Abstract
Colorectal cancer (CRC) is a serious public health issue, and it has the leading incidence and mortality among malignant tumors worldwide. CRC patients with metastasis in the liver, lung or other distant sites always have poor prognosis. Thus, there is an urgent need to discover the underlying mechanisms of metastatic colorectal cancer (mCRC) and to develop optimal therapy for mCRC. Transforming growth factor-β (TGF-β) signaling plays a significant role in various physiologic and pathologic processes, and aberrant TGF-β signal transduction contributes to mCRC progression. In this review, we summarize the alterations of the TGF-β signaling pathway in mCRC patients, the functional mechanisms of TGF-β signaling, its promotion of epithelial–mesenchymal transition, its facilitation of angiogenesis, its suppression of anti-tumor activity of immune cells in the microenvironment and its contribution to stemness of CRC cells. We also discuss the possible applications of TGF-β signaling in mCRC diagnosis, prognosis and targeted therapies in clinical trials. Hopefully, these research advances in TGF-β signaling in mCRC will improve the development of new strategies that can be combined with molecular targeted therapy, immunotherapy and traditional therapies to achieve better efficacy and benefit mCRC patients in the near future.
1. Introduction
Colorectal cancer (CRC) is a type of cancer in which abnormal cells grow out of control in the large intestine. According to global cancer statistics, more than 1.9 million new cases and 935,000 deaths from CRC occurred in 2020. CRC is the third most commonly diagnosed malignancy worldwide and ranks second in cancer-related mortality. In other words, CRC accounts for 10% of all cancer cases and deaths [1]. In the United States, CRC ranked fourth in estimated new cases and second in estimated cancer-related deaths (thus far) in 2022 [2,3]. Due to the prevalence of obesity and lack of exercise in recent decades, the incidence of CRC is on the rise among the entire population in China, and it is currently the fifth leading cause of cancer death there [4]. The accumulation of genetic mutations and environmental risk factors are the main causes of CRC [5].
CRC metastasis is always a thorny problem in clinical situations. At the time of diagnosis, about 20% of CRC patients already have metastasis and 35–45% succumb to recurrence within five years after surgery [6]. The five-year survival rate of stage I-III CRC patients can be as high as 80%, whereas it drops to roughly 13% for patients with stage IV CRC [7]. It is reported that up to 60% of patients with stage IV CRC develop liver metastasis, demonstrating that the liver is the most common site for CRC metastatic spread [8,9]. The lung is the second most common metastatic target organ for CRC. Consequently, although modern surgical techniques and multidisciplinary systematic care have led to significant improvements in survival, long-term remission can only be achieved in 20% of patients with metastasis, and relapse occurred in 60–70% of patients [10,11]. Therefore, there is an urgent need to identify the underlying mechanisms of metastatic colorectal cancer (mCRC) and for new optimal therapeutic strategies for mCRC to be developed.
The transforming growth factor-β (TGF-β) signaling pathway plays a multifaceted role in various biological processes, such as cell growth and differentiation, apoptosis, cell motility, epithelial–mesenchymal transition (EMT), extracellular matrix (ECM) remodeling, angiogenesis and cellular immune responses [12,13]. Therefore, malfunction of the TGF-β signal pathway, either via genetic mutation or misexpression, is associated with many diseases, including cancer, fibrosis, inflammation, cardiovascular diseases, myelodysplastic syndrome, Marfan syndrome, scleroderma, endometriosis and more [14,15,16]. Currently, 33 members in the TGF-β superfamily have been identified in human beings, including three TGF-β isoforms, three activins, nodal, growth and differentiation factor (GDF) and the bone morphogenetic protein (BMP) subfamily, which are involved in various physiologic and pathologic mechanisms [17]. There are three isoforms of TGF-β: TGF-β1, TGF-β2, and TGF-β3. They manifest different expression patterns, bioavailability and physiological functions in organisms, respectively [18]. In addition to these ligands, downstream intracellular effectors, termed SMAD, are a group of proteins that include eight different members in mammalian cells and can transduce extracellular signals to the nucleus [19]. Aberrant signal transduction of TGF-β signaling may lead to a variety of tumors, including esophageal cancer, hepatocellular carcinoma, pancreatic cancer, gastric cancer, CRC, etc. [20]. TGF-β signaling can suppress tumor development by inhibiting cell proliferation and stimulating cell differentiation in the early stages of cancer. However, it induces tumor progression and metastasis in late stages of cancer, which is known as the “TGF-β paradox” [21]. Variation in the TGF-β pathway is also a common event in CRC tumorigenesis and metastasis. When TGF-β or SMAD are mutated, an abnormal TGF-β signaling pathway would contribute to CRC metastasis [22].
The present review mainly focuses on the role of altered TGF-β signaling in mCRC, the mechanisms through which TGF-β affects CRC metastasis and the clinical application of the key components in TGF-β signaling as potential therapeutic targets for mCRC. These research advances will surely shed new light on TGF-β targeting therapy and benefit the mCRC patients in the near future.
2. Alterations in TGF-β Signaling Pathway in mCRC
2.1. TGF-β Signaling Pathway
Research has shown that TGF-β signaling is transduced from cell membrane surface receptors to the nucleus. TGF-β ligands secreted by cells disperses in the matrix in an inactive form, and it can be activated in an integrin-dependent manner [23]. TGF-β1 and TGF-β3 can be activated by avβ6 or avβ8 integrins while TGF-β2 cannot, which implies a different mechanism for TGF-β2 [24]. There are three receptors in the TGF-β signaling pathway: TGFBR1, TGFBR2 and TGFBR3. The TGF-β ligand first binds to the TGFBR2 and induces the formation of a hetero-tetrameric complex of TGFBR2 and TGFBR1 [25,26]. Subsequently, this complex causes the TGFBR2 kinase domain to phosphorylate TGFBR1 in a region of the juxtamembrane domain that is rich in glycine and serine residues. This then activates TGFBR1 and subsequently phosphorylates SMAD2/3 [27,28]. Following this, phosphorylated SMAD2 and SMAD3 can be assembled into complexes with SMAD4 and then translocated to the nucleus where they can regulate the expression of target genes [28]. SMAD7, a negative regulator of the TGF-β pathway, competes with SMAD2/3 for the catalytic site of TGFBR1 phosphorylation and thereby inhibits the phosphorylation of SMAD2/3 [29]. SMAD proteins can be divided into three categories, including the common-mediator SMAD (Co-SMAD), the receptor-regulated SMAD (R-SMAD) and the inhibitory SMAD (I-SMAD). The Co-SMAD (SMAD4) is the central mediator of the TGF-β signaling pathway. The R-SMAD (SMAD1, -2, -3, -5 and -8/9) can be phosphorylated by activated type I receptor kinases. The I-SMAD (SMAD6/7) can competitively inhibit R-SMAD phosphorylation and thereby antagonize TGF-β signaling [19].
In addition to the canonical SMAD-dependent signaling pathway, there are also several non-canonical pathways within the TGF-β superfamily, such as the Rho-associated kinase (ROCK) pathway, the phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) and the mitogen-activated protein kinase (MAPK) pathway [30]. These activated non-canonical SMAD pathways also crosstalk with the canonical SMAD pathway.
2.2. Aberrant TGF-β Pathway Signals in mCRC
It is generally accepted that the occurrence of cancer is accompanied by the accumulation of gene mutations [31]. Mutations in TGF-β receptors and SMAD proteins occur more frequently in CRC resulting in malignant phenotypes, whereas mutations in TGF-β ligands are relatively rare.
With advances in next-generation sequencing technology, variations in TGF-β signaling in mCRC on different levels have become more accessible for clinical investigators. According to the Ingenuity Pathway Analysis, Wnt, PI3K/AKT and TGF-β/SMAD signaling are the most commonly mutated pathways in colorectal cancer metastasis [32]. Carcinoembryonic antigen (CEA) is widely used as a prognostic clinical marker of metastasis, and the TGF-β signaling pathway is significantly enriched in CEA-induced colorectal liver metastases (CRLM) according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis [33]. In a study utilizing targeted next-generation sequencing (NGS) to assess 128 patients with mCRC, alterations of TGF-β pathways were identified in 17% of the mCRC tissues [34]. In another study involving 579 patients undergoing CRLM resection, 11.2% of patients were found to have TGF-β mutations [35]. Furthermore, aberrant DNA-methylation-regulated genes showed enrichment in TGF-β signaling pathway based on data of DNA methylation (GSE90709, GSE77955) downloaded from the Gene Expression Omnibus database [36].
Although most studies utilize genetic and pharmacological strategies to investigate TGF-β signaling of all three isoforms, these three isoforms actually function through distinct mechanisms. The knockout mice of the three isoforms demonstrated non-overlapping defects: TGF-β1-null mice showed inflammatory disease, TGF-β2-null mice exhibit multiple developmental defects in a wide range of organs, while TGF-β3 knockout led to defective palatogenesis [37,38,39]. TGF-β1 is expressed more abundantly in the tumor microenvironment (TME) in various human tumors than the other two isoforms and contributes to resistance to checkpoint blockade therapy [40]. TGF-β2 was shown to be involved in neutrophil recruitment in an organoid model of mCRC [41]. In addition, TGF-β1 and TGF-β3 are reported to both be activated in stroma cells and to contribute to the prometastatic process in CRC [42].
The receptor of the TGF-β signaling pathway is indispensable and its change can lead to abnormalities of the pathway. Reports indicated that TGFBR1*6A can switch TGF-β1 growth-inhibitory functions into growth-stimulatory functions, which significantly increased the invasion of SW48 and DLD-1 cells compared with transfected TGFBR1*9A cell lines [43]. The germline allele-specific expression (ASE) of TGFBR1 increases CRC risk for the Caucasian-dominated population in the United States [44]. In the majority of microsatellite instability (MSI) CRC tumors, the gene encoding TGFBR2 has a very high frequency of uniquely inactivating mutations. According to public databases, tumors harboring TGFBR2 mutations showed a greater degree of vascular invasion than tumors without such mutations, which contributes to tumor progression in MSI-positive CRC [45]. In addition, frameshift mutations of TGFBR2 were present in three quarters of late-stage MSI CRC, and this mutation might mediate CRC progression from the early to late stage [46]. In an MSI CRC model cell line, inactivating frameshift mutations of TGFBR2 can reprogram the protein content and regulate the cytokine secretion profile. These changes are related to tumor angiogenesis, migration, metastasis and immune escape of recipient cells [47]. In the HCT116-TGFBR2 MSI CRC cell line model system, which reflects the inverse situation of the TGFBR2-deficient MSI CRC, sialylated β1-integrin is significantly decreased, and variant sialylation could affect metastasis and migration of CRC cells [48]. Additionally, in a cohort of 184 CRC patients and 307 healthy volunteers, male CRC patients with TGFΒR2-875A genotypes had a lower risk of CRC progression and metastasis compared with CRC patients with TGFBR2-875G [49].
Because SMAD proteins are key factors in the transduction of the classical TGF-β signaling pathway, alterations in SMAD proteins play a crucial role in late stages of CRC by contributing to migration and metastasis. As reported, CRC patients who lose SMAD activity are more likely to have lymph node metastasis resulting in a poor prognosis [50]. As reported in an analysis of exome capture DNA sequencing from 224 participants, both those with tumors and without, SMAD4 and TGFBR2 are two commonly mutated genes. The mutation frequency of SMAD4 and SMAD2 in non-hypermutated tumors is 10%, while that of TGFBR2 in hypermutated tumors (including MSI-high) is 51% [51]. According to the sequencing analysis of SMAD4, SMAD2 and SMAD3 in a group of 744 primary CRC patients and 36 CRC cell lines, the prevalence of SMAD4, SMAD2 and SMAD3 mutations was found to be 8.6%, 3.4% and 4.3% in sporadic CRC, respectively. In addition, the mutation spectra of SMAD2/3 were highly similar to that of SMAD4, and joint biallelic hits in SMAD2/3 were highly frequent and mutually exclusive to SMAD4 mutation, indicating the crucial roles of these three SMAD proteins in the TGF-β signaling pathway [52].
According to a series of high-throughput analyses, SMAD4 was one of the most commonly mutated genes in mCRC, which will now be further discussed. The results of one study that used targeted NGS sequencing involving 123 non-MSI-high mCRC patients showed a 22.8% mutation frequency of SMAD4 [34]. Similarly, in another study of 32 mCRC patients, the SMAD4 mutation frequency rate was approximately 6% [53]. Similar results were achieved for SMAD4 mutation rates (15% vs. 14%) in primary and metastatic CRCs by comparing genetic profiles [54]. In CRC patients, SMAD4 mutation and deletion detected with NGS were significantly associated with invasive-front pathological markers [55]. In 330 early onset (EO) mCRC patients, SMAD4 was recurrently mutated, resulting in aberrance of the TGF-β pathway in 30% of patients [56]. Using samples obtained from 32 patients, Lopez-Gomez et al. found that SMAD4 expression was at similar levels and was positively associated between the formalin-fixed paraffin-embedded (FFPE) mCRC tumor and their matched liver metastases [57]. Moreover, there is an increased frequency of SMAD4 alterations in ovarian metastases from CRC, suggesting that the oncogenic properties conferred by aberrant TGF-β signaling may contribute to CRC metastasis to the ovaries [58].
SMAD7 is a crucial negative regulator of the TGF-β signaling pathway [59]. Reports indicated that the expression of SMAD7 was remarkably lower in mCRC tissues than in non-tumor tissues [60]. Compared with control mice, mice injected with SMAD7-expressing clones had elevated levels of TGFBR2 expression and TGF-β secretion in liver metastases, which could then lead to phosphorylation and nuclear accumulation of SMAD2. In the nude mouse CRC model, ectopic expression of SMAD7 promoted CRC metastasis to the liver in the splenic injection model [61].
Furthermore, TGF-β mutations always occur concurrently with variations in other signaling pathways, demonstrating that the accumulation of these mutations in mCRC has a synergistic effect on CRC metastasis. For example, KRASG12D mutation can induce an EMT-like morphology of tumors when combined with mutations in Tgfbr2-/-. Moreover, KRAS activation promotes liver metastasis when combined with adenomatous polyposis coli (APC) Δ716 and TGFBR2 mutations [62]. In the colon epithelium of a CRC mouse model, combined inactivation of APC and TGFBR2 promoted development of adenocarcinoma in the proximal colon, and gasdermin C expression was upregulated by TGFBR2 mutation, resulting in increased CRC cells proliferation [63]. Based on the mCRC mouse model that harbored a KRASmut allele, conditional null alleles of APC and transformation-related protein 53 (Trp53), the TGF-β pathway was a critical mediator of KRASmut-driven invasiveness, as proven by system-level and functional analysis [64]. Fumagalli et al. found that the accumulation of genetic mutations in the Wnt, epidermal growth factor receptor (EGFR), P53 and TGF-β signaling pathways can drive CRC cells to migrate and grow at distant sites in an orthotopic organoid transplantation model and in engineered human colon tumor organoids [65]. In a Chinese CRLM cohort, CRLM patients with differing primary tumor sites had differences in survival rates, which could be driven by combined variations in the TGF-β, PI3K and RAS signaling pathways [66]. Reports showed patients with mutated SMAD4 had shorter progression-free survival (PFS) than patients with wild-type SMAD4 after receiving anti-EGFR therapy, which may imply a synergistic effect of SMAD4 loss and EGFR in mCRC [67].
In addition, some key proteins or factors can act on the TGF-β signaling pathway and affect CRC progression. Compared with a control group, CRC cells overexpressing Tripartite Motif Containing 25 (TRIM25) exhibit a two-fold higher migration rate. TRIM25 also promotes CRC tumor progression in a nude mice xenograft model by positively regulating the TGF-β signaling pathway [68]. The overexpression of Helicase-like Transcription Factor (HLTF) and activation of Slit2/Robo1 signaling can suppress both CRC cell migration and invasion through the TGF-β/SMAD pathway [69,70]. Prolyl 4-Hydroxylase Subunit Alpha 3 (P4HA3) promotes subcutaneous tumorigenesis in nude mice by upregulating the TGF-β/SMAD signaling pathway, and the knockdown of P4HA3 strongly inhibits the proliferation and invasion abilities of CRC cells [71]. ETS homologous factor could activate the canonical TGF-β pathway through directly upregulating TGF-β1 expression at the transcriptional level and could promote CRC cell proliferation and migration in vitro and in vivo [72]. Glypicans 1 (GPC1) knockdown significantly suppressed levels of TGF-β1 and p-SMAD2, resulting in inhibition of the migration of CRC cells [73]. In vivo experiments showed ZIC2, a protein involved in the advancement of many types of tumors, induced TGF-β1 expression and SMAD3 phosphorylation, resulting in CRLM progression [74].
3. Mechanism of TGF-β Functions in mCRC
CRC metastasis is a dynamic, multistep and multifactorial process, which includes the following successive steps: detachment from the primary CRC site, infiltration into adjacent tissues, invasion into blood/lymphatic circulation, transportation through the circulatory system, intravasation from vasculature and formation of CRC colonies in distant sites. Three critical factors contribute to CRC cells migration (pivotal for early metastasis): regulating the EMT process, stemness and the microenvironment of CRC cells. Additionally, angiogenesis facilitates CRC cell transportation to distal locations. TGF-β signaling contributes to mCRC mainly through the following four mechanisms: promoting EMT, facilitating angiogenesis, creating an immunosuppressive microenvironment and regulating the stemness of mCRC (as shown in Figure 1) [21,28].
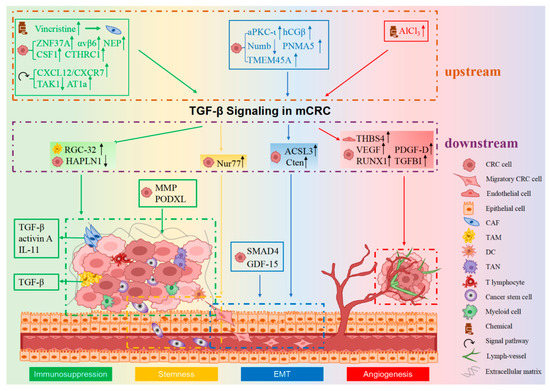
Figure 1.
The mechanism of TGF-β signaling in CRC metastasis. TGF-β mainly affects CRC metastasis in four different ways: EMT, angiogenesis, immunosuppression and stemness. Together, they work to facilitate the metastasis of CRC. Tumor cells undergo the EMT process, acquire a mesenchymal-like phenotype in response to TGF-β signaling and then becoming more invasive and spread to distant sites. TGF-β signaling can mediate the formation of new blood vessels, which can promote intravasation of tumor cells from primary lesions into the blood vessels, resulting in tumor metastasis. In the tumor microenvironment (TME), immune cells such as CAFs and TAMs contribute to the immunosuppressive microenvironment and induce dissemination of tumor cells to distant places through TGF-β signaling. Moreover, TGF-β signaling can regulate CSCs in CRC, further promoting tumor metastasis. The molecules upstream or downstream of TGF-β signaling have been enclosed by dotted lines of different colors, and they function through EMT, angiogenesis, immunosuppression and stemness, respectively (distinguished by four different background colors). The symbols in front of the molecules represent whether it is a chemical, a signal factor or a molecule secreted by CRC cells, epithelial cells or immune cells in the TME. EMT, epithelial-to-mesenchymal transition. The legend for different cell types is shown in the lower right.
3.1. TGF-β Signaling in EMT in mCRC
Epithelial cells undergoing EMT will lose their apicobasal polarity and adhesion, acquire motile mesenchymal characteristics and become more invasive, which contributes to the onset of CRC metastasis [75]. Epithelial and mesenchymal cells can be distinguished by specific molecular markers expressed in cells. For instance, epithelial cells express E-cadherin and cytokeratins, while N-cadherin, Snail, Slug and Vimentin are markers of mesenchymal cells [76]. Cells undergoing the EMT process are distinguished by the loss of E-cadherin expression, a decrease of epithelial cell junctions and cytoskeleton and display a mesenchymal pattern with enhanced cell motility and invasiveness [77].
TGF-β signaling is an essential regulator of the process of EMT. As reported, TGF-β can induce the EMT process by downregulating the expression of tight junction proteins, resulting in weakened tight junctions, which is the key point for EMT induction of TGF-β signaling [78]. SMAD4 is demonstrated to downregulate the expression of Claudin 1, which contributes to CRC metastasis [79]. Most of these observations were made in vitro, although the results of in vivo experiments are more convincing and important. In light of research in human SW480 CRC cells, TGF-β1 can induce Alu RNA expression, the accumulation of which promotes the EMT process, and Alu expression significantly correlates with CRC progression [80]. TGF-β1 upregulates the expression of C-terminal tensin-like (Cten) and EMT markers, and it promotes the cell motility of the CRC cell lines SW620 and HCT116 [81]. TGF-β1 can also induce the upregulation of acyl-CoA synthetases 3 (ACSL3) which produces ATP and reduces NADPH, thus sustaining redox homeostasis and mediating the EMT and metastasis of CRC cells [82]. A functional study indicated that TGF-β can induce SMAD4-dependent EMT followed by apoptosis in HCT-116 and DLD1 CRC cell lines [83]. As reported in CRC cell assays and murine models, acidosis-induced TGF-β2 activation promotes the formation of lipid droplets, which provides energy for cancer cell metastasis and partially promotes EMT [84]. SMAD4 in TGF-β signaling is frequently inactivated in human CRC, and SMAD4 codes for a transcription factor central to canonical TGF-β signaling. Therefore, it is generally understood that EMT will not occur in SMAD4-mutant tumors. However, in SMAD4-mutant CRC cell lines and analyses of human CRC transcriptomes, EMT is not categorically precluded. Possible explanations for this may be that SMAD4-mutant tumors escape the tumor-suppressive function of TGF-β or undergo SMAD4-independent EMT [85]. Moreover, CRC patient tissues exhibited higher GDF-15 expression compared with non-cancerous controls, and in the human CRC cell line LoVo, the overexpression of GDF-15 could upregulate the marker genes of mesenchymal cells. Thus, GDF-15 could lead to EMT and promote CRC cell invasion and migration [86]. Based on the systematic analysis of samples from seven CRC patients, it was found that some potential EMT biomarkers were enriched in TGF-β/Snail and TNF-α/nuclear factor-κB (NF-κB) pathways, and the integrated pathway may be the main axis connecting cancer cells with their TME during EMT [87]. In an immunohistochemical study of 48 resected CRC specimens, SMAD4 was positively linked with the expression of Snail-1, Slug and Twist-1, while it was negatively correlated with E-cadherin expression, implying that SMAD4 promotes the process of EMT [88].
There are also other factors that affect EMT by regulating the TGF-β pathway. For example, atypical protein kinase C-ι (aPKC-ι) knockdown inhibits TGF-β1-induced EMT and cell migration in CRC cells [89]. Furthermore, in 5-fluorouracil (5-FU)-resistant CRC cell lines, knockdown of transmembrane protein 45A (TMEM45A) attenuated multidrug-resistance-enhanced EMT by suppressing the TGF-β/SMAD signaling pathway [90]. In studies utilizing cell line experiments and nude mouse models, Numb expression was negatively correlated with TNM stage and lymph node metastasis, and inhibiting Numb expression promoted the EMT process and the invasion of CRC cells induced by TGF-β [91]. It was found that Paraneoplastic antigen Ma family number 5 (PNMA5) accelerated CRC cell proliferation, invasion and migration in nude mice lung metastasis models, and the knockdown of PNMA5 attenuated TGF-β-induced EMT in CRC cells [92]. As reported in cell assays and mouse xenograft tumors, beta human chorionic gonadotropin (hCGβ) changed expression of EMT-associated genes, and these changes could be reversed by TGFBR1 and TGFBR2 inhibitors, indicating that hCGβ induces EMT in a manner that depends on the TGF-β pathway [93].
3.2. TGF-β Signaling in Angiogenesis in mCRC
Angiogenesis in the TME is a pivotal process that promotes tumor development and metastasis [94]. Newly formed blood vessels can provide oxygen and nutrients to tumor cells as well as allow them to enter into blood circulation and metastasize to distant sites [95].
First, the TGF-β pathway can regulate tumor metastasis by affecting vascular endothelial growth factor (VEGF). In the CRC HCT116 cell line, the upregulation of VEGF expression caused by the absence of SMAD4 enhanced vascular density and promoted the development of metastasis [96]. Additionally, SMAD4 overexpression can inhibit CRC growth by inhibiting VEGF-A and VEGF-C expression in the HCT116 cell line and an promote tumor cell apoptosis in HCT116 cells and nude mouse models [97]. There are primarily two histopathological patterns of vascular changes in CRLM: angiogenic desmoplastic and non-angiogenic replacement [98]. Overexpression of Runt-Related Transcription Factor-1 (RUNX1) in cancer cells of the replacement lesions, which is mediated by TGF-β1 and thrombospondin 1 (TSP1), enhances cell motility to achieve vessel co-option [98]. TGF-β expression is increased in the AlCl3-exposed human CRC cell line HT-29, and this particularly promoted endothelial cell angiogenesis via the induction of VEGF secretion [99]. In an orthotopic mouse model of liver metastasis, the inhibition of TGF-β-induced protein ig-h3 (TGFBI) suppressed angiogenesis of CRC cells and inhibited the progression of CRLM [100]. Second, the synergistic effects of TGF-β and other signal cascades can stimulate angiogenesis by accelerating endothelial cell migration and proliferation [77]. TGF-β can interact with other proteins or pathways to foster angiogenesis in mCRC. The downregulation of platelet-derived growth factor-D (PDGF-D), a downstream signal of TGF-β, inhibited the growth, migration and angiogenesis of CRC cells in vitro and in vivo [101]. Thrombospondin-4 (THBS4), an ECM protein, plays an essential role in the TME and augments the effects of TGF-β1 on angiogenesis [102,103].
3.3. TGF-β Signaling in Immunosuppressive Microenvironment in mCRC
Growing evidence has shown that the TME performs a significant role in tumor initiation, progression and metastasis. The TME comprises non-cancerous cells in the tumor, including cancer-associated fibroblasts (CAFs), endothelial cells, pericytes and different types of immune cells (dendritic cells (DCs), tumor-associated macrophages (TAMs), tumor-associated neutrophils (TANs), natural killer (NK) cells, myeloid cells, T cells, B cells, monocytes etc.), as well as non-cellular components, including ECM and soluble products such as collagen, various cytokines, chemokines and other factors that contribute to CRC metastasis [104,105,106]. Direct cell-to-cell contact between cancer cells and secretion of cytokines in the TME caused crosstalk, resulting in CRC progression and ultimately metastasis. It was reported that the activity of TGF-β signaling in TME cells such as T cells, macrophages, endothelial cells and fibroblasts improved the organ colonization efficiency of CRC cells, while treating the mice with the TGFBR1-spesific inhibitor LY2157299 inhibited CRC metastasis formation [42]. Elevated TGF-β expression levels is an important feature in the TME of CRC, and TGF-β signaling can regulate the development of CRC, form the system structure of tumors and inhibit the activity of anti-tumor immune cells, which results in an immunosuppressive microenvironment [28,107,108].
Here we summarize recent research and find that most of studies focused on CAFs and immune cells such as TAMs, TANs, DCs, T cells, myeloid cells and monocytes. Only a few studies on TGF-β signaling-mediated CRC progression and metastasis were related to collagen (discussed in the CAF section). TGF-β-signaling-related CRC metastasis involving CAFs and immune cells will be further discussed in detail in the following sections.
3.3.1. CAFs
CAFs are the most numerous cells in the TME, and they affect CRC metastasis by regulating TGF-β signaling directly or indirectly [108,109]. TGF-β is mainly produced by CAFs in CRC, and increased TGF-β promotes T cell exclusion and inhibits the effector phenotype acquisition of type 1 T helper cells (TH1). It has been reported that inhibition of TGF-β enhances the cytotoxic T cell response to tumor cells, thus suppressing liver metastasis [110]. TGF-β activates CAFs to secrete activin A, a TGF-β family member, which induces colon epithelial cell migration and EMT, resulting in a more metastatic phenotype of CRC [111]. Wang et al. have recently reported that the activation of C-X-C motif chemokine ligand 12 (CXCL12)/CXCR7 axis drove CRC cells to secrete exosomal miR-146a-5p and miR-155-5p, which could be taken up by CAFs, thus enhancing CAF activation via JAK2-STAT3/NF-κB signaling. CAFs could secrete more inflammatory cytokines, including TGF-β, further promoting EMT and CRC metastasis to the lung in vivo [112]. ZNF37A, which is upregulated in CRC, is reported to facilitate tumor cell metastasis to the lung and liver via the activation of Thrombospondin Type-1 Domain-Containing protein 4 (THSD4)/TGF-β signaling, and increased TGF-β secretion contributes to transforming fibroblasts to CAFs in the TME, further promoting CRC metastasis [113]. Integrin αvβ6 secreted by CRC cells induced the expression of TGF-β, thereby converting fibroblasts into CAFs and promoting CRC metastasis through the stromal cell derived factor-1 (SDF-1)/C-X-C motif chemokine receptor type 4 (CXCR4) axis [114]. Treatment of co-cultured CRC and CAF-like cells with vincristine, which is a chemotherapy drug used widely in mCRC clinical treatment, increased the secretion of TGF-βs, induced EMT and promoted the formation of CAFs, thereby enhancing the invasion and metastasis of CRC [115]. Interleukin-11 (IL-11) secreted by TGF-β-stimulated CAFs is a TGF-β target gene, and it activated GP130/signal transducer and activator of transcription 3 (STAT3) signaling in CRC cells and promoted the initiation of CRC cells to metastasis [42]. Endoglin, a TGF-β family coreceptor produced by CAFs, enhanced CRC cell metastasis to the liver in both zebrafish and mouse models [116]. In addition, it has been demonstrated that tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) secreted by SMAD4-deficient CRC cells promotes fibroblasts to produce BMP2, resulting in CRC cell invasion and metastasis [117].
Moreover, TGF-β1 can be secreted by tumor cells in metastasis. Neutral endopeptidase (NEP) co-culturing human colon cancer cell line SW620 (derived from metastatic tumors) with normal colon fibroblasts induced a significant increase in expression of TGF-β1 in SW620 cells, and this effect could be reversed by deletion of NEP [118]. As reported, TGF-β1 promoted the co-migration of colon cancer cells and CAFs, resulting in enhanced liver metastasis and tumor burden [119]. CAF-derived exosomal microRNA (miR)-17-5p caused CRC cells to secrete TGF-β1 into the TME through RUNX3/MYC/TGF-β1 signaling, which triggered CAFs to release more exosomal miR-17-5p to CRC cells, thus establishing a positive feedback loop for CRC metastasis [120]. In CRC, fibroblasts could be converted to CAFs via IL-1β/TGF-β1 signaling, and both TGF-β-activated kinase 1 (TAK1) and TGFBR1 inhibitors suppressed CRC metastasis and CAF accumulation [121]. Two additional studies revealed that CXCR4/TGF-β1 signaling plays an important role in the transformation of mesenchymal stem cells or hepatic stellate cells into CAFs, further promoting CRLM [122,123].
However, CAFs can also suppress CRC progression in some situations. In a genetically modified metastatic CRC mouse model, depletion of alpha smooth muscle actin (αSMA)+ CAFs resulted in an increase of forkhead box protein 3 (Foxp3)+ regulatory T cells (Tregs) and suppression of CD8+ T cells via BMP4/TGF-β1 paracrine signaling, ultimately promoting CRC invasiveness and lymph node metastasis [124]. A recent study showed that gremlin 1 (GREM1) and the immunoglobulin superfamily contain leucine-rich repeat (ISLR), representing two different types of fibroblast subpopulations that exert opposing roles in the signal transduction of BMP. Neutralization of GREM1 or overexpression of ISLR in fibroblasts could reduce CRC hepatic metastasis [125].
Furthermore, decreased expression of hyaluronan and proteoglycan link protein-1 (HAPLN1) regulated collagen deposition in CRC via the TGF-β signaling pathway, and increased collagen resulted in TME changes and CRC cell proliferation, migration and invasion [126].
3.3.2. Immune Cells
TAMs, one of the most common immune cells in the TME, have been reported as key contributors to promote tumor metastasis [127,128]. Liu et al. found severe TAM infiltration in tumor tissues of mCRC patients, and TAM-derived TGF-β could activate HIF1α/TRIB3/β-catenin/Wnt signaling to enhance CRC progression [129]. GDF-15, secreted by macrophages, is a divergent member of the human TGF-β superfamily, and it can increase expression of EMT genes, thereby promoting the invasion and metastasis of CRC via the ERK1/2/c-Fos signaling pathway [130]. Shimizu et al. found that Kupffer cells, known to be resident hepatic macrophages, released TGF-β1 and promoted liver metastasis of CRC through angiotensin II subtype receptor 1a (AT1a) signaling. Moreover, depletion of Kupffer cells reduced metastatic areas [131]. It has been proven that TGF-β1 secretion of CRC cells upregulated macrophage expression of Response Gene to Complement 32 (RGC-32) and thus enhanced macrophage migration and promoted tumor progression [132]. Recently, Chen et al. found that oxaliplatin-based chemotherapy induced TAM recruitment to release TGF-β, which was mediated by CRC-cell-derived CSF1, resulting in programmed cell death-Ligand 1 (PD-L1) upregulation and an immunosuppressive TME. Inhibition of PD-L1 expression in CRC could make cancer cells sensitive to chemotherapy, reduce CRC lung metastasis and increase infiltration of CD8+ T cells. Both CSF1R+ TAM depletion and TGF-β receptor blockade combined with chemotherapy could inhibit tumor growth significantly [133]. Through specific differentiation, macrophages can be polarized into two different phenotypes: activated M1-type and alternatively activated M2-type. M1-type macrophages inhibit tumor growth and progression, whereas M2-type macrophages induce the progression and metastasis of tumors in CRC [127,134]. Ma et al. found that M2-type macrophages were positively correlated with infiltrating Foxp3+ Tregs in CRC, which may promote the development of CRC via the TGF-β/SMAD signaling pathway [135]. Cai et al. reported that M2-type macrophages that secreted TGF-β promoted EMT by activating the SMAD2,3-4/Snail/E-cadherin signaling pathway, resulting in CRC lung metastasis [136]. Zhang et al. revealed that Collagen Triple Helix Repeat Containing 1 (CTHRC1) secreted by CRC cells induced macrophages to the M2-type through activation of TGF-β signaling, further enhancing CRC liver metastasis [137]. Recently, Li et al. developed a thermosensitive hydrogel called Gel/(regorafenib + NG/LY3200882 (LY)), which could sequentially release regorafenib and LY (a selective TGF-β inhibitor) in tumor cells. Using colorectal tumor-bearing mouse models, they found that Gel/(regorafenib + NG/LY) can effectively inhibit tumor growth and liver metastasis, which was achieved by increasing levels of CD8+ T cells, reducing infiltration of TAMs and myeloid-derived suppressor cells and shifting macrophage polarization from M2-type to M1-type in TME [138].
Apart from CAFs and TAMs, other cellular components, such as TANs, myeloid cells, monocytes, DCs and T cells, in the TME can also affect CRC metastasis through TGF-β signaling. TAN infiltration was demonstrated to be positively correlated with the clinical stage of CRC patients [139]. Anti-TGF-β treatment attenuated tumor growth, which was mediated by inhibition of PI3K/AKT signaling pathways in TANs and TGF-β/SMAD signaling pathways in CRC cells [140]. Activation of epithelial NOTCH1 enhanced epithelial TGF-β2 expression and facilitated liver metastasis of CRC through TAN infiltration, which was mediated by TGF-β signaling. Neutrophil depletion led to increased CD8+ T cells in both primary tumors and livers and decreased metastasis. In addition, blocking TGF-β signaling in neutrophils can effectively reduce CRC metastasis [41]. Using mouse xenograft models, Itatani et al. found that a deficiency of SMAD4 in human CRC cells upregulated CCL15 expression, thus recruiting CCR1+ myeloid cells and promoting liver metastasis of CRC [141]. Furthermore, inflammation is an important driver for CRC development and metastasis. CRC cells treated with lipopolysaccharide-stimulated monocyte conditioned medium showed reduced expression of Growth Factor Independence 1 and enhanced EMT and CRC cell metastatic formation, which might have been mediated by TGF-β signaling [142]. Wang et al. suggested that silencing poly (ADP-ribose) glycohydrolase (PARG) in CT26 cells could suppress liver metastasis of colon carcinoma by suppression of poly (ADP-ribose) polymerase (PARP) and NF-κB and that it could reduce secretion of IL-10 and TGF-β, thus promoting the proliferation and differentiation of DCs and T cells, resulting in inhibition of metastasis by changes in immune function [143]. Treg and T helper 17 (Th17)-related genes seem to contribute greatly to CRC development and progression. Miteva et al. investigated the expression of Treg and Th17-related genes in CRC tissues and found that Foxp3, IL-10 and TGF-β1 expression was increased in CRC metastases in contrast to IL17A and NOS2. Treg and Th17-related gene expression in both primary tumor and regional lymph nodes might provide a suitable microenvironment for accelerating CRC metastasis [144]. The mechanism by which other cells in the TME influence T cells via TGF-β signaling directly or indirectly was covered in the previous section [41,110,124,133,135,138,143].
3.4. TGF-β Signaling in Stemness in mCRC
Most tumors, including CRC, contain a small population of cancer stem cells (CSCs) which are regarded as key contributors to tumor generation, progression, recurrence, metastasis and chemotherapy drug resistance [145,146]. According to recent studies, the TGF-β signaling pathway can affect metastasis of CRC by affecting CSCs in CRC or the stemness of CRC cells.
Mesenchymal stem cells co-cultured with CRC cells showed enhanced invasive ability, which was mediated by increased expression of TGF-β1 and decreased expression of p53, resulting in effective inhibition of CRC metastasis [147]. Reports suggested that TGF-β could convert Nur77’s role from cancer inhibition to cancer promotion, which is associated with CRC stemness, metastasis and oxaliplatin resistance [148]. CSCs have specific markers on their surface. CD51, a novel functional marker for colorectal CSCs, could increase the sphere-forming abilities, tumorigenic capacities and migratory potentials of CRC cells, and it may regulate EMT and chemoresistance through TGF-β/SMAD signaling [149]. In a novel mouse model of CRLM, proteomic analysis revealed that the expression of CRC stem cell markers in CRC cells was elevated compared with the non-metastatic model, and the expression of these markers was regulated negatively by the TGF-β/SMAD4 pathways [150].
3.5. Other Mechanisms of TGF-β in mCRC
In addition to the four mechanisms mentioned above, TGF-β can also affect the metastasis of CRC through some additional mechanisms. TGF-β regulates matrix metalloproteinase (MMP) expression in cancer cells, while MMPs produced by either cancer cells or stroma cells activate latent TGF-β, together facilitating progression of CRC [151]. The expression of TGF-β and the podocalyxin-like (PODXL) protein in CRC cells could increase under radiation and then promote ECM deposition, resulting in cell migration and invasiveness [152]. Bioinformatic analysis and functional characterization indicated that TGF-β and Snail promoted CRC migration by preventing degradation of the non-coding RNA LOC113230-related argininosuccinate synthase 1 (ASS1) [153]. Reports indicate that cancer epithelial cells show a robust outward apical pole throughout the process of dissemination, which is referred to as tumor spheres with inverted polarity (TSIPs). TSIPs form and propagate via the collective apical budding of hypermethylated CRCs downstream of TGF-β signaling, which could drive the formation of peritoneal metastases [154]. Moreover, TrkC, which was overexpressed in CRC, could also increase the ability to form tumor spheroids, thus enhancing the metastatic potential of CRC by activation of AKT and suppression of TGF-β signaling [155]. It has been demonstrated that TGF-β inhibits lymph angiogenesis by inhibiting collagen and calcium-binding EGF domain-1 (CCBE1) expression, and CCBE1 has a pro-tumorigenic role in lymphatic metastasis of CRC [156]. TGF-β2 could enhance the metastatic potential of human CRC cell lines via upregulating the expression of catalase and controlling H2O2 output [157].
4. Potential Application of TGF-β Signaling in Diagnosis and Prognosis of mCRC
Specific molecular biomarkers are significant tools for early diagnosis and prognosis of mCRC, and early prognosis is the most successful and effective method to improve the survival rate of CRC patients [158]. Through integrative clustering of the expression profiles of miRNA-correlated genes and methylation-correlated genes, four molecular subtypes (S-I, S-II, S-III and S-IV) were confirmed in CRC patients from The Cancer Genome Atlas (TCGA) [159].
Due to the poor prognosis of patients with mCRC, it is crucial to make a rapid and accurate diagnosis of mCRC based on specific biomarkers as early as possible. Many reports on applications of TGF-β signaling in the diagnosis of mCRC have been published. According to relative mRNA quantification, the expression of TGF-β1 in CRC distant metastases is significantly increased compared with primary tumor tissues [160]. The increased expression of TGF-β2 is a dependable predictor of lymph node metastasis in CRC patients [161]. Compared with healthy controls, serum levels of GDF-15, a member of the TGF-β superfamily, were remarkably upregulated in mCRC patients and had the same sensitivity as the standard tumor marker CEA, indicating that GDF-15 could be an effective biomarker in mCRC patients [162]. The morphogen nodal is dramatically overexpressed in malignant transformation in CRC, and it could be a potential marker for the consensus molecular subtype 4 (CMS4) subtype of CRC [163].
More reports of TGF-β signaling on prognosis have been shown in mCRC patients. The combined activin and TGF-β ligand expression score was utilized to predict shorter OS in a group of 40 CRC tumors, 10 metastasis and 10 control samples [164]. In EO mCRC patients, mutated TGF-β pathways were found to be associated with unfavorable OS by capture-based targeted sequencing [56]. For the TGFΒ1-509C/T single nucleotide polymorphism (SNP), CRC patients with the TT allele have the shortest median survival, which is due to malignant progression in advanced stages [165]. High levels of TGF-β in blood samples are negatively correlated with PFS in mCRC patients before treatment with regorafenib, and these results suggest that the cytokine signature can distinguish whether patients respond to regorafenib treatment or not [166]. The survival analysis demonstrated that MYC and TGF-β pathway alterations were related to a shorter OS in mCRC patients, and this negative prognostic impact was maintained after receiving an anti-EGFR antibody [34]. Using tumor tissues from 230 mCRC patients treated with oxaliplatin combined with 5-FU chemotherapy, Baraniskin et al. reported that SMAD4 expression was decreased in 34% of mCRC samples, and these patients had a shorter PFS and OS compared with patients in which SMAD4 is stably expressed [167]. In an analysis of multiple gene mutation assessments in 123 regorafenib-treated mCRC patients, researchers detected a SMAD4 mutation in one patient who had long response to regorafenib [168]. In addition, SMAD4-mutated patients performed significantly worse in terms of PFS than those without SMAD4 mutations in a study with 76 regorafenib-treated mCRC patients [169]. Whole exome sequencing analysis of 77 mCRC patients revealed that SMAD4 mutations were significantly correlated with poor prognosis [170]. Patients with SMAD4 mutations developed CRLM and had worse OS after hepatic resection [171]. In general, changes in TGF-β signaling pathways in CRC cause cancer cells to become more aggressive and more likely to metastasize; thus, patients harboring mutations in TGF-β signaling components often have a poor prognosis.
Other members associated with the TGF-β family can also be utilized as prognostic biomarkers in mCRC. Studies imply that high tumor expression of activin A (a homodimer of inhibin beta A) is associated with poor prognosis in patients with CRC, and activin A receptor type 2A (ACVR2A) (a membrane receptor in the TGF-β signaling pathway) depletion plays an important role in CRC distant metastasis and may be recommended as a prognostic biomarker in CRC patients [172,173]. As mentioned in the previous section, the entry level of GDF-15 might be a prognostic factor that is strongly relevant to OS in mCRC patients [162]. Overexpression of inhibin subunit beta B (INHBB) (a protein-coding gene that participates in the synthesis of TGF-β family members) was positively associated with CRC invasion and distant metastasis, suggesting that it could be a potential prognostic biomarker for mCRC [174]. Furthermore, knockdown of high inhibin, beta A (INHBA) in vitro can inhibit CRC cell migration and invasion by inhibiting the TGF-β pathway, and INHBA expression is closely related to poor prognosis in CRC patients [175]. Overexpression of lncRNA-activated by TGF-β (lncRNA-ATB) was significantly associated with CRC metastasis, and lncRNA-ATB expression could be a prognosis biomarker of OS in CRC patients [176]. Additionally, TGFBR2 deficiency is positively correlated with upregulation of miR-31-3p [177], which is a predictive biomarker for the efficacy of anti-EGFR treatment that mCRC patients received [178].
5. Targeting TGF-β Signaling Pathway in mCRC
As we have discussed in previous sections, TGF-β signaling plays a significant role in CRC metastasis by promoting EMT, facilitating angiogenesis, contributing to an immunosuppressive TME, regulating stemness of mCRC cells and other mechanisms. These research achievements led us to explore more strategies targeting TGF-β signaling which may have promising application prospects in mCRC therapy. So far, long non-coding RNAs (lncRNAs), miRNAs, kinase inhibitors and natural compounds are common strategies that have been utilized for TGF-β targeting in clinical trials, and new sequencing techniques could facilitate the development of personalized medicine for mCRC patients. These various factors targeting TGF-β signaling in CRC metastasis are summarized in Table 1.
5.1. LncRNAs
LncRNAs are a class of multifunctional noncoding RNAs whose sizes are greater than 200 nucleotides. Many recent studies have shown that lncRNAs play a critical role in regulating progression and metastasis in CRC [179,180]. In CRC patients, outlier expression of the lncRNA MIR31HG was observed and was characterized by elevated EMT, TGF-β and IFN-α/γ gene expression signatures in pre-clinical models [181]. The lncRNA CTBP1-AS2 increased CRC cell invasion and decreased apoptosis by activating the TGF-β/SMAD2/3 pathway and was closely associated with worse survival rate in CRC patients [182]. Additionally, the lncRNAs TP73-AS1 and MIR503HG inhibited the migration and invasion of CRC cells by inactivating TGF-β1 and downregulating TGF-β2, respectively [183,184]. Transwell assays showed that TGF-β2 overexpression increased cell invasion, while overexpression of the lncRNA HOXC-AS3 could reverse the effect of overexpression of TGF-β2 [185]. According to an experiment in CRC cell lines, silencing of the lncRNA ezrin antisense RNA 1 (lncRNA EZR-AS1) accelerated CRC cell apoptosis and inhibited the migration and EMT of CRC cells by blocking TGF-β signaling [186]. Silencing of the lncRNA MIR22HG promoted CRC cell proliferation and tumor metastasis in vitro and in vivo by competitively interacting with SMAD2 [187]. Moreover, LINC00941 enhanced invasive capacity and accelerated lung metastasis by activating EMT by directly binding SMAD4 and preventing SMAD4 protein degradation in mCRC [188].
5.2. MiRNAs
Increasing evidence has shown that miRNAs that regulate TGF-β signals have significant roles in the progression and metastasis of CRC; they act as oncogenes or tumor suppressors to regulate expression of specific targets [189]. Compared with primary CRC, a series of studies on related miRNAs reported epigenetic alternations in CRLM [36]. MiR-425 and miR-576 were significantly upregulated in CRLM based on GSE81581 and GSE44121 datasets, and the two miRNAs were associated with CRC metastasis by co-participating in inhibition of the TGF-β signaling pathway [190]. It has been proven that upregulation of miR-329 suppresses CRC cell invasion by inhibiting TGF-β1, and low expression of miR-329 is correlated with lymph node metastasis in CRC patients [191]. Upregulated expression of plasma miR-211 and 25, which are relevant to the high expression of TGF-β1 in CRC patients, was positively correlated with lymph node metastasis [192]. In HCT116 colon cancer cells in which kallikrein 6 was knocked down, miR-203 was demonstrated to inhibit migration and invasion of CRC cells by inhibiting the EMT through suppression of TGF-β2 [193].
Some miRNAs function through targeting the TGF-β receptors in mCRC. For instance, downregulation of miR-301a was shown to inhibit CRC migration and invasion both in vitro and in vivo by repressing TGFBR2 protein expression in an analysis containing 48 cases of CRC tissues, adjacent non-tumor tissues and five CRC cell lines [194]. TGFBR2 repression by overexpression of the entire miR-371~373 cluster decreased tumor-initiating potential in tumor-initiating cells [195]. Reports indicated that miR-3191 promoted CRC cells migration and invasion by downregulating TGFBR2 [196]. Artificial overexpression of miR-490-3p inhibited cell migration and invasion in CRC cell lines through the suppression of TGFBR1 and MMP2/9 [197]. CircFAM120B overexpression blocked CRC cell migration and reduced the expression of miR-645. In addition, TGFBR2 was a target of miR-645, whose inhibition suppressed CRC cell migration and can be restored by TGFBR2 knockdown [198]. As reported in the LoVo cell experiment and subcutaneous tumor model, the inhibition of miR-424 suppressed migration and invasion of CRC cells as well as arrested CRC cells at the G0/G1 phase by repressing TGFBR3 [199].
There are also many reports on miRNAs that affect CRC metastasis through SMAD proteins in the TGF-β signaling pathway. Functional studies showed that miR-27a inhibited SMAD2 expression at transcriptional and translational levels and that it promoted colon cancer cell apoptosis and attenuated cell migration [200]. Ectopic expression of miR-140 inhibited EMT partially through downregulating SMAD3, and it enhanced invasive capacities of CRC cells in vitro, while overexpression of miR-140 inhibited the metastasis of CRC in vivo [201]. Studies confirm that miR-20a-5p promoted the invasion and metastasis ability of CRC cells and liver metastasis, as well as accelerated the EMT process by reducing SMAD4 expression, which is slightly controversial compared with most other reports [202]. Furthermore, bioinformatic predictions and experimental validation demonstrated that SMAD7 is a direct target of miR-25 in mCRC, and miR-25 inhibition could promote the migratory ability of CRC cells via the suppression of SMAD7 [203]. High miR-4775 expression promoted CRC cell metastasis and EMT via downregulating SMAD7 and thereby activated the TGF-β pathway both in vitro and in vivo [204]. Wang et al. demonstrated that miR-21-mediated inhibition of SMAD7 accelerated TGF-β-dependent EMT in CRC, indicating that loss or inhibition of SMAD7 could promote CRC metastasis [205]. This implies that the overexpression of circTBL1XR1 enhances the proliferation and migration of CRC cells by binding to miR-424, which inhibits SMAD7 [206]. All these examples show that miRNAs can inhibit SMAD7, promote TGF-β-dependent EMT and contribute to CRC metastasis.
5.3. Kinase Inhibitors
Some kinase inhibitors for the TGF-β signaling pathway have been evaluated in various models for mCRC combination treatment to improve the efficacy of therapy. TGFBR1, TGFBR2 and TGFBR3 mutations were found in mCRC patients who responded to regorafenib, suggesting that the TGF-β signaling pathway may play a leading role in the regorafenib response [207]. While using regorafenib, a novel oral multikinase inhibitor, mCRC patients with SMAD4 mutations or activation of the TGF-β pathway showed a worse PFS, which was demonstrated by NGS-based cancer panel tests [169]. Based on a TGF-β-inducible reporter system, Zhang et, al. showed that the TGF-β receptor kinase inhibitor LY2109761 inhibited CRLM by blocking the tumor promoting function of TGF-β in vivo [208]. In a colon cancer liver metastases murine model, mice were treated with adoptive natural killer cells combined with the TGF-β receptor kinase inhibitor LY2157299, and a significant eradication of liver metastases occurred [209]. A study using human CRC cell lines demonstrated that sitagliptin can inhibit CRC cell metastasis by partially blocking TGF-β1-driven EMT [210]. According to the targeted NGS analysis of tumor samples with pre- and post-cetuximab treatment, the copy number of the SMAD4 gene changed, while the TGF-β signaling pathway had various recurrent mutations [211]. The therapeutic potential of these variants requires further clarification.
Correspondingly, dual treatments with the TGF-β galunisertib (LY2157299) inhibitor and AXL inhibitor prominently reduced migration capabilities of human CRC cell lines [212]. Moreover, melatonin, hyperbaric oxygen and combined treatments inhibited CRC metastasis through a variety of mechanisms, including restraining cancer stemness [213]. However, the application of inhibitors should be taken under careful consideration, as it has been reported that epithelial truncation of TGFBR2 leads to fatal inflammatory diseases and invasive CRC in APC mice (a model of intestinal neoplastic disease). Moreover, APC mice with global suppression of TGF-β signaling present with an overall increase in inflammation and tumor formation, suggesting that CRC patients treated with TGF-β inhibitors may have a worse outcome by enhancing inflammatory responses [214].
5.4. Natural Compounds and Chinese Herbal Formulas
Some natural compounds and Chinese herbal formulas can also be utilized as indirect approaches for targeting TGF-β. In vitro results from transwell and scratch wound assays demonstrated that solasodine inhibited CRC cell invasion and migration, which was strengthened by TGF-β1. Solasodine also attenuated TGF-β1-induced EMT in vivo [215]. A traditional Chinese herbal medicine, Hedyotis diffusa Willd, may develop its anti-metastatic activity by restraining TGF-β/SMAD4 pathway-mediated EMT in 5-FU-resistant CRC cells [216]. In addition, baicalin caused cell cycle arrest in the G1 phase and EMT inhibition through inhibiting the TGF-β/SMAD pathway in CRC RKO and HCT116 cell lines [217]. Celastrol significantly inhibited human CRC cells growth, adhesion and metastasis by repressing the TGF-β1/SMAD signaling pathway [218]. The ethanol extract of Scutellaria barbata D. Don (EESB) significantly reduced the migration ability of HCT-8 cells in a dose-dependent manner. Furthermore, EESB decreased expression of MMPs and proteins involved in PI3K/AKT and TGF-β/SMAD signaling [219]. Mechanistically, metformin can block the activation of TGF-β signaling by INHBA, which is an important ligand of TGF-β signaling. It can then downregulate the activity of the PI3K/AKT pathway, leading to cell cycle arrest and inhibition of the proliferation of CRC [220]. In vitro, ursolic acid inhibited the migration and invasion of human CRC HCT116 and HCT-8 cells by interfering with the TGF-β1/ZEB1/miR-200c signaling network [221]. Compared with control mice, Modified Shenlingbaizhu Decoction (MSD) treatment significantly reduced the size of CRC tumors and the serum content of TGF-β1. Similarly, MSD inhibited CRC cell migration and invasion by limiting TGF-β/SMAD signaling [222]. Qingjie Fuzheng granule (QFG), a traditional Chinese medicine, suppressed the growth, wound-healing abilities and migration of HCT-8 and HCT116 cells. Moreover, QFG decreased the expression of lncRNA ANRIL, TGF-β1, p-SMAD2/3, SMAD4 and N-cadherin in CRC cells, suggesting that QFG inhibits the metastasis of CRC through the TGF-β1/SMAD axis [223]. Combined with the TCGA database results and previous network pharmacology, reports indicated that Fuzheng Xiaojijinzhan might play an anti-CRC metastasis role by inhibiting the TGF-β-Snail1 pathway [224].
Since strong genetic heterogeneity exists in CRC patients, development of personalized medicine for CRC patients is of extraordinary significance and value in clinical trials [225]. Based on large-scale data sharing and analytics, CRC is divided into four CMSs with distinguishing features: CMS1 (microsatellite instability immune, 14%), CMS2 (canonical, 37%), CMS3 (metabolic, 13%) and CMS4 (mesenchymal, 23%). Among them, CMS4 has prominent TGF-β activation, stromal invasion, angiogenesis and an immunosuppressive phenotype [226]. In the past decade, more and more efforts have been made to select the appropriate patient subsets for specific treatment of mCRC. Although the development of novel biological agents for therapies such as VEGF and EGFR has further changed the prospects for the treatments of mCRC, not all patients respond similarly to these therapies, so individualized medical treatments are in great need [227]. Designing personalized medicine targeting TGF-β signaling is definitely a good choice for CMS4 subtypes patients of CRC.

Table 1.
Summary of factors targeting TGF-β signaling and acting on CRC metastasis.
Table 1.
Summary of factors targeting TGF-β signaling and acting on CRC metastasis.
| Types | Targets | Involvements in Metastasis | Clinical Application | References | |
|---|---|---|---|---|---|
| LncRNAs | MIR22HG | TGF-β pathway | Interact with SMAD2 and inhibit EMT | Facilitating immunotherapy in CRC | [187] |
| MIR31HG | TGF-β pathway | Promote CRC cell migration and immunosuppression | Biomarker of cellular state | [181] | |
| EZR-AS1 | TGF-β pathway | Promote CRC cell migration, proliferation and EMT | — | [186] | |
| TP73-AS1 | TGF-β1 | Promote CRC cell migration | Prognosis marker in CRC | [183] | |
| MIR503HG | TGF-β2 | Inhibit CRC cell migration | Prognosis marker in CRC | [184] | |
| HOXC-AS3 | TGF-β2 | Reverse the effect of overexpression of TGF-β2 | — | [185] | |
| CTBP1-AS2 | TGF-β/SMAD2/3 pathway | Promote CRC cell migration and inhibit apoptosis | Prognosis marker in CRC | [182] | |
| LINC00941 | TGF-β/SMAD2/3 pathway | Prevent SMAD4 protein degradation and activate EMT | Prognosis marker in CRC | [188] | |
| miRNAs | miR-425 | PTEN-P53/TGF-β | Inhibit cellular immune function | Shortened overall survival | [190] |
| miR-576 | PTEN-P53/TGF-β | Inhibit cellular immune function | Shortened overall survival | [190] | |
| miR-329 | TGF-β1 | Inhibit CRC cell migration | — | [191] | |
| miR-203 | TGF-β2 | Inhibit EMT | — | [193] | |
| miR-490-3p | TGFBR1 | Inhibit CRC cell migration | Associated with poor prognosis of survival | [197] | |
| miR-301a | TGFBR2 | Promote CRC cell migration | — | [194] | |
| miR-371~373 | TGFBR2 | Decrease tumor-initiating potential of CRC cells | — | [195] | |
| miR-3191 | TGFBR2 | Promote CRC cell migration | — | [196] | |
| miR-645 | TGFBR2 | Promote CRC cell migration and glycolysis | — | [198] | |
| miR-424 | TGFBR3/SMAD7 | Promote CRC cell migration and arrest cell cycle/promote proliferation | — | [199,206] | |
| miR-27a | SMAD2 and SGPP1 | Inhibit CRC cell migration and promote apoptosis | Biomarker for monitoring CRC development and progression | [200] | |
| miR-140 | SMAD3 | Inhibit CRC cell migration | — | [201] | |
| miR-20a-5p | SMAD4 | Promote CRC cell migration and EMT | Predicts poor prognosis in CRC patients | [202] | |
| miR-25 | SMAD7 | Inhibit CRC cell migration | — | [203] | |
| miR-4775 | SMAD7 | Promote CRC cell migration and EMT | Predicts poor survival | [204] | |
| miR-21 | SMAD7 | Accelerate TGF-β dependent EMT | — | [205] | |
| circRNAs | circFAM120B | miR-645 | Inhibit CRC cell migration and glycolysis | — | [198] |
| circTBL1XR1 | miR-424 | Promote CRC cell migration | — | [206] | |
| Kinase Inhibitors | Sitagliptin | TGF-β1 | Inhibit EMT and impair cell cycle | Prevents colon cancer and lung metastasis in animal models and humans | [210] |
| LY2157299 | TGF-β receptor | Mitigate TGF-β driven impairment of NK cell cytotoxicity | Currently in clinical trials for various malignancies | [209] | |
| LY2109761 | TGF-β receptor | Downregulated the phosphorylation of SMAD2 | Applied mostly in preclinical animal experiments | [208] | |
| regorafenib | TGF-β/SMAD4 pathway | — | While using regorafenib, patients with SMAD4 mutation or activation of TGF-β pathway showed a worse PFS | [169] | |
| Natural com-pounds and Chinese herbal formulas | solasodine | TGF-β1 | Inhibit CRC cell stemness and EMT | — | [215] |
| MSD | TGF-β1 | Inhibit CRC cell migration | Reduced the size of CRC tumors in mouse model | [222] | |
| Celastrol | TGF-β1/SMAD pathway | Inhibit CRC cell migration | — | [218] | |
| baicalin | TGF-β/SMAD pathway | Inhibit EMT, stemness and cell cycle | — | [217] | |
| QFG | TGF-β/SMAD pathway | Inhibit CRC cells growth and migration | — | [223] | |
| Hedyotis diffusa Willd | TGF-β/SMAD4 pathway | Inhibit CRC cell migration and EMT | — | [216] | |
| ursolic acid | TGF-β1/ZEB1/ miR-200c | Inhibit CRC cell migration | — | [221] | |
| metformin | TGF-β ligand and PI3K/AKT pathway | Arrest cell cycle and inhibit cell proliferation | — | [220] | |
| EESB | PI3K/AKT and TGF-β/SMAD signaling | Inhibit CRC cell migration and decrease the expression of MMPs | — | [219] | |
| Fuzheng Xiaojijinzhan | TGF-β-Snail1 | Anti-CRC metastasis role | — | [224] |
6. Conclusions and Future Perspective
Metastatic CRC is an intractable disease due to its poor prognosis, high mortality and limited optimal therapies in clinical situations worldwide, even in developed countries. Several key areas in mCRC research include: early identification of metastasis, recognition of specific prognostic and predictive biomarkers, discovery of new molecular targets, development of new drugs and clinical operations. TGF-β represents a conserved signaling pathway that is widely involved in various physiological and pathological processes. In this review, we summarized the changes in the TGF-β signaling pathway in mCRC patients, its functional mechanisms and its possible applications in mCRC diagnosis, prognosis and potential targeted therapies in clinical trials. We explained in detail that TGF-β signaling functions to promote EMT, facilitate angiogenesis, suppress anti-tumor activity of the immune cells in the microenvironment and contribute to stemness of CRC cells in mCRC (as shown in Figure 1). Following these working mechanisms of TGF-β signaling in mCRC, molecular targeting therapies aimed at the different key factors upstream and downstream of TGF-β signaling could be accordingly developed to improve the efficacy and safety of treatments, especially in MCS4 subtypes of mCRC.
Various remaining problems regarding TGF-β signaling in CRC metastasis still need to be clarified. Initially, TGF-β was not considered as a good target for tumor treatments because of its dual roles in tumor early development and late-stage metastasis. However, recent years witnessed an increasing number of new therapies that target TGF-β signaling in CRC metastasis. These include suppressing TGF-β or downstream components of the signaling, blocking crosstalk between TGF-β signaling pathways and other signal pathways and redirecting TGF-β signaling from pro-tumor to anti-tumor functions in CRC metastasis [228]. Additionally, because mCRC is a complicated disease, we should attach importance to not only TGF-β signals from inside but also outside of CRC cells, namely from the stroma cells and microenvironment of mCRC. Hopefully, in-depth and systematic studies in this research field will help us understand more in the future.
Due to the heterogeneity of CRC cells and individual differences among mCRC patients, it is impossible to find an optimal treatment strategy that fits everyone. Thanks to improved knowledge of the molecular mechanisms underlying CRC metastasis, promising advances help us modify traditional treatments. Recent reports demonstrated that targeting TGF-β could be combined with other signal inhibitors such as combinatorial synergy, reverse therapy resistance or sensitize radiotherapy to achieve a sustained therapy response in CRC patients [28,229]. Moreover, since TGF-β signaling is an immunosuppressive regulator in the TME of mCRC, there is great potentials in combining TGF-β targeting with immunotherapy agents to enhance the efficacy and benefit for patients. In order to move forward, applications of NGS and genetic profiling in clinical trials will help us characterize the molecular subtypes of TGF-β signaling in mCRC patients, and appropriate personalized medicine specifically targeting TGF-β can be definitely and smoothly translated into mCRC treatments.
Author Contributions
Writing—original draft preparation, X.L., Y.W. and T.T.; writing—review and editing, X.L., Y.W. and T.T.; supervision and funding acquisition, T.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Beijing (grant number 5222019).
Acknowledgments
We thank Juan Liu and Tongtong Cui for reviewing and editing the manuscript. We also thank Angela Papierski for proofreading the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 5-FU | 5-fluorouracil |
| αSMA | alpha smooth muscle actin |
| ACSL3 | acyl-CoA synthetases 3 |
| ACVR2A | activin A receptor type 2A |
| AKT | protein kinase B |
| APC | adenomatous polyposis coli |
| aPKC-ι | atypical protein kinase C-ι |
| ASE | allele-specific expression |
| ASS1 | argininosuccinate synthase 1 |
| AT1a | angiotensin II subtype receptor 1a |
| BMP | bone morphogenetic protein |
| CAFs | cancer-associated fibroblasts |
| CCBE1 | collagen and calcium-binding EGF domain-1 |
| CEA | Carcinoembryonic antigen |
| CMS | consensus molecular subtype |
| Co-SMAD | common-mediator SMAD |
| CRC | colorectal cancer |
| CRLM | colorectal liver metastases |
| CSCs | cancer stem cells |
| Cten | C-terminal tensin-like |
| CTHRC1 | collagen triple helix repeat containing 1 |
| CXCL12 | C-X-C motif chemokine ligand 12 |
| CXCR4 | C-X-C motif chemokine receptor type 4 |
| DCs | dendritic cells |
| ECM | extracellular matrix |
| EESB | ethanol extract of Scutellaria barbata D. Don |
| EGFR | epidermal growth factor receptor |
| EMT | epithelial-mesenchymal transition |
| EO | early onset |
| FFPE | formalin-fixed paraffin-embedded |
| Foxp3 | forkhead box protein 3 |
| GDF | growth and differentiation factor |
| GPC1 | Glypicans 1 |
| GREM1 | gremlin 1 |
| HAPLN1 | hyaluronan and proteoglycan link protein-1 |
| hCGβ | beta human chorionic gonadotropin |
| HLTF | helicase-like transcription factor |
| IL-11 | interleukin-11 |
| INHBA | inhibin, beta A |
| INHBB | inhibin subunit beta B |
| ISLR | immunoglobulin superfamily containing leucine-rich repeat |
| I-SMAD | inhibitory SMAD |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| lncRNA-ATB | lncRNA-activated by TGF-β |
| lncRNA EZR-AS1 | lncRNA ezrin antisense RNA 1 |
| lncRNAs | long non-coding RNAs |
| MAPK | mitogen-activated protein kinase |
| mCRC | metastatic colorectal cancer |
| MiR | microRMA |
| MMPs | matrix metalloproteinases |
| MSD | Modified Shenlingbaizhu Decoction |
| MSI | microsatellite instability |
| NEP | neutral endopeptidase |
| NF-κB | nuclear factor-κB |
| NGS | next-generation sequencing |
| OS | overall survival |
| PARG | poly (ADP-ribose) glycohydrolase |
| PDGF-D | platelet-derived growth factor-D |
| PD-L1 | programmed cell death-Ligand 1 |
| PFS | progression-free survival |
| P4HA3 | prolyl 4-hydroxylase subunit alpha 3 |
| PI3K | phosphoinositide 3-kinase |
| PNMA5 | Paraneoplastic antigen Ma family number 5 |
| PODXL | podocalyxin-like |
| QFG | Qingjie Fuzheng granule |
| RGC-32 | response gene to complement 32 |
| ROCK | Rho-associated kinase |
| R-SMAD | receptor-regulated SMAD |
| RUNX | runt related transcription factor |
| SDF-1 | stromal cell derived factor-1 |
| SNP | single nucleotide polymorphism |
| STAT3 | signal transducer and activator of transcription 3 |
| TAK1 | TGF-β-activated kinase 1 |
| TAMs | tumor-associated macrophages |
| TANs | tumor-associated neutrophils |
| TCGA | The Cancer Genome Atlas |
| TGF-β | transforming growth factor-β |
| TGFBI | TGF-β-induced protein ig-h3 |
| TH1 | type 1 T-helper cell |
| Th17 | T helper 17 |
| THBS4 | thrombospondin-4 |
| THSD4 | thrombospondin type-1 domain-containing protein 4 |
| TME | tumor microenvironment |
| TMEM45A | transmembrane protein 45A |
| TRAIL | tumor necrosis factor-related apoptosis-inducing ligand |
| Tregs | regulatory T cells |
| TRIM25 | tripartite motif containing 25 |
| Trp53 | transformation related protein 53 |
| TSIPs | tumor spheres with inverted polarity |
| TSP1 | thrombospondin 1 |
| VEGF | vascular endothelial growth factor |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Dong, X.; Li, H.; Cao, M.; Sun, D.; He, S.; Yang, F.; Yan, X.; Zhang, S.; Li, N.; et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 2022, 135, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Hutter, C.M.; Chang-Claude, J.; Slattery, M.L.; Pflugeisen, B.M.; Lin, Y.; Duggan, D.; Nan, H.; Lemire, M.; Rangrej, J.; Figueiredo, J.C.; et al. Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer Res. 2012, 72, 2036–2044. [Google Scholar] [CrossRef] [PubMed]
- Tauriello, D.V.; Calon, A.; Lonardo, E.; Batlle, E. Determinants of metastatic competency in colorectal cancer. Mol. Oncol. 2017, 11, 97–119. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef]
- Oki, E.; Ando, K.; Nakanishi, R.; Sugiyama, M.; Nakashima, Y.; Kubo, N.; Kudou, K.; Saeki, H.; Nozoe, T.; Emi, Y.; et al. Recent advances in treatment for colorectal liver metastasis. Ann. Gastroenterol. Surg. 2018, 2, 167–175. [Google Scholar] [CrossRef]
- Zarour, L.R.; Anand, S.; Billingsley, K.G.; Bisson, W.H.; Cercek, A.; Clarke, M.F.; Coussens, L.M.; Gast, C.E.; Geltzeiler, C.B.; Hansen, L.; et al. Colorectal Cancer Liver Metastasis: Evolving Paradigms and Future Directions. Cell Mol. Gastroenterol. Hepatol. 2017, 3, 163–173. [Google Scholar] [CrossRef]
- Modest, D.P.; Pant, S.; Sartore-Bianchi, A. Treatment sequencing in metastatic colorectal cancer. Eur. J. Cancer 2019, 109, 70–83. [Google Scholar] [CrossRef]
- Jones, R.P.; Jackson, R.; Dunne, D.F.; Malik, H.Z.; Fenwick, S.W.; Poston, G.J.; Ghaneh, P. Systematic review and meta-analysis of follow-up after hepatectomy for colorectal liver metastases. Br. J. Surg. 2012, 99, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Akhurst, R.J.; Balmain, A. TGF-beta signaling in tumor suppression and cancer progression. Nat. Genet. 2001, 29, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Marinelli Busilacchi, E.; Costantini, A.; Mancini, G.; Tossetta, G.; Olivieri, J.; Poloni, A.; Viola, N.; Butini, L.; Campanati, A.; Goteri, G.; et al. Nilotinib Treatment of Patients Affected by Chronic Graft-versus-Host Disease Reduces Collagen Production and Skin Fibrosis by Downmodulating the TGF-beta and p-SMAD Pathway. Biol. Blood Marrow Transpl. 2020, 26, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Akhurst, R.J.; Hata, A. Targeting the TGFbeta signalling pathway in disease. Nat. Rev. Drug Discov. 2012, 11, 790–811. [Google Scholar] [CrossRef]
- Goteri, G.; Altobelli, E.; Tossetta, G.; Zizzi, A.; Avellini, C.; Licini, C.; Lorenzi, T.; Castellucci, M.; Ciavattini, A.; Marzioni, D. High temperature requirement A1, transforming growth factor beta1, phosphoSmad2 and Ki67 in eutopic and ectopic endometrium of women with endometriosis. Eur. J. Histochem. 2015, 59, 2570. [Google Scholar] [CrossRef]
- Pardali, E.; Ten Dijke, P. TGFbeta signaling and cardiovascular diseases. Int. J. Biol. Sci. 2012, 8, 195–213. [Google Scholar] [CrossRef]
- Morikawa, M.; Derynck, R.; Miyazono, K. TGF-beta and the TGF-beta Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb. Perspect. Biol. 2016, 8, a021873. [Google Scholar] [CrossRef]
- Tauriello, D.V.; Sancho, E.; Batlle, E. Overcoming TGFβ-mediated immune evasion in cancer. Nat. Rev. Cancer 2022, 22, 25–44. [Google Scholar] [CrossRef]
- Attisano, L.; Wrana, J.L. Signal transduction by the TGF-beta superfamily. Science 2002, 296, 1646–1647. [Google Scholar] [CrossRef]
- Gough, N.R.; Xiang, X.; Mishra, L. TGF-beta Signaling in Liver, Pancreas, and Gastrointestinal Diseases and Cancer. Gastroenterology 2021, 161, 434–452.e15. [Google Scholar] [CrossRef]
- Haque, S.; Morris, J.C. Transforming growth factor-beta: A therapeutic target for cancer. Hum. Vacc. Immunother. 2017, 13, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Jung, B.; Staudacher, J.J.; Beauchamp, D. Transforming Growth Factor beta Superfamily Signaling in Development of Colorectal Cancer. Gastroenterology 2017, 152, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Robertson, I.B.; Rifkin, D.B. Regulation of the Bioavailability of TGF-beta and TGF-beta-Related Proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a021907. [Google Scholar] [CrossRef] [PubMed]
- Justin, P.A.; Daniel, B.R.; John, S.M. The integrin alphaVbeta6 binds and activates latent TGFbeta3. FEBS Lett. 2002, 511, 65–68. [Google Scholar]
- Nagaraj, N.S.; Datta, P.K. Targeting the transforming growth factor-beta signaling pathway in human cancer. Expert Opin. Investig. Drugs 2010, 19, 77–91. [Google Scholar] [CrossRef]
- Gu, S.; Feng, X.H. TGF-beta signaling in cancer. Acta Biochim. Biophys. Sin. 2018, 50, 941–949. [Google Scholar] [CrossRef]
- Heldin, C.H.; Moustakas, A. Signaling Receptors for TGF-beta Family Members. Cold Spring Harb. Perspect. Biol. 2016, 8, a022053. [Google Scholar] [CrossRef]
- Colak, S.; Ten Dijke, P. Targeting TGF-beta Signaling in Cancer. Trends Cancer 2017, 3, 56–71. [Google Scholar] [CrossRef]
- Nakao, A.; Afrakhte, M.; Morén, A.; Nakayama, T.; Christian, J.L.; Heuchel, R.; Itoh, S.; Kawabata, M.; Heldin, N.E.; Heldin, C.H.; et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature 1997, 389, 631–635. [Google Scholar] [CrossRef]
- Derynck, R.; Muthusamy, B.P.; Saeteurn, K.Y. Signaling pathway cooperation in TGF-beta-induced epithelial-mesenchymal transition. Curr. Opin. Cell Biol. 2014, 31, 56–66. [Google Scholar] [CrossRef]
- Croce, C.M. Oncogenes and cancer. N. Engl. J. Med. 2008, 358, 502–511. [Google Scholar] [CrossRef]
- Fang, W.; Radovich, M.; Zheng, Y.; Fu, C.Y.; Zhao, P.; Mao, C.; Zheng, Y.; Zheng, S. Druggable alterations detected by Ion Torrent in metastatic colorectal cancer patients. Oncol. Lett. 2014, 7, 1761–1766. [Google Scholar] [CrossRef] [PubMed]
- Bajenova, O.; Gorbunova, A.; Evsyukov, I.; Rayko, M.; Gapon, S.; Bozhokina, E.; Shishkin, A.; O’Brien, S.J. The Genome-Wide Analysis of Carcinoembryonic Antigen Signaling by Colorectal Cancer Cells Using RNA Sequencing. PLoS ONE 2016, 11, e0161256. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Lin, P.C.; Su, W.C.; Chan, R.H.; Chen, P.C.; Lin, B.W.; Shen, M.R.; Chen, S.H.; Yeh, Y.M. Association between Altered Oncogenic Signaling Pathways and Overall Survival of Patients with Metastatic Colorectal Cancer. Diagnostics 2021, 11, 2308. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, Y.; Kopetz, S.; Kwong, L.; Xiao, L.; Morris, J.S.; Tran Cao, H.S.; Tzeng, C.D.; Chun, Y.S.; Lee, J.E.; Vauthey, J.N. Genomic Sequencing and Insight into Clinical Heterogeneity and Prognostic Pathway Genes in Patients with Metastatic Colorectal Cancer. J. Am. Coll. Surg. 2021, 233, 272–284.e13. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, H.; Sun, L.; Shen, S.; Zhou, Q.; Yuan, Y.; Xing, C. Epigenetic Alternations of MicroRNAs and DNA Methylation Contribute to Liver Metastasis of Colorectal Cancer. Dig. Dis. Sci. 2019, 64, 1523–1534. [Google Scholar] [CrossRef]
- Kulkarni, A.B.; Huh, C.G.; Becker, D.; Geiser, A.; Lyght, M.; Flanders, K.C.; Roberts, A.B.; Sporn, M.B.; Ward, J.M.; Karlsson, S. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc. Natl. Acad. Sci. USA 1993, 90, 770–774. [Google Scholar] [CrossRef]
- Sanford, L.P.; Ormsby, I.; Gittenberger-de Groot, A.C.; Sariola, H.; Friedman, R.; Boivin, G.P.; Cardell, E.L.; Doetschman, T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development 1997, 124, 2659–2670. [Google Scholar] [CrossRef]
- Kaartinen, V.; Voncken, J.W.; Shuler, C.; Warburton, D.; Bu, D.; Heisterkamp, N.; Groffen, J. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat. Genet. 1995, 11, 415–421. [Google Scholar] [CrossRef]
- Martin, C.J.; Datta, A.; Littlefield, C.; Kalra, A.; Chapron, C.; Wawersik, S.; Dagbay, K.B.; Brueckner, C.T.; Nikiforov, A.; Danehy, F.T.J.; et al. Selective inhibition of TGFβ1 activation overcomes primary resistance to checkpoint blockade therapy by altering tumor immune landscape. Sci. Transl. Med. 2020, 12, eaay8456. [Google Scholar] [CrossRef]
- Jackstadt, R.; van Hooff, S.R.; Leach, J.D.; Cortes-Lavaud, X.; Lohuis, J.O.; Ridgway, R.A.; Wouters, V.M.; Roper, J.; Kendall, T.J.; Roxburgh, C.S.; et al. Epithelial NOTCH Signaling Rewires the Tumor Microenvironment of Colorectal Cancer to Drive Poor-Prognosis Subtypes and Metastasis. Cancer Cell 2019, 36, 319–336.e7. [Google Scholar] [CrossRef] [PubMed]
- Calon, A.; Espinet, E.; Palomo-Ponce, S.; Tauriello, D.V.; Iglesias, M.; Cespedes, M.V.; Sevillano, M.; Nadal, C.; Jung, P.; Zhang, X.H.; et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 2012, 22, 571–584. [Google Scholar] [CrossRef]
- Zhou, R.; Huang, Y.; Cheng, B.; Wang, Y.; Xiong, B. TGFBR1*6A is a potential modifier of migration and invasion in colorectal cancer cells. Oncol. Lett. 2018, 15, 3971–3976. [Google Scholar] [CrossRef] [PubMed]
- Valle, L.; Serena-Acedo, T.; Liyanarachchi, S.; Hampel, H.; Comeras, I.; Li, Z.; Zeng, Q.; Zhang, H.T.; Pennison, M.J.; Sadim, M.; et al. Germline allele-specific expression of TGFBR1 confers an increased risk of colorectal cancer. Science 2008, 321, 1361–1365. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Peralta, A.M.; Nejda, N.; Oliart, S.; Medina, V.; Azcoita, M.M.; Gonzalez-Aguilera, J.J. Significance of mutations in TGFBR2 and BAX in neoplastic progression and patient outcome in sporadic colorectal tumors with high-frequency microsatellite instability. Cancer Genet. Cytogenet. 2005, 157, 18–24. [Google Scholar] [CrossRef]
- Yashiro, M.; Hirakawa, K.; Boland, C.R. Mutations in TGFbeta-RII and BAX mediate tumor progression in the later stages of colorectal cancer with microsatellite instability. BMC Cancer 2010, 10, 303. [Google Scholar] [CrossRef]
- Fricke, F.; Lee, J.; Michalak, M.; Warnken, U.; Hausser, I.; Suarez-Carmona, M.; Halama, N.; Schnolzer, M.; Kopitz, J.; Gebert, J. TGFBR2-dependent alterations of exosomal cargo and functions in DNA mismatch repair-deficient HCT116 colorectal cancer cells. Cell Commun. Signal. 2017, 15, 14. [Google Scholar] [CrossRef]
- Lee, J.; Ballikaya, S.; Schonig, K.; Ball, C.R.; Glimm, H.; Kopitz, J.; Gebert, J. Transforming growth factor beta receptor 2 (TGFBR2) changes sialylation in the microsatellite unstable (MSI) Colorectal cancer cell line HCT116. PLoS ONE 2013, 8, e57074. [Google Scholar] [CrossRef]
- Stanilov, N.; Grigorova, A.; Velikova, T.; Stanilova, S.A. Genetic variation of TGF-BetaR2 as a protective genotype for the development of colorectal cancer in men. World J. Gastrointest. Oncol. 2021, 13, 1766–1780. [Google Scholar] [CrossRef]
- Xie, W.; Rimm, D.L.; Lin, Y.; Shih, W.J.; Reiss, M. Loss of Smad signaling in human colorectal cancer is associated with advanced disease and poor prognosis. Cancer J. 2003, 9, 302–312. [Google Scholar] [CrossRef]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Fleming, N.I.; Jorissen, R.N.; Mouradov, D.; Christie, M.; Sakthianandeswaren, A.; Palmieri, M.; Day, F.; Li, S.; Tsui, C.; Lipton, L.; et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013, 73, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.; Cooke, L.; Mahadevan, D. Next generation sequencing identifies ‘interactome’ signatures in relapsed and refractory metastatic colorectal cancer. J. Gastrointest. Oncol. 2017, 8, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Park, H.Y.; Hwang, D.Y.; Han, H.S. High Concordance of Genomic Profiles between Primary and Metastatic Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 5561. [Google Scholar] [CrossRef] [PubMed]
- Oyanagi, H.; Shimada, Y.; Nagahashi, M.; Ichikawa, H.; Tajima, Y.; Abe, K.; Nakano, M.; Kameyama, H.; Takii, Y.; Kawasaki, T.; et al. SMAD4 alteration associates with invasive-front pathological markers and poor prognosis in colorectal cancer. Histopathology 2019, 74, 873–882. [Google Scholar] [CrossRef]
- Xu, T.; Zhang, Y.; Zhang, J.; Qi, C.; Liu, D.; Wang, Z.; Li, Y.; Ji, C.; Li, J.; Lin, X.; et al. Germline Profiling and Molecular Characterization of Early Onset Metastatic Colorectal Cancer. Front. Oncol. 2020, 10, 568911. [Google Scholar] [CrossRef]
- Lopez-Gomez, M.; Moreno-Rubio, J.; Suarez-Garcia, I.; Cejas, P.; Madero, R.; Casado, E.; Jimenez, A.M.; Sereno, M.; Gomez-Raposo, C.; Zambrana, F.; et al. Gene expression differences in primary colorectal tumors and matched liver metastases: Chemotherapy related or tumoral heterogeneity? Clin. Transl. Oncol. 2015, 17, 322–329. [Google Scholar] [CrossRef]
- Ganesh, K.; Shah, R.H.; Vakiani, E.; Nash, G.M.; Skottowe, H.P.; Yaeger, R.; Cercek, A.; Lincoln, A.; Tran, C.; Segal, N.H.; et al. Clinical and genetic determinants of ovarian metastases from colorectal cancer. Cancer 2017, 123, 1134–1143. [Google Scholar] [CrossRef]
- Abd El-Fattah, A.A.; Sadik, N.A.H.; Shaker, O.G.; Mohamed Kamal, A. Single Nucleotide Polymorphism in SMAD7 and CHI3L1 and Colorectal Cancer Risk. Mediat. Inflamm. 2018, 2018, 9853192. [Google Scholar] [CrossRef]
- Rosic, J.; Dragicevic, S.; Miladinov, M.; Despotovic, J.; Bogdanovic, A.; Krivokapic, Z.; Nikolic, A. SMAD7 and SMAD4 expression in colorectal cancer progression and therapy response. Exp. Mol. Pathol. 2021, 123, 104714. [Google Scholar] [CrossRef]
- Halder, S.K.; Rachakonda, G.; Deane, N.G.; Datta, P.K. Smad7 induces hepatic metastasis in colorectal cancer. Br. J. Cancer 2008, 99, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Sakai, E.; Nakayama, M.; Oshima, H.; Kouyama, Y.; Niida, A.; Fujii, S.; Ochiai, A.; Nakayama, K.I.; Mimori, K.; Suzuki, Y.; et al. Combined Mutation of Apc, Kras, and Tgfbr2 Effectively Drives Metastasis of Intestinal Cancer. Cancer Res. 2018, 78, 1334–1346. [Google Scholar] [CrossRef] [PubMed]
- Miguchi, M.; Hinoi, T.; Shimomura, M.; Adachi, T.; Saito, Y.; Niitsu, H.; Kochi, M.; Sada, H.; Sotomaru, Y.; Ikenoue, T.; et al. Gasdermin C Is Upregulated by Inactivation of Transforming Growth Factor beta Receptor Type II in the Presence of Mutated Apc, Promoting Colorectal Cancer Proliferation. PLoS ONE 2016, 11, e0166422. [Google Scholar] [CrossRef] [PubMed]
- Boutin, A.T.; Liao, W.T.; Wang, M.; Hwang, S.S.; Karpinets, T.V.; Cheung, H.; Chu, G.C.; Jiang, S.; Hu, J.; Chang, K.; et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev. 2017, 31, 370–382. [Google Scholar] [CrossRef]
- Fumagalli, A.; Drost, J.; Suijkerbuijk, S.J.; van Boxtel, R.; de Ligt, J.; Offerhaus, G.J.; Begthel, H.; Beerling, E.; Tan, E.H.; Sansom, O.J.; et al. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc. Natl. Acad. Sci. USA 2017, 114, E2357–E2364. [Google Scholar] [CrossRef]
- Wang, H.W.; Yan, X.L.; Wang, L.J.; Zhang, M.H.; Yang, C.H.; Wei, L.; Jin, K.M.; Bao, Q.; Li, J.; Wang, K.; et al. Characterization of genomic alterations in Chinese colorectal cancer patients with liver metastases. J. Transl. Med. 2021, 19, 313. [Google Scholar] [CrossRef]
- Mehrvarz Sarshekeh, A.; Advani, S.; Overman, M.J.; Manyam, G.; Kee, B.K.; Fogelman, D.R.; Dasari, A.; Raghav, K.; Vilar, E.; Manuel, S.; et al. Association of SMAD4 mutation with patient demographics, tumor characteristics, and clinical outcomes in colorectal cancer. PLoS ONE 2017, 12, e0173345. [Google Scholar]
- Sun, N.; Xue, Y.; Dai, T.; Li, X.; Zheng, N. Tripartite motif containing 25 promotes proliferation and invasion of colorectal cancer cells through TGF-beta signaling. Biosci. Rep. 2017, 37, BSR20170805. [Google Scholar] [CrossRef]
- Yao, Y.; Zhou, Z.; Li, L.; Li, J.; Huang, L.; Li, J.; Qi, C.; Zheng, L.; Wang, L.; Zhang, Q.Q. Activation of Slit2/Robo1 Signaling Promotes Tumor Metastasis in Colorectal Carcinoma through Activation of the TGF-beta/Smads Pathway. Cells 2019, 8, 635. [Google Scholar] [CrossRef]
- Liu, L.; Liu, H.; Zhou, Y.; He, J.; Liu, Q.; Wang, J.; Zeng, M.; Yuan, D.; Tan, F.; Zhou, Y.; et al. HLTF suppresses the migration and invasion of colorectal cancer cells via TGFbeta/SMAD signaling in vitro. Int. J. Oncol. 2018, 53, 2780–2788. [Google Scholar]
- Zhou, H.; Zou, J.; Shao, C.; Zhou, A.; Yu, J.; Chen, S.; Xu, C. Prolyl 4-hydroxylase subunit alpha 3 facilitates human colon cancer growth and metastasis through the TGF-beta/Smad signaling pathway. Pathol. Res. Pract. 2022, 230, 153749. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ai, M.; Nie, M.; Zhao, L.; Deng, G.; Hu, S.; Han, Y.; Zeng, W.; Wang, Y.; Yang, M.; et al. EHF promotes colorectal carcinoma progression by activating TGF-beta1 transcription and canonical TGF-beta signaling. Cancer Sci. 2020, 111, 2310–2324. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Chen, S.; Shi, W.; Su, X.; Wu, H.; Liu, M. GPC1 promotes the growth and migration of colorectal cancer cells through regulating the TGF-beta1/SMAD2 signaling pathway. PLoS ONE 2022, 17, e0269094. [Google Scholar]
- Liu, F.; Shi, Z.; Bao, W.; Zheng, J.; Chen, K.; Lin, Z.; Song, H.N.; Luo, X.; Dong, Q.; Jiang, L.; et al. ZIC2 promotes colorectal cancer growth and metastasis through the TGF-beta signaling pathway. Exp. Cell Res. 2022, 415, 113118. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.; Datta, P.K. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers 2017, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Dedhar, S.; Kalluri, R.; Thompson, E.W. The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J. Cell Biol. 2006, 172, 973–981. [Google Scholar] [CrossRef]
- Seoane, J.; Gomis, R.R. TGF-beta Family Signaling in Tumor Suppression and Cancer Progression. Cold Spring Harb. Perspect. Biol. 2017, 9, a022277. [Google Scholar] [CrossRef]
- Tossetta, G.; Paolinelli, F.; Avellini, C.; Salvolini, E.; Ciarmela, P.; Lorenzi, T.; Emanuelli, M.; Toti, P.; Giuliante, R.; Gesuita, R.; et al. IL-1beta and TGF-beta weaken the placental barrier through destruction of tight junctions: An in vivo and in vitro study. Placenta 2014, 35, 509–516. [Google Scholar] [CrossRef]
- Shiou, S.R.; Singh, A.B.; Moorthy, K.; Datta, P.K.; Washington, M.K.; Beauchamp, R.D.; Dhawan, P. Smad4 regulates claudin-1 expression in a transforming growth factor-beta-independent manner in colon cancer cells. Cancer Res. 2007, 67, 1571–1579. [Google Scholar] [CrossRef]
- Di Ruocco, F.; Basso, V.; Rivoire, M.; Mehlen, P.; Ambati, J.; De Falco, S.; Tarallo, V. Alu RNA accumulation induces epithelial-to-mesenchymal transition by modulating miR-566 and is associated with cancer progression. Oncogene 2018, 37, 627–637. [Google Scholar] [CrossRef]
- Asiri, A.; Raposo, T.P.; Alfahed, A.; Ilyas, M. TGFbeta1-induced cell motility but not cell proliferation is mediated through Cten in colorectal cancer. Int. J. Exp. Pathol. 2018, 99, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Cheng, C.; Tan, Y.; Jiang, N.; Liao, C.; Liao, W.; Cao, Y.; Luo, X. Acyl-CoA synthetase long-chain 3-mediated fatty acid oxidation is required for TGFbeta1-induced epithelial-mesenchymal transition and metastasis of colorectal carcinoma. Int. J. Biol. Sci. 2022, 18, 2484–2496. [Google Scholar] [CrossRef] [PubMed]
- Siraj, A.K.; Pratheeshkumar, P.; Divya, S.P.; Parvathareddy, S.K.; Bu, R.; Masoodi, T.; Kong, Y.; Thangavel, S.; Al-Sanea, N.; Ashari, L.H.; et al. TGFbeta-induced SMAD4-dependent Apoptosis Proceeded by EMT in CRC. Mol. Cancer Ther. 2019, 18, 1312–1322. [Google Scholar] [CrossRef] [PubMed]
- Corbet, C.; Bastien, E.; Santiago de Jesus, J.P.; Dierge, E.; Martherus, R.; Vander Linden, C.; Doix, B.; Degavre, C.; Guilbaud, C.; Petit, L.; et al. TGFbeta2-induced formation of lipid droplets supports acidosis-driven EMT and the metastatic spreading of cancer cells. Nat. Commun. 2020, 11, 454. [Google Scholar] [CrossRef] [PubMed]
- Frey, P.; Devisme, A.; Rose, K.; Schrempp, M.; Freihen, V.; Andrieux, G.; Boerries, M.; Hecht, A. SMAD4 mutations do not preclude epithelial-mesenchymal transition in colorectal cancer. Oncogene 2022, 41, 824–837. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Zhang, M.; Zhang, Z.; Jiang, L.; Li, L. GDF15 promotes epithelial-to-mesenchymal transition in colorectal [corrected]. Artif. Cell. Nanomed. Biotechnol. 2018, 46 (Suppl. 2), 652–658. [Google Scholar] [CrossRef]
- Li, H.; Zhong, A.; Li, S.; Meng, X.; Wang, X.; Xu, F.; Lai, M. The integrated pathway of TGFbeta/Snail with TNFalpha/NFkappaB may facilitate the tumor-stroma interaction in the EMT process and colorectal cancer prognosis. Sci. Rep. 2017, 7, 4915. [Google Scholar] [CrossRef]
- Ioannou, M.; Kouvaras, E.; Papamichali, R.; Samara, M.; Chiotoglou, I.; Koukoulis, G. Smad4 and epithelial-mesenchymal transition proteins in colorectal carcinoma: An immunohistochemical study. J. Mol. Histol. 2018, 49, 235–244. [Google Scholar] [CrossRef]
- Du, G.S.; Qiu, Y.; Wang, W.S.; Peng, K.; Zhang, Z.C.; Li, X.S.; Xiao, W.D.; Yang, H. Knockdown on aPKC-iota inhibits epithelial-mesenchymal transition, migration and invasion of colorectal cancer cells through Rac1-JNK pathway. Exp. Mol. Pathol. 2019, 107, 57–67. [Google Scholar] [CrossRef]
- Zhu, M.; Jiang, B.; Yan, D.; Wang, X.; Ge, H.; Sun, Y. Knockdown of TMEM45A overcomes multidrug resistance and epithelial-mesenchymal transition in human colorectal cancer cells through inhibition of TGF-beta signalling pathway. Clin. Exp. Pharmacol. Physiol. 2020, 47, 503–516. [Google Scholar] [CrossRef]
- Yang, Y.; Li, L.; He, H.; Shi, M.; He, L.; Liang, S.; Qi, J.; Chen, W. Numb inhibits migration and promotes proliferation of colon cancer cells via RhoA/ROCK signaling pathway repression. Exp. Cell Res. 2022, 411, 113004. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhang, X.; Meng, F.; Zeng, F.; Liu, W.; He, X. PNMA5 accelerated cellular proliferation, invasion and migration in colorectal cancer. Am. J. Transl. Res. 2022, 14, 2231–2243. [Google Scholar] [PubMed]
- Kawamata, F.; Nishihara, H.; Homma, S.; Kato, Y.; Tsuda, M.; Konishi, Y.; Wang, L.; Kohsaka, S.; Liu, C.; Yoshida, T.; et al. Chorionic Gonadotropin-beta Modulates Epithelial-Mesenchymal Transition in Colorectal Carcinoma Metastasis. Am. J. Pathol. 2018, 188, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J. Recent Advances in the Development of TGF-β Signaling Inhibitors for Anticancer Therapy. J. Cancer Prev. 2020, 25, 213–222. [Google Scholar] [CrossRef]
- Akhurst, R.J.; Derynck, R. TGF-beta signaling in cancer--a double-edged sword. Trends Cell Biol. 2001, 11, S44–S51. [Google Scholar]
- Papageorgis, P.; Cheng, K.; Ozturk, S.; Gong, Y.; Lambert, A.W.; Abdolmaleky, H.M.; Zhou, J.R.; Thiagalingam, S. Smad4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res. 2011, 71, 998–1008. [Google Scholar] [CrossRef]
- Li, X.; Li, X.; Lv, X.; Xiao, J.; Liu, B.; Zhang, Y. Smad4 Inhibits VEGF-A and VEGF-C Expressions via Enhancing Smad3 Phosphorylation in Colon Cancer. Anat. Rec. 2017, 300, 1560–1569. [Google Scholar] [CrossRef]
- Rada, M.; Kapelanski-Lamoureux, A.; Petrillo, S.; Tabaries, S.; Siegel, P.; Reynolds, A.R.; Lazaris, A.; Metrakos, P. Runt related transcription factor-1 plays a central role in vessel co-option of colorectal cancer liver metastases. Commun. Biol. 2021, 4, 950. [Google Scholar] [CrossRef]
- Jeong, C.H.; Kwon, H.C.; Cheng, W.N.; Kim, D.H.; Choi, Y.; Han, S.G. Aluminum exposure promotes the metastatic proclivity of human colorectal cancer cells through matrix metalloproteinases and the TGF-beta/Smad signaling pathway. Food Chem. Toxicol. 2020, 141, 111402. [Google Scholar] [CrossRef]
- Chiavarina, B.; Costanza, B.; Ronca, R.; Blomme, A.; Rezzola, S.; Chiodelli, P.; Giguelay, A.; Belthier, G.; Doumont, G.; Van Simaeys, G.; et al. Metastatic colorectal cancer cells maintain the TGFbeta program and use TGFBI to fuel angiogenesis. Theranostics 2021, 11, 1626–1640. [Google Scholar] [CrossRef]
- Chen, J.; Yuan, W.; Wu, L.; Tang, Q.; Xia, Q.; Ji, J.; Liu, Z.; Ma, Z.; Zhou, Z.; Cheng, Y.; et al. PDGF-D promotes cell growth, aggressiveness, angiogenesis and EMT transformation of colorectal cancer by activation of Notch1/Twist1 pathway. Oncotarget 2017, 8, 9961–9973. [Google Scholar] [CrossRef] [PubMed]
- Muppala, S.; Frolova, E.; Xiao, R.; Krukovets, I.; Yoon, S.; Hoppe, G.; Vasanji, A.; Plow, E.; Stenina-Adognravi, O. Proangiogenic Properties of Thrombospondin-4. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1975–1986. [Google Scholar] [CrossRef] [PubMed]
- Muppala, S.; Xiao, R.; Krukovets, I.; Verbovetsky, D.; Yendamuri, R.; Habib, N.; Raman, P.; Plow, E.; Stenina-Adognravi, O. Thrombospondin-4 mediates TGF-beta-induced angiogenesis. Oncogene 2017, 36, 5189–5198. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Turley, S.J.; Akhurst, R.J. TGFβ biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 2021, 18, 9–34. [Google Scholar] [CrossRef]
- Tommelein, J.; Verset, L.; Boterberg, T.; Demetter, P.; Bracke, M.; De Wever, O. Cancer-associated fibroblasts connect metastasis-promoting communication in colorectal cancer. Front. Oncol. 2015, 5, 63. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef]
- Calon, A.; Lonardo, E.; Berenguer-Llergo, A.; Espinet, E.; Hernando-Momblona, X.; Iglesias, M.; Sevillano, M.; Palomo-Ponce, S.; Tauriello, D.V.; Byrom, D.; et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015, 47, 320–329. [Google Scholar] [CrossRef]
- Itatani, Y.; Kawada, K.; Sakai, Y. Transforming Growth Factor-beta Signaling Pathway in Colorectal Cancer and Its Tumor Microenvironment. Int. J. Mol. Sci. 2019, 20, 5822. [Google Scholar] [CrossRef]
- Koliaraki, V.; Pallangyo, C.K.; Greten, F.R.; Kollias, G. Mesenchymal Cells in Colon Cancer. Gastroenterology 2017, 152, 964–979. [Google Scholar] [CrossRef]
- Tauriello, D.V.F.; Palomo-Ponce, S.; Stork, D.; Berenguer-Llergo, A.; Badia-Ramentol, J.; Iglesias, M.; Sevillano, M.; Ibiza, S.; Canellas, A.; Hernando-Momblona, X.; et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018, 554, 538–543. [Google Scholar] [CrossRef]
- Bauer, J.; Emon, M.A.B.; Staudacher, J.J.; Thomas, A.L.; Zessner-Spitzenberg, J.; Mancinelli, G.; Krett, N.; Saif, M.T.; Jung, B. Increased stiffness of the tumor microenvironment in colon cancer stimulates cancer associated fibroblast-mediated prometastatic activin A signaling. Sci. Rep. 2020, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, X.; Song, Y.; Si, M.; Sun, Y.; Liu, X.; Cui, S.; Qu, X.; Yu, X. Exosomal miR-146a-5p and miR-155-5p promote CXCL12/CXCR7-induced metastasis of colorectal cancer by crosstalk with cancer-associated fibroblasts. Cell Death Dis. 2022, 13, 380. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Huang, Z.; Chen, H.-N.; Qin, S.; Chen, Y.; Jiang, J.; Zhang, Z.; Luo, M.; Ye, Q.; Xie, N.; et al. ZNF37A promotes tumor metastasis through transcriptional control of THSD4/TGF-β axis in colorectal cancer. Oncogene 2021, 40, 3394–3407. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Zou, X.; Xia, W.; Gao, H.; Li, Z.; Liu, N.; Xu, Z.; Gao, C.; He, Z.; Niu, W.; et al. Integrin αvβ6 plays a bi-directional regulation role between colon cancer cells and cancer-associated fibroblasts. Biosci. Rep. 2018, 38, BSR20180243. [Google Scholar] [CrossRef]
- Wawro, M.E.; Sobierajska, K.; Ciszewski, W.M.; Niewiarowska, J. Nonsteroidal Anti-Inflammatory Drugs Prevent Vincristine-Dependent Cancer-Associated Fibroblasts Formation. Int. J. Mol. Sci. 2019, 20, 1941. [Google Scholar] [CrossRef]
- Paauwe, M.; Schoonderwoerd, M.J.A.; Helderman, R.F.C.P.; Harryvan, T.J.; Groenewoud, A.; van Pelt, G.W.; Bor, R.; Hemmer, D.M.; Versteeg, H.H.; Snaar-Jagalska, B.E.; et al. Endoglin Expression on Cancer-Associated Fibroblasts Regulates Invasion and Stimulates Colorectal Cancer Metastasis. Clin. Cancer Res. 2018, 24, 6331–6344. [Google Scholar] [CrossRef]
- Ouahoud, S.; Voorneveld, P.W.; van der Burg, L.R.A.; de Jonge-Muller, E.S.M.; Schoonderwoerd, M.J.A.; Paauwe, M.; de Vos, T.; de Wit, S.; van Pelt, G.W.; Mesker, W.E.; et al. Bidirectional tumor/stroma crosstalk promotes metastasis in mesenchymal colorectal cancer. Oncogene 2020, 39, 2453–2466. [Google Scholar] [CrossRef]
- Mizerska-Kowalska, M.; Sawa-Wejksza, K.; Slawinska-Brych, A.; Kandefer-Szerszen, M.; Zdzisinska, B. Neutral endopeptidase depletion decreases colon cancer cell proliferation and TGF-beta1 synthesis in indirect co-cultures with normal colon fibroblasts. Clin. Transl. Oncol. 2021, 23, 1405–1414. [Google Scholar] [CrossRef]
- Gonzalez-Zubeldia, I.; Dotor, J.; Redrado, M.; Bleau, A.-M.; Manrique, I.; de Aberasturi, A.L.; Villalba, M.; Calvo, A. Co-migration of colon cancer cells and CAFs induced by TGFβ1 enhances liver metastasis. Cell Tissue Res. 2015, 359, 829–839. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S.; Lai, Q.; Fang, Y.; Wu, C.; Liu, Y.; Li, Q.; Wang, X.; Gu, C.; Chen, J.; et al. Cancer-associated fibroblasts-derived exosomal miR-17-5p promotes colorectal cancer aggressive phenotype by initiating a RUNX3/MYC/TGF-β1 positive feedback loop. Cancer Lett. 2020, 491, 22–35. [Google Scholar] [CrossRef]
- Guillén Díaz-Maroto, N.; Sanz-Pamplona, R.; Berdiel-Acer, M.; Cimas, F.J.; García, E.; Gonçalves-Ribeiro, S.; Albert, N.; Garcia-Vicién, G.; Capella, G.; Moreno, V.; et al. Noncanonical TGFβ Pathway Relieves the Blockade of IL1β/TGFβ-Mediated Crosstalk between Tumor and Stroma: TGFBR1 and TAK1 Inhibition in Colorectal Cancer. Clin. Cancer Res. 2019, 25, 4466–4479. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.-X.; Gong, W.-Z.; Zhou, K.; Xiao, Z.-G.; Hou, F.-T.; Huang, T.; Zhang, L.; Dong, H.-Y.; Zhang, W.-L.; Liu, Y.; et al. CXCR4/TGF-β1 mediated hepatic stellate cells differentiation into carcinoma-associated fibroblasts and promoted liver metastasis of colon cancer. Cancer Biol. Ther. 2020, 21, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.-X.; Xiao, Z.-G.; Huang, T.; Fang, Z.-X.; Liu, Y.; Huang, Z.-C. CXCR4/TGF-β1 mediated self-differentiation of human mesenchymal stem cells to carcinoma-associated fibroblasts and promoted colorectal carcinoma development. Cancer Biol. Ther. 2020, 21, 248–257. [Google Scholar] [CrossRef] [PubMed]
- McAndrews, K.M.; Vazquez-Arreguin, K.; Kwak, C.; Sugimoto, H.; Zheng, X.; Li, B.; Kirtley, M.L.; LeBleu, V.S.; Kalluri, R. alphaSMA(+) fibroblasts suppress Lgr5(+) cancer stem cells and restrain colorectal cancer progression. Oncogene 2021, 40, 4440–4452. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Gieniec, K.A.; Wright, J.A.; Wang, T.; Asai, N.; Mizutani, Y.; Lida, T.; Ando, R.; Suzuki, N.; Lannagan, T.R.M.; et al. The Balance of Stromal BMP Signaling Mediated by GREM1 and ISLR Drives Colorectal Carcinogenesis. Gastroenterology 2021, 160, 1224–1239.e30. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, X.; Marshall, J.E.; Gong, M.; Zhao, Y.; Dua, K.; Hansbro, P.M.; Xu, J.; Liu, G. Loss of Hyaluronan and Proteoglycan Link Protein-1 Induces Tumorigenesis in Colorectal Cancer. Front. Oncol. 2021, 11, 754240. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tian, T.; Zhang, J. Tumor-Associated Macrophages (TAMs) in Colorectal Cancer (CRC): From Mechanism to Therapy and Prognosis. Int. J. Mol. Sci. 2021, 22, 8470. [Google Scholar] [CrossRef]
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat Rev. Drug Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, W.; Wang, J.; Si, T.; Xing, W. Tumor-associated macrophage-derived transforming growth factor-β promotes colorectal cancer progression through HIF1-TRIB3 signaling. Cancer Sci. 2021, 112, 4198–4207. [Google Scholar] [CrossRef]
- Ding, Y.; Hao, K.; Li, Z.; Ma, R.; Zhou, Y.; Zhou, Z.; Wei, M.; Liao, Y.; Dai, Y.; Yang, Y.; et al. c-Fos separation from Lamin A/C by GDF15 promotes colon cancer invasion and metastasis in inflammatory microenvironment. J. Cell. Physiol. 2020, 235, 4407–4421. [Google Scholar] [CrossRef]
- Shimizu, Y.; Amano, H.; Ito, Y.; Betto, T.; Yamane, S.; Inoue, T.; Nishizawa, N.; Matsui, Y.; Kamata, M.; Nakamura, M.; et al. Angiotensin II subtype 1a receptor signaling in resident hepatic macrophages induces liver metastasis formation. Cancer Sci. 2017, 108, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wang, B.; Zhang, Z.; Zhang, W.; Liu, Y. Response gene to complement 32 expression in macrophages augments paracrine stimulation-mediated colon cancer progression. Cell Death Dis. 2019, 10, 776. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.W.; Hung, W.Z.; Chiang, S.F.; Chen, W.T.; Ke, T.W.; Liang, J.A.; Huang, C.Y.; Yang, P.C.; Huang, K.C.; Chao, K.S.C. Dual inhibition of TGFbeta signaling and CSF1 CSF1R reprograms tumor-infiltrating macrophages and improves response to chemotherapy via suppressing PD-L1. Cancer Lett. 2022, 543, 215795. [Google Scholar] [CrossRef]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.-A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Gao, Y.; Chen, Y.; Liu, J.; Yang, C.; Bao, C.; Wang, Y.; Feng, Y.; Song, X.; Qiao, S. M2-Type Macrophages Induce Tregs Generation by Activating the TGF-beta/Smad Signalling Pathway to Promote Colorectal Cancer Development. OncoTargets Ther. 2021, 14, 5391–5402. [Google Scholar] [CrossRef]
- Cai, J.; Xia, L.; Li, J.; Ni, S.; Song, H.; Wu, X. Tumor-Associated Macrophages Derived TGF-β–Induced Epithelial to Mesenchymal Transition in Colorectal Cancer Cells through Smad2,3-4/Snail Signaling Pathway. Cancer Res. Treat. 2019, 51, 252–266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-L.; Hu, L.-P.; Yang, Q.; Qin, W.-T.; Wang, X.; Xu, C.-J.; Tian, G.-A.; Yang, X.-M.; Yao, L.-L.; Zhu, L.; et al. CTHRC1 promotes liver metastasis by reshaping infiltrated macrophages through physical interactions with TGF-β receptors in colorectal cancer. Oncogene 2021, 40, 3959–3973. [Google Scholar] [CrossRef]
- Li, Z.; Xu, W.; Yang, J.; Wang, J.; Wang, J.; Zhu, G.; Li, D.; Ding, J.; Sun, T. A Tumor Microenvironments-Adapted Polypeptide Hydrogel/Nanogel Composite Boosts Antitumor Molecularly Targeted Inhibition and Immunoactivation. Adv. Mater. 2022, 34, e2200449. [Google Scholar] [CrossRef]
- Rao, H.-L.; Chen, J.-W.; Li, M.; Xiao, Y.-B.; Fu, J.; Zeng, Y.-X.; Cai, M.-Y.; Xie, D. Increased intratumoral neutrophil in colorectal carcinomas correlates closely with malignant phenotype and predicts patients’ adverse prognosis. PLoS ONE 2012, 7, e30806. [Google Scholar] [CrossRef]
- Qin, F.; Liu, X.; Chen, J.; Huang, S.; Wei, W.; Zou, Y.; Liu, X.; Deng, K.; Mo, S.; Chen, J.; et al. Anti-TGF-β attenuates tumor growth via polarization of tumor associated neutrophils towards an anti-tumor phenotype in colorectal cancer. J. Cancer 2020, 11, 2580–2592. [Google Scholar] [CrossRef]
- Itatani, Y.; Kawada, K.; Fujishita, T.; Kakizaki, F.; Hirai, H.; Matsumoto, T.; Iwamoto, M.; Inamoto, S.; Hatano, E.; Hasegawa, S.; et al. Loss of SMAD4 from colorectal cancer cells promotes CCL15 expression to recruit CCR1+ myeloid cells and facilitate liver metastasis. Gastroenterology 2013, 145, 1064–1075.e11. [Google Scholar] [CrossRef] [PubMed]
- Xing, W.; Xiao, Y.; Lu, X.; Zhu, H.; He, X.; Huang, W.; Lopez, E.S.; Wong, J.; Ju, H.; Tian, L.; et al. GFI1 downregulation promotes inflammation-linked metastasis of colorectal cancer. Cell Death Differ. 2017, 24, 929–943. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.Q.; Tang, Y.; Li, Q.S.; Xiao, M.; Li, M.; Sheng, Y.T.; Yang, Y.; Wang, Y.L. PARG regulates the proliferation and differentiation of DCs and T cells via PARP/NFkappaB in tumour metastases of colon carcinoma. Oncol. Rep. 2019, 41, 2657–2666. [Google Scholar]
- Miteva, L.D.; Stanilov, N.S.; Cirovski Gcapital Em, C.; Stanilova, S.A. Upregulation of Treg-Related Genes in Addition with IL6 Showed the Significant Role for the Distant Metastasis in Colorectal Cancer. Cancer Microenviron. 2017, 10, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Zeuner, A.; Todaro, M.; Stassi, G.; De Maria, R. Colorectal cancer stem cells: From the crypt to the clinic. Cell Stem Cell 2014, 15, 692–705. [Google Scholar] [CrossRef]
- Oh, I.R.; Raymundo, B.; Kim, M.; Kim, C.W. Mesenchymal stem cells co-cultured with colorectal cancer cells showed increased invasive and proliferative abilities due to its altered p53/TGF-beta1 levels. Biosci. Biotechnol. Biochem. 2020, 84, 256–267. [Google Scholar] [CrossRef]
- Niu, B.; Liu, J.; Lv, B.; Lin, J.; Li, X.; Wu, C.; Jiang, X.; Zeng, Z.; Zhang, X.K.; Zhou, H. Interplay between transforming growth factor-beta and Nur77 in dual regulations of inhibitor of differentiation 1 for colonic tumorigenesis. Nat. Commun. 2021, 12, 2809. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, B.; Wu, H.; Cai, J.; Sui, X.; Wang, Y.; Li, H.; Qiu, Y.; Wang, T.; Chen, Z.; et al. CD51 correlates with the TGF-beta pathway and is a functional marker for colorectal cancer stem cells. Oncogene 2017, 36, 1351–1363. [Google Scholar] [CrossRef]
- Fujishita, T.; Kojima, Y.; Kajino-Sakamoto, R.; Mishiro-Sato, E.; Shimizu, Y.; Hosoda, W.; Yamaguchi, R.; Taketo, M.M.; Aoki, M. The cAMP/PKA/CREB and TGF-β/SMAD4 pathways regulate stemness and metastatic potential in colorectal cancer cells. Cancer Res. 2022, 82, 4179–4190. [Google Scholar] [CrossRef]
- Krstic, J.; Santibanez, J.F. Transforming growth factor-beta and matrix metalloproteinases: Functional interactions in tumor stroma-infiltrating myeloid cells. Sci. World J. 2014, 2014, 521754. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kong, J.S.; Lee, S.S.; Kim, A. Radiation-Induced Overexpression of TGFbeta and PODXL Contributes to Colorectal Cancer Cell Radioresistance through Enhanced Motility. Cells 2021, 10, 2087. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Yang, Y.; Li, M.; Chu, Y.; Song, H.; Zhang, J.; Zhang, D.; Zhang, Q.; Xu, Y.; Wang, J.; et al. Snail enhances arginine synthesis by inhibiting ubiquitination-mediated degradation of ASS1. EMBO Rep. 2021, 22, e51780. [Google Scholar] [CrossRef] [PubMed]
- Zajac, O.; Raingeaud, J.; Libanje, F.; Lefebvre, C.; Sabino, D.; Martins, I.; Roy, P.; Benatar, C.; Canet-Jourdan, C.; Azorin, P.; et al. Tumour spheres with inverted polarity drive the formation of peritoneal metastases in patients with hypermethylated colorectal carcinomas. Nat. Cell Biol. 2018, 20, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Suh, K.W.; Hong, S.; Jin, W. TrkC promotes colorectal cancer growth and metastasis. Oncotarget 2017, 8, 41319–41333. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Chen, W.; Cui, X.; Huang, Z.; Wen, D.; Yang, Y.; Yu, W.; Cui, L.; Liu, C.Y. CCBE1 promotes tumor lymphangiogenesis and is negatively regulated by TGFbeta signaling in colorectal cancer. Theranostics 2020, 10, 2327–2341. [Google Scholar] [CrossRef] [PubMed]
- Haidar, M.; Metheni, M.; Batteux, F.; Langsley, G. TGF-beta2, catalase activity, H2O2 output and metastatic potential of diverse types of tumour. Free Radic. Biol. Med. 2019, 134, 282–287. [Google Scholar] [CrossRef]
- Gutierrez, A.; Demond, H.; Brebi, P.; Ili, C.G. Novel Methylation Biomarkers for Colorectal Cancer Prognosis. Biomolecules 2021, 11, 1722. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Wang, D.; Feng, M.; Wu, X. Epigenetically regulated gene expression profiles reveal four molecular subtypes with prognostic and therapeutic implications in colorectal cancer. Brief. Bioinform. 2021, 22, bbaa309. [Google Scholar] [CrossRef]
- Stanilova, S.; Stanilov, N.; Julianov, A.; Manolova, I.; Miteva, L. Transforming growth factor-beta1 gene promoter -509C/T polymorphism in association with expression affects colorectal cancer development and depends on gender. PLoS ONE 2018, 13, e0201775. [Google Scholar] [CrossRef]
- Tu, Y.; Han, J.; Dong, Q.; Chai, R.; Li, N.; Lu, Q.; Xiao, Z.; Guo, Y.; Wan, Z.; Xu, Q. TGF-beta2 is a Prognostic Biomarker Correlated with Immune Cell Infiltration in Colorectal Cancer: A STROBE-compliant article. Medicine 2020, 99, e23024. [Google Scholar] [CrossRef] [PubMed]
- Vocka, M.; Langer, D.; Fryba, V.; Petrtyl, J.; Hanus, T.; Kalousova, M.; Zima, T.; Petruzelka, L. Growth/differentiation factor 15 (GDF-15) as new potential serum marker in patients with metastatic colorectal cancer. Cancer Biomark. 2018, 21, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, S.; Cao, H.; Li, X.; Rong, Y.; Liu, G.; Du, H.; Shen, H. Increasing Embryonic Morphogen Nodal Expression Suggests Malignant Transformation in Colorectal Lesions and as a Potential Marker for CMS4 Subtype of Colorectal Cancer. Pathol. Oncol. Res. 2021, 27, 587029. [Google Scholar] [CrossRef]
- Staudacher, J.J.; Bauer, J.; Jana, A.; Tian, J.; Carroll, T.; Mancinelli, G.; Ozden, O.; Krett, N.; Guzman, G.; Kerr, D.; et al. Activin signaling is an essential component of the TGF-beta induced pro-metastatic phenotype in colorectal cancer. Sci. Rep. 2017, 7, 5569. [Google Scholar] [CrossRef] [PubMed]
- Gulubova, M.; Aleksandrova, E.; Vlaykova, T. Promoter polymorphisms in TGFB1 and IL10 genes influence tumor dendritic cells infiltration, development and prognosis of colorectal cancer. J. Genet. Med. 2018, 20, e3005. [Google Scholar] [CrossRef] [PubMed]
- Ricci, V.; Granetto, C.; Falletta, A.; Paccagnella, M.; Abbona, A.; Fea, E.; Fabozzi, T.; Lo Nigro, C.; Merlano, M.C. Circulating cytokines and outcome in metastatic colorectal cancer patients treated with regorafenib. World J. Gastrointest. Oncol. 2020, 12, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Baraniskin, A.; Munding, J.; Schulmann, K.; Meier, D.; Porschen, R.; Arkenau, H.T.; Graeven, U.; Schmiegel, W.; Tannapfel, A.; Reinacher-Schick, A. Prognostic value of reduced SMAD4 expression in patients with metastatic colorectal cancer under oxaliplatin-containing chemotherapy: A translational study of the AIO colorectal study group. Clin. Colorectal Cancer 2011, 10, 24–29. [Google Scholar] [CrossRef]
- Martinelli, E.; Sforza, V.; Cardone, C.; Capasso, A.; Nappi, A.; Martini, G.; Napolitano, S.; Rachiglio, A.M.; Normanno, N.; Cappabianca, S.; et al. Clinical outcome and molecular characterisation of chemorefractory metastatic colorectal cancer patients with long-term efficacy of regorafenib treatment. ESMO Open 2017, 2, e000177. [Google Scholar] [CrossRef]
- Lee, M.S.; Cho, H.J.; Hong, J.Y.; Lee, J.; Park, S.H.; Park, J.O.; Park, Y.S.; Lim, H.Y.; Kang, W.K.; Cho, Y.B.; et al. Clinical and molecular distinctions in patients with refractory colon cancer who benefit from regorafenib treatment. Ther. Adv. Med. Oncol. 2020, 12, 1758835920965842. [Google Scholar] [CrossRef]
- D’Agay, M.G.; Galland, L.; Tharin, Z.; Truntzer, C.; Ghiringhelli, F. Utility of exome sequencing in routine care for metastatic colorectal cancer. Mol. Clin. Oncol. 2021, 15, 229. [Google Scholar] [CrossRef]
- Mizuno, T.; Cloyd, J.M.; Vicente, D.; Omichi, K.; Chun, Y.S.; Kopetz, S.E.; Maru, D.; Conrad, C.; Tzeng, C.D.; Wei, S.H.; et al. SMAD4 gene mutation predicts poor prognosis in patients undergoing resection for colorectal liver metastases. Eur. J. Surg. Oncol. 2018, 44, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, C.; Hu, D.; Li, J.; Yu, H.; Lin, X.; Chen, Y.; Zhuang, Y.; Li, Q.; Zheng, X.; Yang, C. Downregulation of Activin A Receptor Type 2A Is Associated with Metastatic Potential and Poor Prognosis of Colon Cancer. J. Cancer 2018, 9, 3626–3633. [Google Scholar] [CrossRef] [PubMed]
- Daitoku, N.; Miyamoto, Y.; Hiyoshi, Y.; Tokunaga, R.; Sakamoto, Y.; Sawayama, H.; Ishimoto, T.; Baba, Y.; Yoshida, N.; Baba, H. Activin A promotes cell proliferation, invasion and migration and predicts poor prognosis in patients with colorectal cancer. Oncol. Rep. 2022, 47, 107. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Xie, A.; Cao, Q.; Li, X.; Chen, J. INHBB Is a Novel Prognostic Biomarker Associated with Cancer-Promoting Pathways in Colorectal Cancer. Biomed. Res. Int. 2020, 2020, 6909672. [Google Scholar] [CrossRef]
- He, Z.; Liang, J.; Wang, B. Inhibin, beta A regulates the transforming growth factor-beta pathway to promote malignant biological behaviour in colorectal cancer. Cell Biochem. Funct. 2020, 39, 258–266. [Google Scholar] [CrossRef]
- Liu, X.; Wang, C. Long non-coding RNA ATB is associated with metastases and promotes cell invasion in colorectal cancer via sponging miR-141-3p. Exp. Ther. Med. 2020, 20, 261. [Google Scholar] [CrossRef]
- Fricke, F.; Mussack, V.; Buschmann, D.; Hausser, I.; Pfaffl, M.W.; Kopitz, J.; Gebert, J. TGFBR2dependent alterations of microRNA profiles in extracellular vesicles and parental colorectal cancer cells. Int. J. Oncol. 2019, 55, 925–937. [Google Scholar]
- Nosho, K.; Igarashi, H.; Nojima, M.; Ito, M.; Maruyama, R.; Yoshii, S.; Naito, T.; Sukawa, Y.; Mikami, M.; Sumioka, W.; et al. Association of microRNA-31 with BRAF mutation, colorectal cancer survival and serrated pathway. Carcinogenesis 2014, 35, 776–783. [Google Scholar] [CrossRef]
- Schwarzmueller, L.; Bril, O.; Vermeulen, L.; Leveille, N. Emerging Role and Therapeutic Potential of lncRNAs in Colorectal Cancer. Cancers 2020, 12, 3843. [Google Scholar] [CrossRef]
- Wang, D.Z.; Chen, G.Y.; Li, Y.F.; Zhang, N.W. Comprehensive analysis of long non-coding RNA and mRNA expression profile in rectal cancer. Chin. Med. J. 2020, 133, 1312–1321. [Google Scholar] [CrossRef]
- Eide, P.W.; Eilertsen, I.A.; Sveen, A.; Lothe, R.A. Long noncoding RNA MIR31HG is a bona fide prognostic marker with colorectal cancer cell-intrinsic properties. Int. J. Cancer 2019, 144, 2843–2853. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yue, W.; Li, M.; Jiang, Z.; Hou, Z.; Liu, W.; Ma, N.; Gan, W.; Li, Y.; Zhou, T.; et al. Downregulating Long Non-coding RNAs CTBP1-AS2 Inhibits Colorectal Cancer Development by Modulating the miR-93-5p/TGF-beta/SMAD2/3 Pathway. Front. Oncol. 2021, 11, 626620. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Jin, Y.; Li, Y. LncRNA TP73-AS1 Activates TGF-beta1 to Promote the Migration and Invasion of Colorectal Cancer Cell. Cancer Manag. Res. 2019, 11, 10523–10529. [Google Scholar] [CrossRef] [PubMed]
- Chuo, D.; Liu, F.; Chen, Y.; Yin, M. LncRNA MIR503HG is downregulated in Han Chinese with colorectal cancer and inhibits cell migration and invasion mediated by TGF-beta2. Gene 2019, 713, 143960. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.T.; Chen, H.P.; Yu, S.Y.; Zhao, S.P. LncRNA HOXC-AS3 overexpression inhibits TGF-beta2-induced colorectal cancer cell migration and invasion by sponging miR-1269. Hum. Exp. Toxicol. 2022, 41, 9603271221093630. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, N.; Wang, F.; Zhang, S.; Ding, J. Silencing of lncRNA EZR-AS1 inhibits proliferation, invasion, and migration of colorectal cancer cells through blocking transforming growth factor beta signaling. Biosci. Rep. 2019, 39, BSR20191199. [Google Scholar] [CrossRef]
- Xu, J.; Shao, T.; Song, M.; Xie, Y.; Zhou, J.; Yin, J.; Ding, N.; Zou, H.; Li, Y.; Zhang, J. MIR22HG acts as a tumor suppressor via TGFbeta/SMAD signaling and facilitates immunotherapy in colorectal cancer. Mol. Cancer 2020, 19, 51. [Google Scholar] [CrossRef]
- Wu, N.; Jiang, M.; Liu, H.; Chu, Y.; Wang, D.; Cao, J.; Wang, Z.; Xie, X.; Han, Y.; Xu, B. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF-beta/SMAD2/3 signaling pathway. Cell Death Differ. 2021, 28, 219–232. [Google Scholar] [CrossRef]
- Balacescu, O.; Sur, D.; Cainap, C.; Visan, S.; Cruceriu, D.; Manzat-Saplacan, R.; Muresan, M.S.; Balacescu, L.; Lisencu, C.; Irimie, A. The Impact of miRNA in Colorectal Cancer Progression and Its Liver Metastases. Int. J. Mol. Sci. 2018, 19, 3711. [Google Scholar] [CrossRef]
- Hu, X.; Chen, Q.; Guo, H.; Li, K.; Fu, B.; Chen, Y.; Zhao, H.; Wei, M.; Li, Y.; Wu, H. Identification of Target PTEN-Based miR-425 and miR-576 as Potential Diagnostic and Immunotherapeutic Biomarkers of Colorectal Cancer With Liver Metastasis. Front. Oncol. 2021, 11, 657984. [Google Scholar] [CrossRef]
- Li, B.; Huang, M.; Liu, M.; Wen, S.; Sun, F. MicroRNA329 serves a tumor suppressive role in colorectal cancer by directly targeting transforming growth factor beta1. Mol. Med. Rep. 2017, 16, 3825–3832. [Google Scholar] [CrossRef] [PubMed]
- Radwan, E.; Shaltout, A.S.; Mansor, S.G.; Shafik, E.A.; Abbas, W.A.; Shehata, M.R.; Ali, M. Evaluation of circulating microRNAs-211 and 25 as diagnostic biomarkers of colorectal cancer. Mol. Biol. Rep. 2021, 48, 4601–4610. [Google Scholar] [CrossRef]
- Sells, E.; Pandey, R.; Chen, H.; Skovan, B.A.; Cui, H.; Ignatenko, N.A. Specific microRNA-mRNA Regulatory Network of Colon Cancer Invasion Mediated by Tissue Kallikrein-Related Peptidase 6. Neoplasia 2017, 19, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, T.; Jin, R.; Zhao, H.; Hu, J.; Feng, B.; Zang, L.; Zheng, M.; Wang, M. MicroRNA-301a promotes migration and invasion by targeting TGFBR2 in human colorectal cancer. J. Exp. Clin. Cancer Res. 2014, 33, 113. [Google Scholar] [CrossRef] [PubMed]
- Ullmann, P.; Rodriguez, F.; Schmitz, M.; Meurer, S.K.; Qureshi-Baig, K.; Felten, P.; Ginolhac, A.; Antunes, L.; Frasquilho, S.; Zugel, N.; et al. The miR-371 approximately 373 Cluster Represses Colon Cancer Initiation and Metastatic Colonization by Inhibiting the TGFBR2/ID1 Signaling Axis. Cancer Res. 2018, 78, 3793–3808. [Google Scholar] [CrossRef]
- He, H.; Zhao, X.; Zhu, Z.; Du, L.; Chen, E.; Liu, S.; Li, Q.; Dong, J.; Yang, J.; Lei, L. MicroRNA-3191 promotes migration and invasion by downregulating TGFBR2 in colorectal cancer. J. Biochem. Mol. Toxicol. 2019, 33, e22308. [Google Scholar] [CrossRef]
- Xu, X.; Chen, R.; Li, Z.; Huang, N.; Wu, X.; Li, S.; Li, Y.; Wu, S. MicroRNA-490-3p inhibits colorectal cancer metastasis by targeting TGFbetaR1. BMC Cancer 2015, 15, 1023. [Google Scholar] [CrossRef]
- Yu, Y.; Lei, X. CircFAM120B Blocks the Development of Colorectal Cancer by Activating TGF-Beta Receptor II Expression via Targeting miR-645. Front. Cell Dev. Biol. 2021, 9, 682543. [Google Scholar] [CrossRef]
- Zhang, N.; Li, L.; Luo, J.; Tan, J.; Hu, W.; Li, Z.; Wang, X.; Ye, T. Inhibiting microRNA-424 in bone marrow mesenchymal stem cells-derived exosomes suppresses tumor growth in colorectal cancer by upregulating TGFBR3. Arch. Biochem. Biophys. 2021, 709, 108965. [Google Scholar] [CrossRef]
- Bao, Y.; Chen, Z.; Guo, Y.; Feng, Y.; Li, Z.; Han, W.; Wang, J.; Zhao, W.; Jiao, Y.; Li, K.; et al. Tumor suppressor microRNA-27a in colorectal carcinogenesis and progression by targeting SGPP1 and Smad2. PLoS ONE 2014, 9, e105991. [Google Scholar] [CrossRef]
- Li, J.; Zou, K.; Yu, L.; Zhao, W.; Lu, Y.; Mao, J.; Wang, B.; Wang, L.; Fan, S.; Song, B.; et al. MicroRNA-140 Inhibits the Epithelial-Mesenchymal Transition and Metastasis in Colorectal Cancer. Mol. Ther. Nucl. Acids 2018, 10, 426–437. [Google Scholar] [CrossRef]
- Cheng, D.; Zhao, S.; Tang, H.; Zhang, D.; Sun, H.; Yu, F.; Jiang, W.; Yue, B.; Wang, J.; Zhang, M.; et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget 2016, 7, 45199–45213. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zou, C.; Zou, C.; Han, Z.; Xiao, H.; Wei, H.; Wang, W.; Zhang, L.; Zhang, X.; Tang, Q.; et al. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett. 2013, 335, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Sun, H.; Jiang, W.; Mi, Y.; Zhang, D.; Wen, Y.; Cheng, D.; Tang, H.; Wu, S.; Yu, Y.; et al. miR-4775 promotes colorectal cancer invasion and metastasis via the Smad7/TGFbeta-mediated epithelial to mesenchymal transition. Mol. Cancer 2017, 16, 12. [Google Scholar] [CrossRef]
- Wang, H.; Nie, L.; Wu, L.; Liu, Q.; Guo, X. NR2F2 inhibits Smad7 expression and promotes TGF-beta-dependent epithelial-mesenchymal transition of CRC via transactivation of miR-21. Biochem. Biophys. Res. Commun. 2017, 485, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Li, N. CircTBL1XR1/miR-424 axis regulates Smad7 to promote the proliferation and metastasis of colorectal cancer. J. Gastrointest. Oncol. 2020, 11, 918–931. [Google Scholar] [CrossRef]
- De Summa, S.; Danza, K.; Pilato, B.; Matera, G.; Fasano, R.; Calabrese, A.; Lacalamita, R.; Silvestris, N.; Tommasi, S.; Argentiero, A.; et al. A Promising Role of TGF-beta Pathway in Response to Regorafenib in Metastatic Colorectal Cancer: A Case Report. Medicina 2021, 57, 1241. [Google Scholar] [CrossRef]
- Zhang, B.H.; Wang, C.; Dong, W.; Chen, X.; Leng, C.; Luo, X.; Dong, S.L.; Yin, P.; Zhang, B.X.; Datta, P.K.; et al. A novel approach for monitoring TGF-beta signaling in vivo in colon cancer. Carcinogenesis 2021, 42, 631–639. [Google Scholar] [CrossRef]
- Otegbeye, F.; Ojo, E.; Moreton, S.; Mackowski, N.; Lee, D.A.; de Lima, M.; Wald, D.N. Inhibiting TGF-beta signaling preserves the function of highly activated, in vitro expanded natural killer cells in AML and colon cancer models. PLoS ONE 2018, 13, e0191358. [Google Scholar]
- Varela-Calvino, R.; Rodriguez-Quiroga, M.; Dias Carvalho, P.; Martins, F.; Serra-Roma, A.; Vazquez-Iglesias, L.; Paez de la Cadena, M.; Velho, S.; Cordero, O.J. The mechanism of sitagliptin inhibition of colorectal cancer cell lines’ metastatic functionalities. IUBMB Life 2021, 73, 761–773. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, K.; Cho, S.H.; Chun, S.M.; Tak, E.; Hong, Y.S.; Kim, J.E.; Kim, T.W. Longitudinal change of genetic variations in cetuximab-treated metastatic colorectal cancer. Cancer Genet. 2021, 258–259, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, D.; Blauensteiner, B.; Matrone, N.; Belli, V.; Mohr, T.; Vitiello, P.P.; Martini, G.; Poliero, L.; Cardone, C.; Napolitano, S.; et al. Dual inhibition of TGFbeta and AXL as a novel therapy for human colorectal adenocarcinoma with mesenchymal phenotype. Med. Oncol. 2021, 38, 24. [Google Scholar] [CrossRef]
- Li, Y.C.; Chen, C.H.; Chang, C.L.; Chiang, J.Y.; Chu, C.H.; Chen, H.H.; Yip, H.K. Melatonin and hyperbaric oxygen therapies suppress colorectal carcinogenesis through pleiotropic effects and multifaceted mechanisms. Int. J. Biol. Sci. 2021, 17, 3728–3744. [Google Scholar] [CrossRef] [PubMed]
- Principe, D.R.; DeCant, B.; Staudacher, J.; Vitello, D.; Mangan, R.J.; Wayne, E.A.; Mascariñas, E.; Diaz, A.M.; Bauer, J.; McKinney, R.D.; et al. Loss of TGFβ signaling promotes colon cancer progression and tumor-associated inflammation. Oncotarget 2017, 8, 3826–3839. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.W.; Wu, C.E.; Zhou, J.Y.; Zhao, Z.M.; Liu, C.L.; Shen, J.Y.; Cai, H.; Liu, S.L. Solasodine reverses stemness and epithelial-mesenchymal transition in human colorectal cancer. Biochem. Biophys. Res. Commun. 2018, 505, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Yan, Z.; Chen, W.; Peng, J.; Feng, J.; Li, Q.; Jin, Y.; Lin, J. Hedyotis diffusa Willd suppresses metastasis in 5fluorouracilresistant colorectal cancer cells by regulating the TGFbeta signaling pathway. Mol. Med. Rep. 2017, 16, 7752–7758. [Google Scholar] [CrossRef]
- Yang, B.; Bai, H.; Sa, Y.; Zhu, P.; Liu, P. Inhibiting EMT, stemness and cell cycle involved in baicalin-induced growth inhibition and apoptosis in colorectal cancer cells. J. Cancer 2020, 11, 2303–2317. [Google Scholar] [CrossRef]
- Jiang, Z.; Cao, Q.; Dai, G.; Wang, J.; Liu, C.; Lv, L.; Pan, J. Celastrol inhibits colorectal cancer through TGF-beta1/Smad signaling. OncoTargets Ther. 2019, 12, 509–518. [Google Scholar] [CrossRef]
- Jin, Y.; Chen, W.; Yang, H.; Yan, Z.; Lai, Z.; Feng, J.; Peng, J.; Lin, J. Scutellaria barbata D. Don inhibits migration and invasion of colorectal cancer cells via suppression of PI3K/AKT and TGF-beta/Smad signaling pathways. Exp. Ther. Med. 2017, 14, 5527–5534. [Google Scholar]
- Xiao, Q.; Xiao, J.; Liu, J.; Liu, J.; Shu, G.; Yin, G. Metformin suppresses the growth of colorectal cancer by targeting INHBA to inhibit TGF-beta/PI3K/AKT signaling transduction. Cell Death Dis. 2022, 13, 202. [Google Scholar] [CrossRef]
- Zhang, L.; Cai, Q.Y.; Liu, J.; Peng, J.; Chen, Y.Q.; Sferra, T.J.; Lin, J.M. Ursolic acid suppresses the invasive potential of colorectal cancer cells by regulating the TGF-beta1/ZEB1/miR-200c signaling pathway. Oncol. Lett. 2019, 18, 3274–3282. [Google Scholar] [PubMed]
- Dai, Y.; Wang, H.; Sun, R.; Diao, J.; Ma, Y.; Shao, M.; Xu, Y.; Zhang, Q.; Gao, Z.; Zeng, Z.; et al. Modified Shenlingbaizhu Decoction represses the pluripotency of colorectal cancer stem cells by inhibiting TGF-beta mediated EMT program. Phytomedicine 2022, 103, 154234. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, J.; Lin, S.; Tan, J.; Huang, B.; Lin, J. Qingjie Fuzheng Granule Inhibited the Migration and Invasion of Colorectal Cancer Cells by Regulating the lncRNA ANRIL/let-7a/TGF-beta1/Smad Axis. Evid. Based Compl. Altern. Med. 2020, 2020, 5264651. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, J.X.; Wu, Y.; Lv, L.L.; Ying, H.F.; Zhu, W.H.; Xu, J.Y.; Ruan, M.; Guo, Y.; Zhu, W.R.; et al. The mechanism of FZXJJZ decoction suppresses colorectal liver metastasis via the VDR/TGF-beta/Snail1 signaling pathways based on network pharmacology-TCGA data-transcriptomics analysis. J. Ethnopharmacol. 2022, 287, 114904. [Google Scholar] [CrossRef]
- Roerink, S.F.; Sasaki, N.; Lee-Six, H.; Young, M.D.; Alexandrov, L.B.; Behjati, S.; Mitchell, T.J.; Grossmann, S.; Lightfoot, H.; Egan, D.A.; et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 2018, 556, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Yu, I.S.; Cheung, W.Y. Metastatic Colorectal Cancer in the Era of Personalized Medicine: A More Tailored Approach to Systemic Therapy. Can. J. Gastroenterol. Hepatol. 2018, 2018, 9450754. [Google Scholar] [CrossRef]
- Xie, F.; Ling, L.; van Dam, H.; Zhou, F.; Zhang, L. TGF-beta signaling in cancer metastasis. Acta Biochim. Biophys. Sin. 2018, 50, 121–132. [Google Scholar] [CrossRef]
- Binefa, G.; Rodriguez-Moranta, F.; Teule, A.; Medina-Hayas, M. Colorectal cancer: From prevention to personalized medicine. World J. Gastroenterol. 2014, 20, 6786–6808. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).