea Resistance and Developmental Leaf Senescence. Plant Physiol. 2020, 182, 1161–1181. [Google Scholar] [CrossRef]
- Thomas, H.; Ougham, H.J.; Wagstaff, C.; Stead, A.D. Defining senescence and death. J. Exp. Bot. 2003, 54, 1127–1132. [Google Scholar] [CrossRef]
- Jiao, B.; Wang, J.; Zhu, X.; Zeng, L.; Li, Q.; He, Z. A novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice. Mol. Plant 2011, 5, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.; Liu, S.; Luan, X.; Xie, X.; Hsieh, T.; Zhang, X. Mutation in a putative glycosyltransferase-like gene causes programmed cell death and early leaf senescence in rice. Rice 2019, 12, 7. [Google Scholar] [CrossRef]
- Ramiro, D.A.; Melotto-Passarin, D.M.; Barbosa Mde, A.; Santos, F.D.; Gomez, S.G.; Massola Júnior, N.S.; Lam, E.; Carrer, H. Expression of Arabidopsis Bax Inhibitor-1 in transgenic sugarcane confers drought tolerance. Plant Biotechnol. J. 2016, 9, 1826–1837. [Google Scholar] [CrossRef]
- Lincoln, J.E.; Sanchez, J.P.; Zumstein, K.; Gilchrist, D.G. Plant and animal PR1 family members inhibit programmed cell death and suppress bacterial pathogens in plant tissues. Mol. Plant Pathol. 2018, 19, 2111–2123. [Google Scholar] [CrossRef]
- Lam, E. Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Biol. 2004, 5, 305–315. [Google Scholar] [CrossRef]
- Daneva, A.; Gao, Z.; Van Durme, M.; Nowack, M.K. Functions and regulation of programmed Cell death in Plant Development. Annu. Rev. Cell Dev. Biol. 2016, 32, 441–468. [Google Scholar] [CrossRef]
- Gilroy, E.M.; Hein, I.; van der Hoorn, R.; Boevink, P.C.; Venter, E.; McLellan, H.; Kaffarnik, F.; Hrubikova, K.; Shaw, J.; Holeva, M.; et al. Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J. 2007, 52, 1–13. [Google Scholar] [CrossRef]
- Lin, C.-H.; Pan, Y.-C.; Ye, N.-H.; Shih, Y.-T.; Liu, F.-W.; Chen, C.-Y. LsGRP1, a class II glycine-rich protein of Lilium, confers plant resistance via mediating innate immune activation and inducing fungal programmed cell death. Mol. Plant Pathol. 2020, 21, 1149–1166. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2020, 90, 856–867. [Google Scholar] [CrossRef]
- Heyno, E.; Klose, C.; Krieger-Liszkay, A. Origin of cadmium-induced reactive oxygen species production: Mitochondrial electron transfer versus plasma membrane NADPH oxidase. New Phytol. 2008, 179, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Chai, T.; Zhou, J.; Liu, J.; Xing, D. LSD1 and HY5 antagonistically regulate red light induced-programmed cell death in Arabidopsis. Front. Plant Sci. 2015, 6, 292. [Google Scholar] [CrossRef]
- Chen, C.; Galon, Y.; Rahmati Ishka, M.; Malihi, S.; Shimanovsky, V.; Twito, S.; Rath, A.; Vatamaniuk, O.K.; Miller, G. Ascorbate peroxidasa6 delays the onset of age-dependent leaf senescence. Plant Physiol. 2021, 185, 441–456. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, G.; Liu, S.; An, Q.; Zheng, Q.; Li, B.; Li, Z. Comparative changes in the antioxidant system in the flag leaf of early and normally senescing near-isogenic lines of wheat (Triticum aestivum L.). Plant Cell Rep. 2014, 33, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, T.; Feng, H.; Qiu, T.; Li, Z.; Yang, J.; Peng, Y.-L.; Zhao, W. OsNBL1, a multi-organelle localized protein, plays essential roles in rice senescence, disease resistance, and salt tolerance. Rice 2021, 14, 10. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef]
- Basu, S.; Kumari, S.; Kumar, P.; Kumar, G.; Rajwanshi, R. Redox imbalance impedes photosynthetic activity in rice by disrupting cellular membrane integrity and induces programmed cell death under submergence. Physiol. Plant 2021, 172, 1764–1778. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Han, X.; Hu, H.; Chang, M.; Ding, L.; Xiang, H.; Chen, Y.; Li, Y. 2D vanadium carbide MXenzyme to alleviate ROS-mediated inflammatory and neurodegenerative diseases. Nat. Commun. 2021, 12, 2203. [Google Scholar] [CrossRef]
- You, J.; Zong, W.; Hu, H.; Li, X.; Xiao, J.; Xiong, L. A stress-responsive NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant Physiol. 2014, 166, 2100–2114. [Google Scholar] [CrossRef]
- Dong, W.; Wang, M.; Xu, F.; Quan, T.; Peng, K.; Xiao, L.; Xia, G. Wheat oxophytodienoate reductase gene TaOPR1 confers salinity tolerance via enhancement of abscisic acid signaling and reactive oxygen species scavenging. Plant Physiol. 2013, 161, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Huda, K.M.; Banu, M.S.; Garg, B.; Tula, S.; Tuteja, R.; Tuteja, N. OsACA6, a P-type IIB Ca²⁺ ATPase promotes salinity and drought stress tolerance in tobacco by ROS scavenging and enhancing the expression of stress-responsive genes. Plant J. 2013, 76, 997–1015. [Google Scholar] [CrossRef]
- Doorn, W.; Woltering, E.J. Senescence and programmed cell death: Substance or semantics? J. Exp. Bot. 2004, 55, 2147–2153. [Google Scholar] [CrossRef]
- Niu, F.; Cui, X.; Zhao, P.; Sun, M.; Yang, B.; Deyholos, M.K.; Li, Y.; Zhao, X.; Jiang, Y.Q. WRKY42 transcription factor positively regulates leaf senescence through modulating SA and ROS synthesis in Arabidopsis thaliana. Plant J. 2020, 104, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Chu, Y.; Chen, H.; Li, X.; Wu, Q.; Jin, L.; Wang, G.; Huang, J. Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA-mediated regulatory pathway and ROS scavenging. PLoS Genet. 2018, 14, e1007662. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Qi, Y.; Xu, J.; Dai, X.; Chen, J.; Dong, C.H.; Xiang, F. Arabidopsis WRKY71 regulates ethylene-mediated leaf senescence by directly activating EIN2, ORE1 and ACS2 genes. Plant J. 2021, 107, 1819–1836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, Y.; Wang, H.-L.; Kan, C.; Li, Z.; Yang, X.; Yin, W.; Xia, X.; Nam, H.G.; Li, Z.; et al. Verticillium dahliae secretory effector PevD1 induces leaf senescence by promoting ORE1-mediated ethylene biosynthesis. Mol. Plant 2021, 14, 1901–1917. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Piao, W.; Kim, S.H.; Lee, B.D.; An, G.; Sakuraba, Y.; Paek, N.C. Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J. Exp. Bot. 2019, 70, 2699–2715. [Google Scholar] [CrossRef]
- Han, M.; Kim, C.Y.; Lee, J.; Lee, S.K.; Jeon, J.S. OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol. Cells 2014, 37, 532–539. [Google Scholar] [CrossRef]
- Jan, A.; Maruyama, K.; Todaka, D.; Kidokoro, S.; Abo, M.; Yoshimura, E.; Shinozaki, K.; Nakashima, K.; Yamaguchi-Shinozaki, K. A CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 2013, 161, 1202–1216. [Google Scholar] [CrossRef] [PubMed]
- Uji, Y.; Akimitsu, K.; Gomi, K. Identification of OsMYC2-regulated senescence-associated genes in rice. Planta 2017, 245, 1241–1246. [Google Scholar] [CrossRef]
- Ciftci-Yilmaz, S.; Mittler, R. The zinc finger network of plants. Cell Mol. Life Sci. 2008, 65, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Matthews, J.M.; Sunde, M. Zinc fingers-folds for many occasions. IUBMB Life 2002, 54, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.M. Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 2005, 15, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Gamsjaeger, R.; Liew, C.K.; Loughlin, F.E.; Crossley, M.; Mackay, J.P. Sticky fingers: Zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 2007, 32, 63–70. [Google Scholar] [CrossRef]
- Kong, Z.; Li, M.; Yang, W.; Xu, W.; Xue, Y. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 2006, 141, 1376–1388. [Google Scholar] [CrossRef]
- Lu, G.; Casaretto, J.A.; Ying, S.; Mahmood, K.; Liu, F.; Bi, Y.M.; Rothstein, S.J. Overexpression of OsGATA12 regulates chlorophyll content, delays plant senescence and improves rice yield under high density planting. Plant Mol. Biol. 2017, 94, 215–227. [Google Scholar] [CrossRef]
- Shim, Y.; Kang, K.; An, G.; Paek, N.C. Rice DNA-binding one zinc finger 24 (OsDOF24) delays leaf senescence in a jasmonate-mediated pathway. Plant Cell Physiol. 2019, 60, 2065–2076. [Google Scholar] [CrossRef]
- Huang, J.; Wang, M.; Yan, J.; Wang, Q.; Huang, X.; Zhang, H. Stress repressive expression of rice SRZ1 and characterization of plant SRZ gene family. Plant Sci. 2008, 174, 227–235. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Yang, X.; Gong, P.; Li, K.; Huang, F.; Cheng, F.; Pan, G. A single cytosine deletion in the OsPLS1 gene encoding vacuolar-type H+-ATPase subunit A1 leads to premature leaf senescence and seed dormancy in rice. J. Exp. Bot. 2018, 67, 2761–2776. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, T.; Peng, H.; Luo, S.; Tan, J.; Jiang, K.; Heng, Y.; Zhang, X.; Guo, X.; Zheng, J.; et al. Rice premature leaf senescence 2, encoding a glycosyltransferase (GT), is involved in leaf senescence. Front. Plant Sci. 2018, 9, 560. [Google Scholar] [CrossRef]
- Song, G.; Kwon, C.T.; Kim, S.H.; Shim, Y.; Lim, C.; Koh, H.J.; An, G.; Kang, K.; Paek, N.C. The rice spotted leaf 4 (SPL4) encodes a plant spastin that inhibits ROS accumulation in leaf development and functions in leaf senescence. Front. Plant Sci. 2015, 9, 1925. [Google Scholar] [CrossRef]
- Lee, H.I.; Lee, J.W.; Yang, T.C.; Kang, S.O.; Hoffman, B.M. ENDOR and ESEEM investigation of the Ni-containing superoxide dismutase. J. Biol. Inorg. Chem. 2010, 15, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.-Y.; Wang, P.-C.; Chen, J.; Song, C.-P. Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J. Integr. Plant Biol. 2008, 50, 1318–1326. [Google Scholar] [CrossRef] [PubMed]
- Khanna-Chopra, R. Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 2012, 249, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, H.-L.; Li, Z.; Guo, H. Genetic network between leaf senescence and plant immunity: Crucial regulatory nodes and new insights. Plants 2020, 9, 495. [Google Scholar] [CrossRef]
- Yang, S.; Fang, G.; Zhang, A.; Ruan, B.; Jiang, H.; Ding, S. Rice early senescence 2, encoding an inositol polyphosphate kinase, is involved in leaf senescence. BMC Plant Biol. 2020, 20, 393. [Google Scholar] [CrossRef]
- Leng, Y.; Yang, Y.; Ren, D.; Huang, L.; Dai, L.; Wang, Y.; Chen, L.; Tu, Z.; Gao, Y.; Li, X.; et al. A rice pectate lyase-like gene is required for plant growth and leaf senescence. Plant Physiol. 2017, 174, 1151–1166. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, Y.; Bi, Z.; Liu, Q.; Xu, T.; Yu, N.; Cao, Y.; Zhu, A.; Wu, W.; Zhan, X.; et al. Impaired function of the calcium-dependent protein kinase, OsCPK12, leads to early senescence in rice (Oryza sativa L.). Front. Plant Sci. 2019, 10, 52. [Google Scholar] [CrossRef]
- Wei, Q.; Yan, Z.; Xiong, Y.; Fang, Z. Altered Expression of OsAAP3 Influences Rice Lesion Mimic and Leaf Senescence by Regulating Arginine Transport and Nitric Oxide Pathway. Int. J. Mol. Sci. 2021, 22, 2181. [Google Scholar] [CrossRef] [PubMed]
- Shim, K.C.; Kim, S.H.; Jeon, Y.A.; Lee, H.S.; Adeva, C.; Kang, J.W.; Kim, H.J.; Tai, T.H.; Ahn, S.N. A RING-Type E3 Ubiquitin Ligase, OsGW2, Controls Chlorophyll Content and Dark-Induced Senescence in Rice. Int. J. Mol. Sci. 2020, 21, 1704. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Kwasniewski, M.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Xue, G.-P.; Mueller-Roeber, B. ORS1, an H₂O₂-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant 2011, 4, 346–360. [Google Scholar] [CrossRef]
- Hung, K.T.; Kao, C.H. Hydrogen peroxide is necessary for abscisic acid-induced senescence of rice leaves. J. Plant Physiol. 2004, 161, 1347–1357. [Google Scholar] [CrossRef]
- Hu, S.; Yu, Y.; Chen, Q.; Mu, G.; Shen, Z.; Zheng, L. OsMYB45 plays an important role in rice resistance to cadmium stress. Plant Sci. 2017, 264, 1–8. [Google Scholar] [CrossRef]
- Zheng, Y.; Xu, J.; Wang, F.; Tang, Y.; Wei, Z.; Ji, Z.; Wang, C.; Zhao, K. Mutation types of CYP71P1 cause different phenotypes of mosaic spot lesion and premature leaf senescence in rice. Front. Plant Sci. 2021, 12, 641300. [Google Scholar] [CrossRef]
- Park, J.; Bae, S.; Kim, J.S. Cas-Designer: A web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 2015, 31, 4014–4016. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, K. A high-efficiency Agrobacterium-mediated transformation system of rice (Oryza sativa L.). Methods Mol. Biol. 2012, 847, 51–57. [Google Scholar] [CrossRef]
- Qiao, Y.; Jiang, W.; Lee, J.; Park, B.; Choi, M.S.; Piao, R.; Woo, M.O.; Roh, J.H.; Han, L.; Paek, N.C.; et al. SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol. 2015, 185, 258–274. [Google Scholar] [CrossRef] [PubMed]
- Han, S.H.; Sakuraba, Y.; Koh, H.J.; Paek, N.C. Leaf variegation in the rice zebra2 mutant is caused by photoperiodic accumulation of tetra-cis-lycopene and singlet oxygen. Mol. Cells. 2012, 33, 87–97. [Google Scholar] [CrossRef]
- Yin, Z.; Chen, J.; Zeng, L.; Goh, M.; Leung, H.; Khush, G.S.; Wang, G.L. Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol. Plant Microbe Interact. 2000, 13, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhu, K.; Wang, Z.; Zhang, H.; Gu, J.; Liu, L.; Yang, J.; Zhang, J. Brassinosteroids function in spikelet differentiation and degeneration in rice. J. Integr. Plant Biol. 2019, 61, 943–963. [Google Scholar] [CrossRef]
- Wellburn, A.R. The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Zheng, S.; Li, J.; Ma, L.; Wang, H.; Zhou, H.; Ni, E.; Jiang, D.; Liu, Z.; Zhuang, C. OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers. Proc. Natl. Acad. Sci. USA 2019, 116, 7549–7558. [Google Scholar] [CrossRef]
- Ren, D.; Yu, H.; Rao, Y.; Xu, Q.; Zhou, T.; Hu, J.; Zhang, Y.; Zhang, G.; Zhu, L.; Gao, Z.; et al. “Two-floret spikelet” as a novel resource has the potential to increase rice yield. Plant Biotechnol J. 2018, 16, 351–353. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Li, N.; Hu, Z.; Liu, H.; Hu, Y.; Tan, Y.; Sun, Q.; Liu, X.; Xiao, L.; Wang, W.; et al. Mutation of Leaf Senescence 1 Encoding a C2H2 Zinc Finger Protein Induces ROS Accumulation and Accelerates Leaf Senescence in Rice. Int. J. Mol. Sci. 2022, 23, 14464. https://doi.org/10.3390/ijms232214464
Zhang C, Li N, Hu Z, Liu H, Hu Y, Tan Y, Sun Q, Liu X, Xiao L, Wang W, et al. Mutation of Leaf Senescence 1 Encoding a C2H2 Zinc Finger Protein Induces ROS Accumulation and Accelerates Leaf Senescence in Rice. International Journal of Molecular Sciences. 2022; 23(22):14464. https://doi.org/10.3390/ijms232214464
Chicago/Turabian StyleZhang, Chao, Ni Li, Zhongxiao Hu, Hai Liu, Yuanyi Hu, Yanning Tan, Qiannan Sun, Xiqin Liu, Langtao Xiao, Weiping Wang, and et al. 2022. "Mutation of Leaf Senescence 1 Encoding a C2H2 Zinc Finger Protein Induces ROS Accumulation and Accelerates Leaf Senescence in Rice" International Journal of Molecular Sciences 23, no. 22: 14464. https://doi.org/10.3390/ijms232214464
APA StyleZhang, C., Li, N., Hu, Z., Liu, H., Hu, Y., Tan, Y., Sun, Q., Liu, X., Xiao, L., Wang, W., & Wang, R. (2022). Mutation of Leaf Senescence 1 Encoding a C2H2 Zinc Finger Protein Induces ROS Accumulation and Accelerates Leaf Senescence in Rice. International Journal of Molecular Sciences, 23(22), 14464. https://doi.org/10.3390/ijms232214464





