Isosorbide Fatty Acid Diesters Have Synergistic Anti-Inflammatory Effects in Cytokine-Induced Tissue Culture Models of Atopic Dermatitis
Abstract
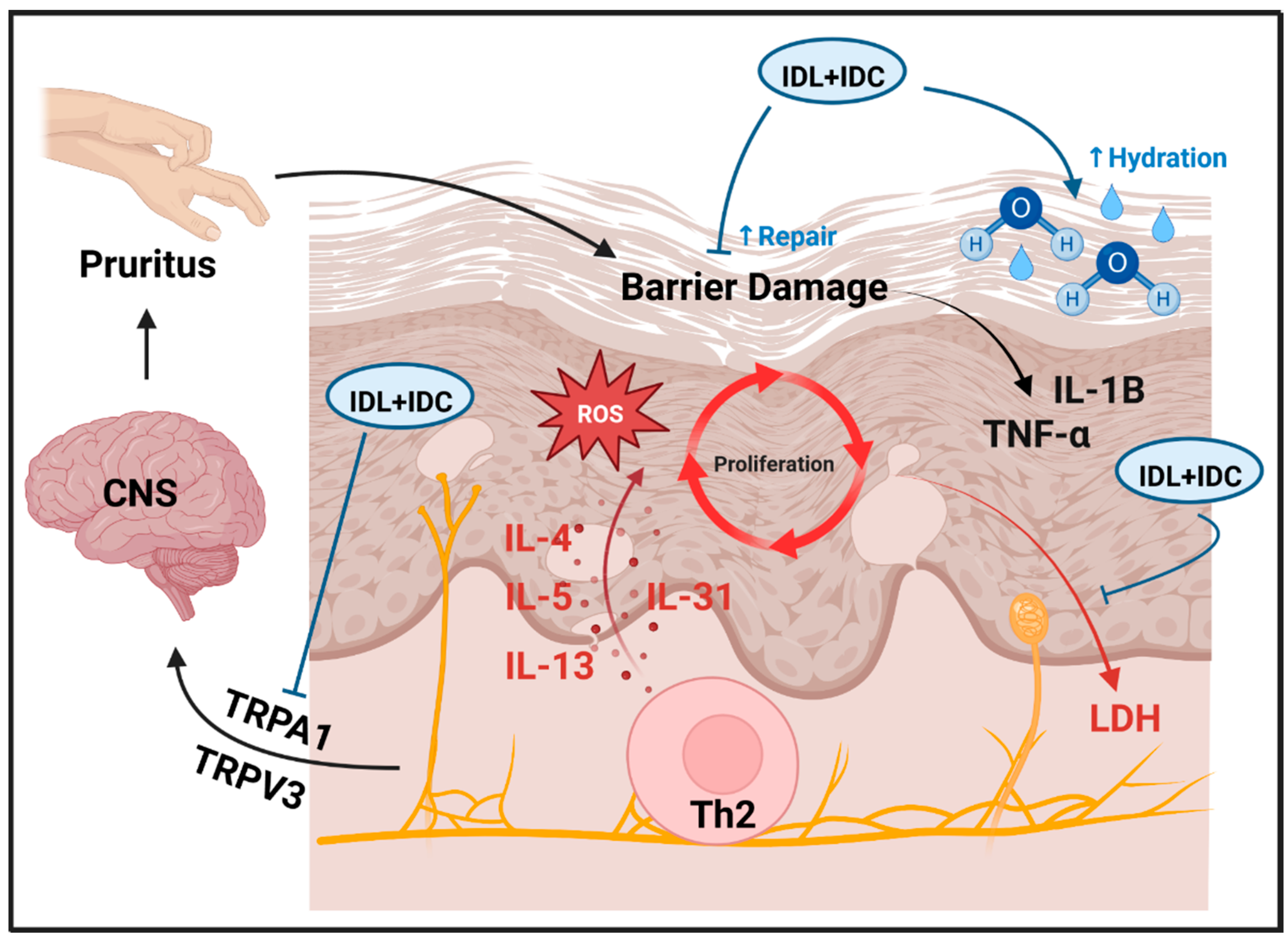
1. Introduction
2. Results
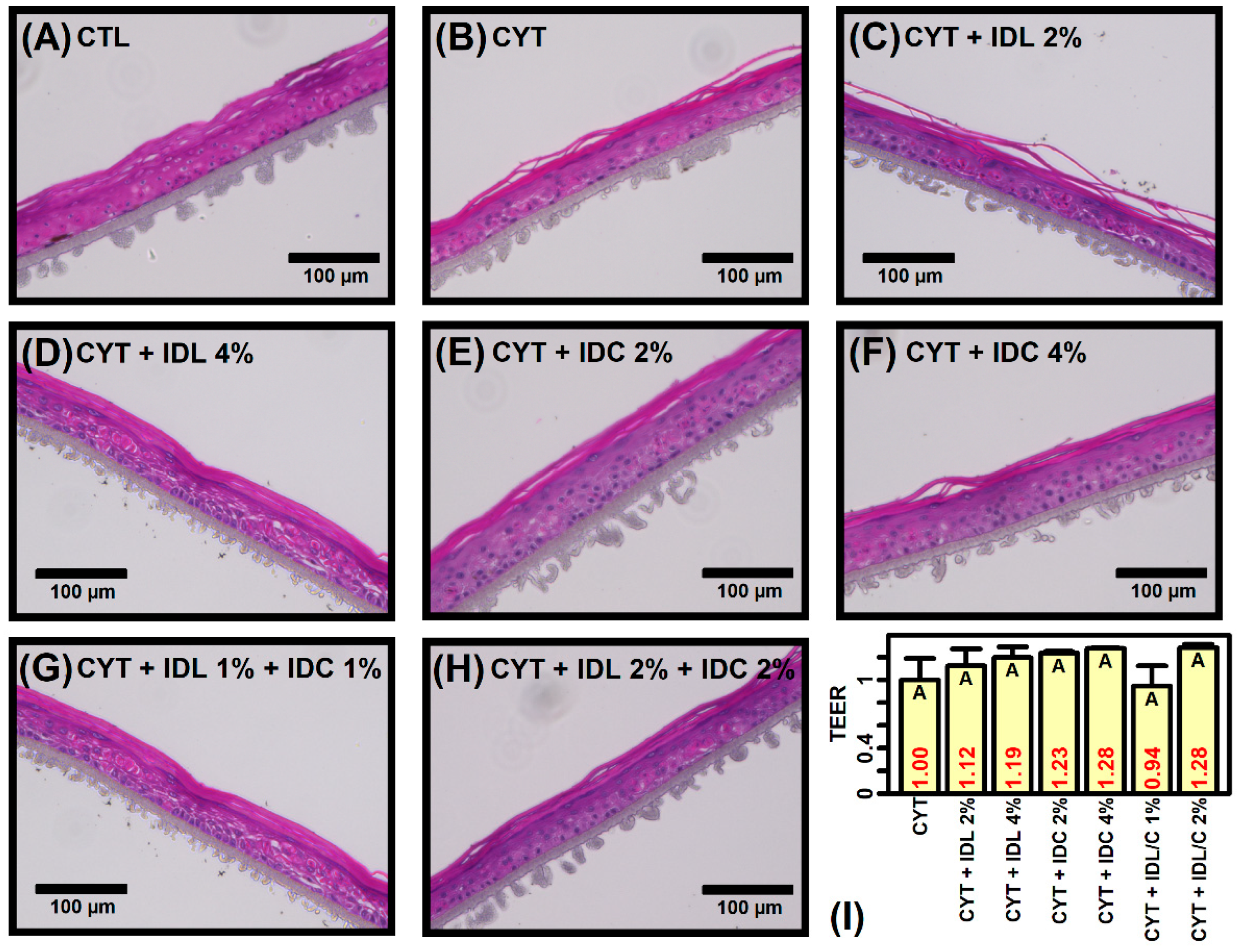
2.1. IDL and IDC Prevent Cytokine-Induced Disruption of Epidermal Morphology
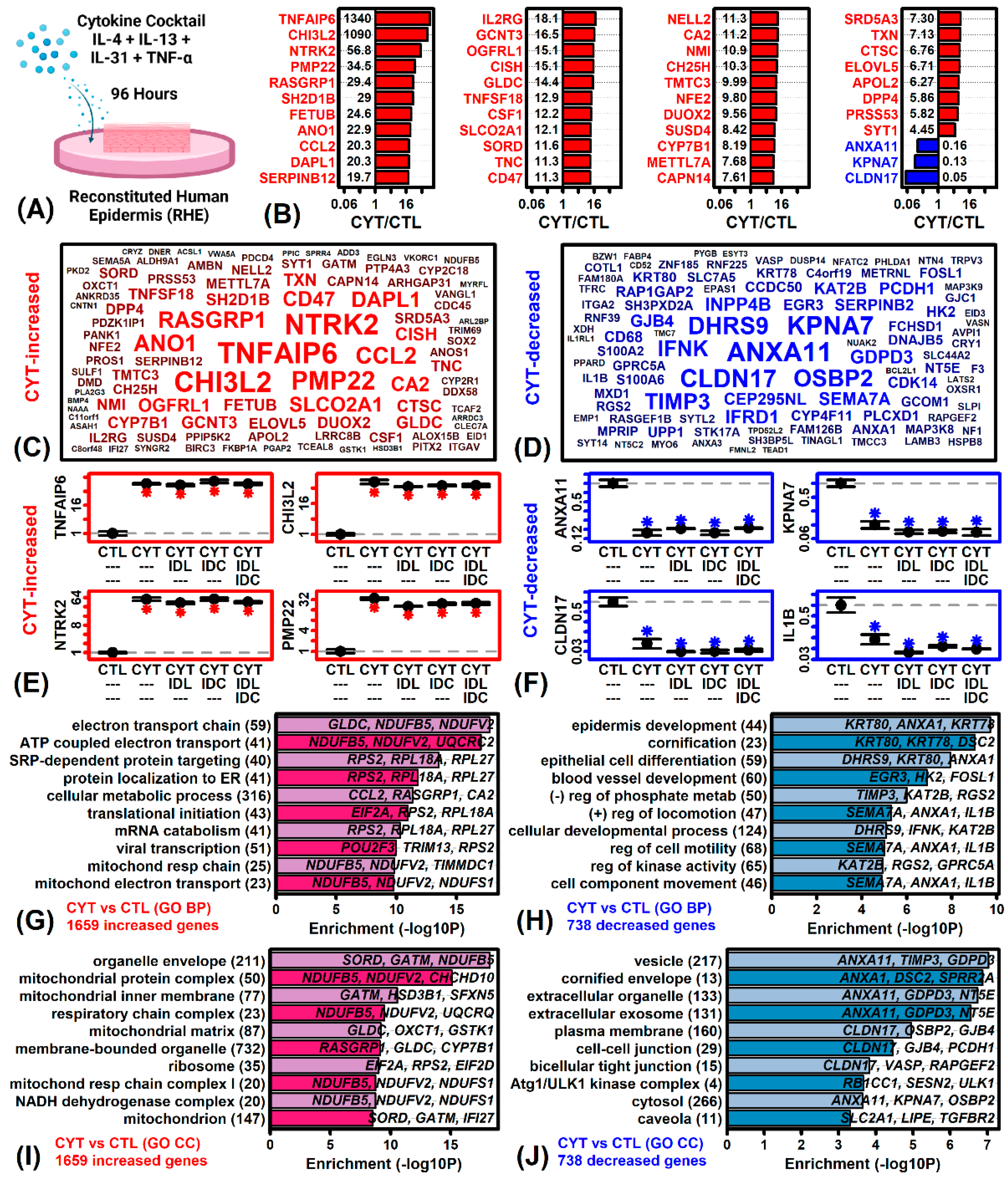
2.2. Cytokines Induce Inflammatory and Mitochondrial Gene Expression but Repress Epidermal Development and KC Differentiation Pathways
2.3. The Cytokine Cocktail Induces an AD-like Gene Expression Response in RHE Tissue Cultures
2.4. IDL, IDC and IDL + IDC Up-Regulate Cell Cycle Genes and Decrease Expression of Genes Associated with Development and Differentiation
2.5. IDL, IDC and IDL + IDC Increase Expression of Basal Layer and Early KC Differentiation Genes but Repress Expression of Genes Associated with Late KC Differentiation
2.6. IDL, IDC and IDL + IDC Increase Expression of Genes Associated with Late but Not Early Interphase
2.7. IDL, IDC and IDL + IDC Oppose Gene Expression Responses Associated with TNF-α but Not Th2 Cytokines (IL-4, IL-13, IL-31)
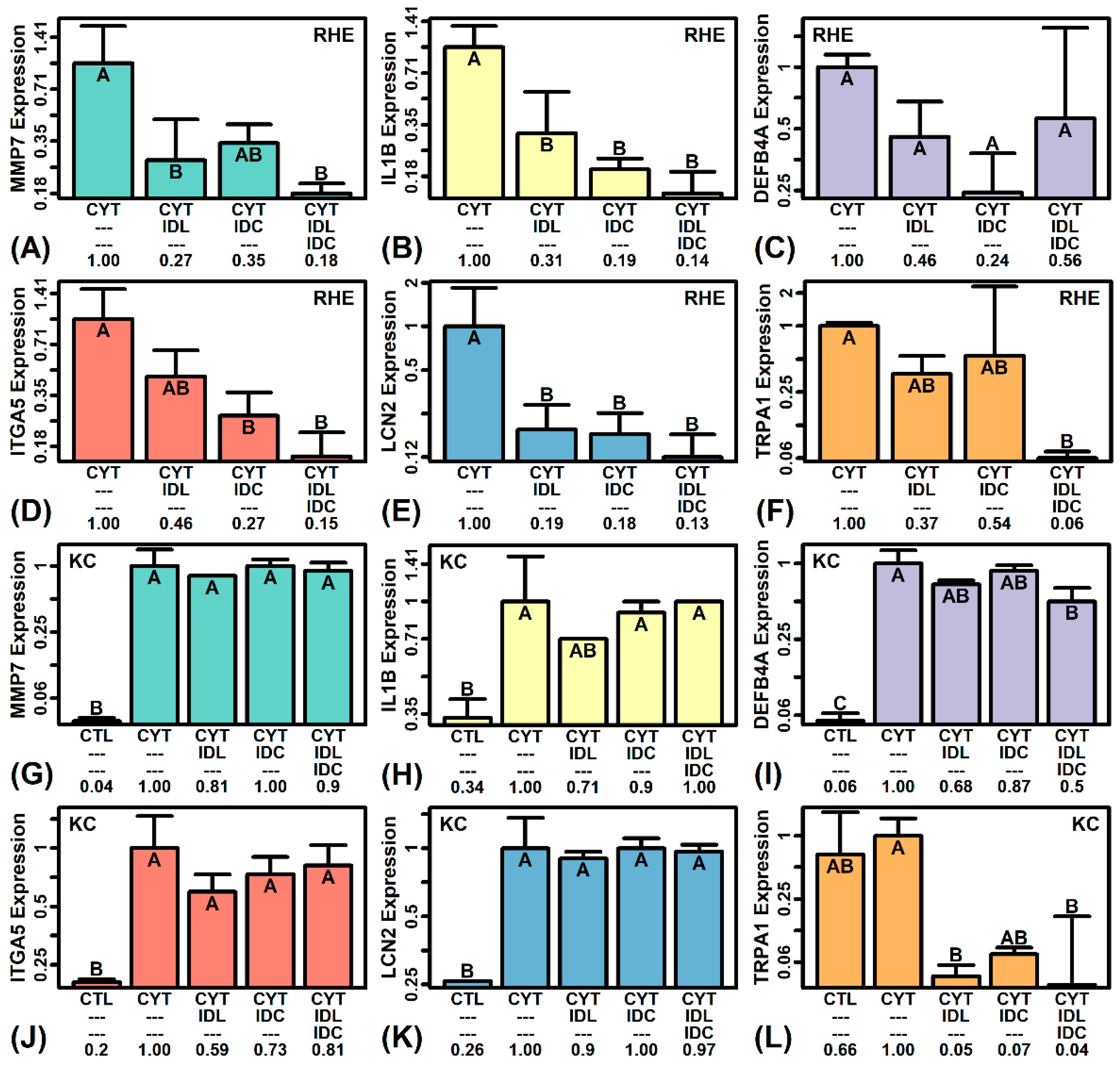
2.8. IDL and IDC Synergistically Repress Expression of Pro-Inflammatory Mediators (IL1B, ITGA5, TRPA1)
2.9. The IDL + IDC Anti-Inflammatory Response More Closely Resembles That of a Calcineurin Inhibitor Rather Than Corticosteroid
2.10. IDL, IDC and IDL + IDC Alter the Expression of Pruritus-Associated Genes
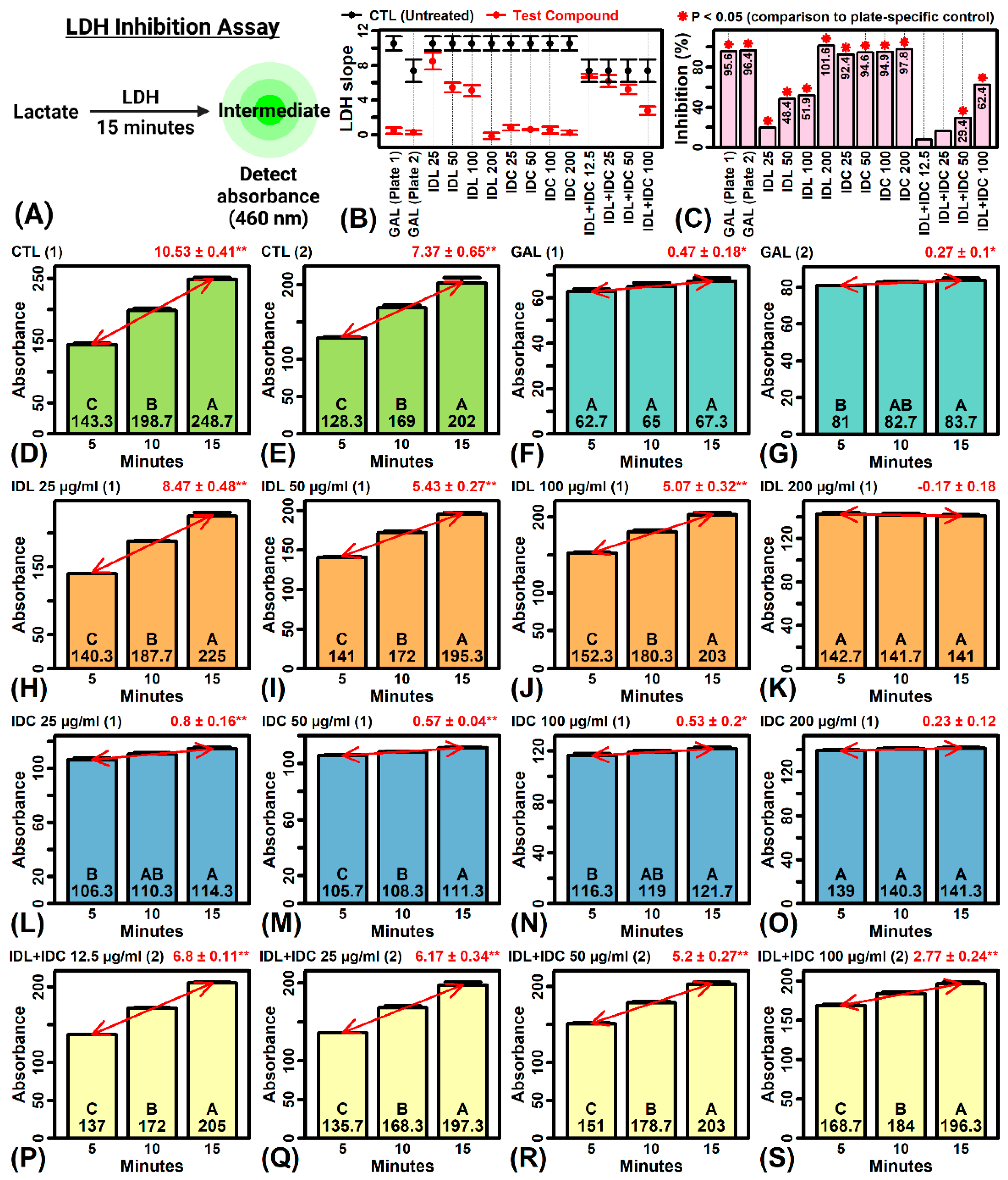
2.11. IDC Decreases LDH Secretion by more Than 50% in Cytokine-Treated Skin Explants
2.12. IDL and IDC Inhibit LDH Activity
3. Discussion
4. Materials and Methods
4.1. Test Materials
4.2. RHE Tissue Culture
4.3. Microarray Analyses
4.4. Differential Expression Analyses
4.5. Cytokine-Stimulated Skin Explants
4.6. LDH Detection Assay
4.7. TSLP Assays
4.8. LDH Inhibition Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ständer, S. Atopic Dermatitis. N. Engl. J. Med. 2021, 384, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- Appiah, M.M.; Haft, M.A.; Kleinman, E.; Laborada, J.; Lee, S.; Loop, L.; Geng, B.; Eichenfield, L.F. Atopic dermatitis: Review of comorbidities and therapeutics. Ann. Allergy Asthma Immunol. Off. Publ. Am. Coll. Allergy Asthma Immunol. 2022, 129, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, A.; Nomura, T.; Rerknimitr, P.; Seidel, J.A.; Honda, T.; Kabashima, K. The interplay between genetic and environmental factors in the pathogenesis of atopic dermatitis. Immunol. Rev. 2017, 278, 246–262. [Google Scholar] [CrossRef]
- Wollenberg, A.; Barbarot, S.; Bieber, T.; Christen-Zaech, S.; Deleuran, M.; Fink-Wagner, A.; Gieler, U.; Girolomoni, G.; Lau, S.; Muraro, A.; et al. Consensus-based European guidelines for treatment of atopic eczema (atopic dermatitis) in adults and children: Part II. J. Eur. Acad. Dermatol. Venereol. JEADV 2018, 32, 850–878. [Google Scholar] [CrossRef] [PubMed]
- Mawazi, S.M.; Ann, J.; Othman, N.; Khan, J.; Alolayan, S.O.; Kaleemullah, M.J.C. A Review of Moisturizers; History, Preparation, Characterization and Applications. Cosmetics 2022, 9, 61. [Google Scholar] [CrossRef]
- Pons-Guiraud, A. Dry skin in dermatology: A complex physiopathology. J. Eur. Acad. Dermatol. Venereol. JEADV 2007, 21 (Suppl. S2), 1–4. [Google Scholar] [CrossRef] [PubMed]
- Elias, P.M.; Wakefield, J.S.; Man, M.Q. Moisturizers versus Current and Next-Generation Barrier Repair Therapy for the Management of Atopic Dermatitis. Ski. Pharmacol. Physiol. 2019, 32, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Quadri, M.; Lotti, R.; Bonzano, L.; Ciardo, S.; Guanti, M.B.; Pellacani, G.; Pincelli, C.; Marconi, A. A Novel Multi-Action Emollient Plus Cream Improves Skin Barrier Function in Patients with Atopic Dermatitis: In vitro and Clinical Evidence. Ski. Pharmacol. Physiol. 2021, 34, 8–18. [Google Scholar] [CrossRef]
- Hou, D.D.; Di, Z.H.; Qi, R.Q.; Wang, H.X.; Zheng, S.; Hong, Y.X.; Guo, H.; Chen, H.D.; Gao, X.H. Sea Buckthorn (Hippophaë rhamnoides L.) Oil Improves Atopic Dermatitis-Like Skin Lesions via Inhibition of NF-κB and STAT1 Activation. Ski. Pharmacol. Physiol. 2017, 30, 268–276. [Google Scholar] [CrossRef]
- Darmstadt, G.L.; Mao-Qiang, M.; Chi, E.; Saha, S.K.; Ziboh, V.A.; Black, R.E.; Santosham, M.; Elias, P.M. Impact of topical oils on the skin barrier: Possible implications for neonatal health in developing countries. Acta Paediatr. 2002, 91, 546–554. [Google Scholar] [CrossRef]
- Darmstadt, G.L.; Badrawi, N.; Law, P.A.; Ahmed, S.; Bashir, M.; Iskander, I.; Al Said, D.; El Kholy, A.; Husein, M.H.; Alam, A.; et al. Topically applied sunflower seed oil prevents invasive bacterial infections in preterm infants in Egypt: A randomized, controlled clinical trial. Pediatr. Infect. Dis. J. 2004, 23, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Bojanowski, K.; Swindell, W.R.; Cantor, S.; Chaudhuri, R.K. Isosorbide Di-(Linoleate/Oleate) Stimulates Prodifferentiation Gene Expression to Restore the Epidermal Barrier and Improve Skin Hydration. J. Investig. Dermatol. 2021, 141, 1416–1427.e12. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, R.K.; Bojanowski, K. Improvement of hydration and epidermal barrier function in human skin by a novel compound isosorbide dicaprylate. Int. J. Cosmet. Sci. 2017, 39, 518–526. [Google Scholar] [CrossRef] [PubMed]
- Eyerich, K.; Brown, S.J.; Perez White, B.E.; Tanaka, R.J.; Bissonette, R.; Dhar, S.; Bieber, T.; Hijnen, D.J.; Guttman-Yassky, E.; Irvine, A.; et al. Human and computational models of atopic dermatitis: A review and perspectives by an expert panel of the International Eczema Council. J. Allergy Clin. Immunol. 2019, 143, 36–45. [Google Scholar] [CrossRef]
- De Vuyst, E.; Salmon, M.; Evrard, C.; Lambert de Rouvroit, C.; Poumay, Y. Atopic Dermatitis Studies through In Vitro Models. Front. Med. 2017, 4, 119. [Google Scholar] [CrossRef]
- Danso, M.O.; van Drongelen, V.; Mulder, A.; van Esch, J.; Scott, H.; van Smeden, J.; El Ghalbzouri, A.; Bouwstra, J.A. TNF-α and Th2 cytokines induce atopic dermatitis-like features on epidermal differentiation proteins and stratum corneum lipids in human skin equivalents. J. Investig. Dermatol. 2014, 134, 1941–1950. [Google Scholar] [CrossRef]
- Kamsteeg, M.; Bergers, M.; de Boer, R.; Zeeuwen, P.L.; Hato, S.V.; Schalkwijk, J.; Tjabringa, G.S. Type 2 helper T-cell cytokines induce morphologic and molecular characteristics of atopic dermatitis in human skin equivalent. Am. J. Pathol. 2011, 178, 2091–2099. [Google Scholar] [CrossRef]
- Kim, H.J.; Baek, J.; Lee, J.R.; Roh, J.Y.; Jung, Y. Optimization of Cytokine Milieu to Reproduce Atopic Dermatitis-related Gene Expression in HaCaT Keratinocyte Cell Line. Immune Netw. 2018, 18, e9. [Google Scholar] [CrossRef]
- De Vuyst, É.; Giltaire, S.; Lambert de Rouvroit, C.; Malaisse, J.; Mound, A.; Bourtembourg, M.; Poumay, Y.; Nikkels, A.F.; Chrétien, A.; Salmon, M. Methyl-β-cyclodextrin concurs with interleukin (IL)-4, IL-13 and IL-25 to induce alterations reminiscent of atopic dermatitis in reconstructed human epidermis. Exp. Dermatol. 2018, 27, 435–437. [Google Scholar] [CrossRef]
- Bogiatzi, S.I.; Fernandez, I.; Bichet, J.C.; Marloie-Provost, M.A.; Volpe, E.; Sastre, X.; Soumelis, V. Cutting Edge: Proinflammatory and Th2 cytokines synergize to induce thymic stromal lymphopoietin production by human skin keratinocytes. J. Immunol. 2007, 178, 3373–3377. [Google Scholar] [CrossRef]
- Bernard, F.X.; Morel, F.; Camus, M.; Pedretti, N.; Barrault, C.; Garnier, J.; Lecron, J.C. Keratinocytes under Fire of Proinflammatory Cytokines: Bona Fide Innate Immune Cells Involved in the Physiopathology of Chronic Atopic Dermatitis and Psoriasis. J. Allergy 2012, 2012, 718725. [Google Scholar] [CrossRef] [PubMed]
- Rouaud-Tinguely, P.; Boudier, D.; Marchand, L.; Barruche, V.; Bordes, S.; Coppin, H.; Roth, M.P.; Closs, B. From the morphological to the transcriptomic characterization of a compromised three-dimensional in vitro model mimicking atopic dermatitis. Br. J. Dermatol. 2015, 173, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Beken, B.; Serttas, R.; Yazicioglu, M.; Turkekul, K.; Erdogan, S. Quercetin Improves Inflammation, Oxidative Stress, and Impaired Wound Healing in Atopic Dermatitis Model of Human Keratinocytes. Pediatr. Allergy Immunol. Pulmonol. 2020, 33, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.S.; Seong, K.H.; Kim, C.D.; Seo, S.J.; Park, B.C.; Kim, M.H.; Hong, S.P. Adiponectin Attenuates the Inflammation in Atopic Dermatitis-Like Reconstructed Human Epidermis. Ann. Dermatol. 2019, 31, 186–195. [Google Scholar] [CrossRef]
- Ewald, D.A.; Malajian, D.; Krueger, J.G.; Workman, C.T.; Wang, T.; Tian, S.; Litman, T.; Guttman-Yassky, E.; Suárez-Fariñas, M. Meta-analysis derived atopic dermatitis (MADAD) transcriptome defines a robust AD signature highlighting the involvement of atherosclerosis and lipid metabolism pathways. BMC Med. Genom. 2015, 8, 60. [Google Scholar] [CrossRef]
- Lopez-Pajares, V.; Qu, K.; Zhang, J.; Webster, D.E.; Barajas, B.C.; Siprashvili, Z.; Zarnegar, B.J.; Boxer, L.D.; Rios, E.J.; Tao, S.; et al. A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev. Cell 2015, 32, 693–706. [Google Scholar] [CrossRef]
- Mizuno, H.; Nakanishi, Y.; Ishii, N.; Sarai, A.; Kitada, K. A signature-based method for indexing cell cycle phase distribution from microarray profiles. BMC Genom. 2009, 10, 137. [Google Scholar] [CrossRef]
- Swindell, W.R.; Xing, X.; Stuart, P.E.; Chen, C.S.; Aphale, A.; Nair, R.P.; Voorhees, J.J.; Elder, J.T.; Johnston, A.; Gudjonsson, J.E. Heterogeneity of inflammatory and cytokine networks in chronic plaque psoriasis. PLoS ONE 2012, 7, e34594. [Google Scholar] [CrossRef]
- Kleinman, E.; Laborada, J.; Metterle, L.; Eichenfield, L.F. What’s New in Topicals for Atopic Dermatitis? Am. J. Clin. Dermatol. 2022, 23, 595–603. [Google Scholar] [CrossRef]
- Sideris, N.; Paschou, E.; Bakirtzi, K.; Kiritsi, D.; Papadimitriou, I.; Tsentemeidou, A.; Sotiriou, E.; Vakirlis, E. New and Upcoming Topical Treatments for Atopic Dermatitis: A Review of the Literature. J. Clin. Med. 2022, 11, 4974. [Google Scholar] [CrossRef]
- Umar, B.U.; Rahman, S.; Dutta, S.; Islam, T.; Nusrat, N.; Chowdhury, K.; Binti Wan Ahmad Fakuradzi, W.F.S.; Haque, M. Management of Atopic Dermatitis: The Role of Tacrolimus. Cureus 2022, 14, e28130. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.M.; Scherer, A.; Wanke, C.; Bräutigam, M.; Bongiovanni, S.; Letzkus, M.; Staedtler, F.; Kehren, J.; Zuehlsdorf, M.; Schwarz, T.; et al. Gene expression is differently affected by pimecrolimus and betamethasone in lesional skin of atopic dermatitis. Allergy 2012, 67, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Ahn, C.; Huang, W. Clinical Presentation of Atopic Dermatitis. Adv. Exp. Med. Biol. 2017, 1027, 39–46. [Google Scholar] [PubMed]
- Köhler, S.; Gargano, M.; Matentzoglu, N.; Carmody, L.C.; Lewis-Smith, D.; Vasilevsky, N.A.; Danis, D.; Balagura, G.; Baynam, G.; Brower, A.M.; et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 2021, 49, D1207–D1217. [Google Scholar] [CrossRef] [PubMed]
- Manerba, M.; Vettraino, M.; Fiume, L.; Di Stefano, G.; Sartini, A.; Giacomini, E.; Buonfiglio, R.; Roberti, M.; Recanatini, M. Galloflavin (CAS 568-80-9): A novel inhibitor of lactate dehydrogenase. ChemMedChem 2012, 7, 311–317. [Google Scholar] [CrossRef]
- Elias, P.M. Optimizing emollient therapy for skin barrier repair in atopic dermatitis. Ann. Allergy Asthma Immunol. Off. Publ. Am. Coll. Allergy Asthma Immunol. 2022, 128, 505–511. [Google Scholar] [CrossRef]
- Oyoshi, M.K.; He, R.; Kumar, L.; Yoon, J.; Geha, R.S. Cellular and molecular mechanisms in atopic dermatitis. Adv. Immunol. 2009, 102, 135–226. [Google Scholar]
- Pastar, Z.; Lipozencić, J.; Ljubojević, S. Etiopathogenesis of atopic dermatitis—An overview. Acta Dermatovenerol. Croat. ADC 2005, 13, 54–62. [Google Scholar]
- Sugaya, M. The Role of Th17-Related Cytokines in Atopic Dermatitis. Int. J. Mol. Sci. 2020, 21, 1314. [Google Scholar] [CrossRef]
- Kamijo, H.; Miyagaki, T.; Hayashi, Y.; Akatsuka, T.; Watanabe-Otobe, S.; Oka, T.; Shishido-Takahashi, N.; Suga, H.; Sugaya, M.; Sato, S. Increased IL-26 Expression Promotes T Helper Type 17- and T Helper Type 2-Associated Cytokine Production by Keratinocytes in Atopic Dermatitis. J. Investig. Dermatol. 2020, 140, 636–644.e2. [Google Scholar] [CrossRef]
- Tsoi, L.C.; Rodriguez, E.; Stölzl, D.; Wehkamp, U.; Sun, J.; Gerdes, S.; Sarkar, M.K.; Hübenthal, M.; Zeng, C.; Uppala, R.; et al. Progression of acute-to-chronic atopic dermatitis is associated with quantitative rather than qualitative changes in cytokine responses. J. Allergy Clin. Immunol. 2020, 145, 1406–1415. [Google Scholar] [CrossRef] [PubMed]
- Fania, L.; Moretta, G.; Antonelli, F.; Scala, E.; Abeni, D.; Albanesi, C.; Madonna, S. Multiple Roles for Cytokines in Atopic Dermatitis: From Pathogenic Mediators to Endotype-Specific Biomarkers to Therapeutic Targets. Int. J. Mol. Sci. 2022, 23, 2684. [Google Scholar] [CrossRef] [PubMed]
- Leman, G.; Pavel, P.; Hermann, M.; Crumrine, D.; Elias, P.M.; Minzaghi, D.; Goudounèche, D.; Roshardt Prieto, N.M.; Cavinato, M.; Wanner, A.; et al. Mitochondrial Activity Is Upregulated in Nonlesional Atopic Dermatitis and Amenable to Therapeutic Intervention. J. Investig. Dermatol. 2022, 142, 2623–2634.e12. [Google Scholar] [CrossRef] [PubMed]
- Hoffjan, S.; Stemmler, S. Unravelling the complex genetic background of atopic dermatitis: From genetic association results towards novel therapeutic strategies. Arch. Dermatol. Res. 2015, 307, 659–670. [Google Scholar] [CrossRef]
- Stemmler, S.; Hoffjan, S. Trying to understand the genetics of atopic dermatitis. Mol. Cell. Probes 2016, 30, 374–385. [Google Scholar] [CrossRef]
- Bussmann, C.; Weidinger, S.; Novak, N. Genetics of atopic dermatitis. JDDG J. Der Dtsch. Dermatol. Ges. 2011, 9, 670–676. [Google Scholar] [CrossRef]
- Elias, P.M. Primary role of barrier dysfunction in the pathogenesis of atopic dermatitis. Exp. Dermatol. 2018, 27, 847–851. [Google Scholar] [CrossRef]
- Wood, L.C.; Jackson, S.M.; Elias, P.M.; Grunfeld, C.; Feingold, K.R. Cutaneous barrier perturbation stimulates cytokine production in the epidermis of mice. J. Clin. Investig. 1992, 90, 482–487. [Google Scholar] [CrossRef]
- Wood, L.C.; Stalder, A.K.; Liou, A.; Campbell, I.L.; Grunfeld, C.; Elias, P.M.; Feingold, K.R. Barrier disruption increases gene expression of cytokines and the 55 kD TNF receptor in murine skin. Exp. Dermatol. 1997, 6, 98–104. [Google Scholar] [CrossRef]
- Wood, L.C.; Feingold, K.R.; Sequeira-Martin, S.M.; Elias, P.M.; Grunfeld, C. Barrier function coordinately regulates epidermal IL-1 and IL-1 receptor antagonist mRNA levels. Exp. Dermatol. 1994, 3, 56–60. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, Y.-F.; Song, S.-S.; Wu, L.-L.; Chen, Y.; Li, X.-Y.; Ju, M. Alleviating skin barrier disruption, skin inflammation and pruritus: A moisturizing spray containing β-glucan and panthenol. Dermatol. Venereol. 2022. [Google Scholar] [CrossRef]
- Wood, L.C.; Elias, P.M.; Sequeira-Martin, S.M.; Grunfeld, C.; Feingold, K.R. Occlusion lowers cytokine mRNA levels in essential fatty acid-deficient and normal mouse epidermis, but not after acute barrier disruption. J. Investig. Dermatol. 1994, 103, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Mauro, T.M.; Dang, E.; Wang, G.; Hu, L.Z.; Yu, C.; Jeong, S.; Feingold, K.; Elias, P.M.; Lv, C.Z.; et al. Topical applications of an emollient reduce circulating pro-inflammatory cytokine levels in chronically aged humans: A pilot clinical study. J. Eur. Acad. Dermatol. Venereol. JEADV 2019, 33, 2197–2201. [Google Scholar] [CrossRef]
- Man, M.Q.; Wood, L.; Elias, P.M.; Feingold, K.R. Cutaneous barrier repair and pathophysiology following barrier disruption in IL-1 and TNF type I receptor deficient mice. Exp. Dermatol. 1999, 8, 261–266. [Google Scholar] [CrossRef]
- Sawada, E.; Yoshida, N.; Sugiura, A.; Imokawa, G. Th1 cytokines accentuate but Th2 cytokines attenuate ceramide production in the stratum corneum of human epidermal equivalents: An implication for the disrupted barrier mechanism in atopic dermatitis. J. Dermatol. Sci. 2012, 68, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.E.; Howell, M.D.; Guttman-Yassky, E.; Gilleaudeau, P.M.; Cardinale, I.R.; Boguniewicz, M.; Krueger, J.G.; Leung, D.Y. TNF-alpha downregulates filaggrin and loricrin through c-Jun N-terminal kinase: Role for TNF-alpha antagonists to improve skin barrier. J. Investig. Dermatol. 2011, 131, 1272–1279. [Google Scholar] [CrossRef]
- Kirschner, N.; Poetzl, C.; von den Driesch, P.; Wladykowski, E.; Moll, I.; Behne, M.J.; Brandner, J.M. Alteration of tight junction proteins is an early event in psoriasis: Putative involvement of proinflammatory cytokines. Am. J. Pathol. 2009, 175, 1095–1106. [Google Scholar] [CrossRef]
- Kasai, H.; Kawasaki, H.; Fukushima-Nomura, A.; Yasuda-Sekiguchi, F.; Amagai, M.; Ebihara, T.; Tanese, K. Stratification of atopic dermatitis patients by patterns of response to proactive therapy with topical tacrolimus: Low serum IgE levels and inadequately controlled disease activity at the start of treatment predict its failure. Ann. Med. 2021, 53, 2205–2214. [Google Scholar] [CrossRef]
- Mizuno, M.; Horiguchi, G.; Teramukai, S.; Ichiyama, S.; Ito, M.; Hoashi, T.; Kanda, N.; Saeki, H. Association study of transition of laboratory marker levels and transition of disease activity of atopic dermatitis patients treated with dupilumab. Australas. J. Dermatol. 2021, 62, e504–e509. [Google Scholar] [CrossRef]
- Gohar, M.K.; Atta, A.H.; Nasr, M.M.; Hussein, D.N. Serum Thymus and Activation Regulated Chemokine (TARC), IL- 18 and IL-18 Gene Polymorphism as Associative Factors with Atopic Dermatitis. Egypt. J. Immunol. 2017, 24, 9–22. [Google Scholar]
- Thijs, J.; Krastev, T.; Weidinger, S.; Buckens, C.F.; de Bruin-Weller, M.; Bruijnzeel-Koomen, C.; Flohr, C.; Hijnen, D. Biomarkers for atopic dermatitis: A systematic review and meta-analysis. Curr. Opin. Allergy Clin. Immunol. 2015, 15, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Mizawa, M.; Yamaguchi, M.; Ueda, C.; Makino, T.; Shimizu, T. Stress evaluation in adult patients with atopic dermatitis using salivary cortisol. BioMed. Res. Int. 2013, 2013, 138027. [Google Scholar] [CrossRef] [PubMed]
- Kou, K.; Aihara, M.; Matsunaga, T.; Chen, H.; Taguri, M.; Morita, S.; Fujita, H.; Yamaguchi, Y.; Kambara, T.; Ikezawa, Z. Association of serum interleukin-18 and other biomarkers with disease severity in adults with atopic dermatitis. Arch. Dermatol. Res. 2012, 304, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Morishima, Y.; Kawashima, H.; Takekuma, K.; Hoshika, A. Changes in serum lactate dehydrogenase activity in children with atopic dermatitis. Pediatr. Int. Off. J. Jpn. Pediatr. Soc. 2010, 52, 171–174. [Google Scholar] [CrossRef]
- Kato, A.; Kamata, M.; Ito, M.; Uchida, H.; Nagata, M.; Fukaya, S.; Hayashi, K.; Fukuyasu, A.; Tanaka, T.; Ishikawa, T.; et al. Higher baseline serum lactate dehydrogenase level is associated with poor effectiveness of dupilumab in the long term in patients with atopic dermatitis. J. Dermatol. 2020, 47, 1013–1019. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.E.; Nahm, D.H. Real Clinical Practice Data of Monthly Dupilumab Therapy in Adult Patients With Moderate-to-Severe Atopic Dermatitis: Clinical Efficacy and Predictive Markers for a Favorable Clinical Response. Allergy Asthma Immunol. Res. 2021, 13, 733–745. [Google Scholar] [CrossRef]
- Jang, D.H.; Heo, S.J.; Jung, H.J.; Park, M.Y.; Seo, S.J.; Ahn, J. Retrospective Study of Dupilumab Treatment for Moderate to Severe Atopic Dermatitis in Korea: Efficacy and Safety of Dupilumab in Real-World Practice. J. Clin. Med. 2020, 9, 1982. [Google Scholar] [CrossRef]
- Olesen, C.M.; Holm, J.G.; Nørreslet, L.B.; Serup, J.V.; Thomsen, S.F.; Agner, T. Treatment of atopic dermatitis with dupilumab: Experience from a tertiary referral centre. J. Eur. Acad. Dermatol. Venereol. JEADV 2019, 33, 1562–1568. [Google Scholar] [CrossRef]
- Mukai, H.; Noguchi, T.; Kamimura, K.; Nishioka, K.; Nishiyama, S. Significance of elevated serum LDH (lactate dehydrogenase) activity in atopic dermatitis. J. Dermatol. 1990, 17, 477–481. [Google Scholar] [CrossRef]
- Komatsu, N.; Saijoh, K.; Kuk, C.; Liu, A.C.; Khan, S.; Shirasaki, F.; Takehara, K.; Diamandis, E.P. Human tissue kallikrein expression in the stratum corneum and serum of atopic dermatitis patients. Exp. Dermatol. 2007, 16, 513–519. [Google Scholar] [CrossRef]
- Koguchi-Yoshioka, H.; Watanabe, R.; Matsumura, Y.; Ishitsuka, Y.; Inoue, S.; Furuta, J.; Nakamura, Y.; Okiyama, N.; Matsuzaka, T.; Shimano, H.; et al. Serum lactate dehydrogenase level as a possible predictor of treatment preference in psoriasis. J. Dermatol. Sci. 2021, 103, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Hanai, J.; Ren, J.G.; Kats, L.; Burgess, K.; Bhargava, P.; Signoretti, S.; Billiard, J.; Duffy, K.J.; Grant, A.; et al. Targeting lactate dehydrogenase—A inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell. Metab. 2014, 19, 795–809. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, T.; Carstens, E. Neural processing of itch. Neuroscience 2013, 250, 697–714. [Google Scholar] [CrossRef] [PubMed]
- Naert, R.; López-Requena, A.; Talavera, K. TRPA1 Expression and Pathophysiology in Immune Cells. Int. J. Mol. Sci. 2021, 22, 11460. [Google Scholar] [CrossRef]
- Atoyan, R.; Shander, D.; Botchkareva, N.V. Non-neuronal expression of transient receptor potential type A1 (TRPA1) in human skin. J. Investig. Dermatol. 2009, 129, 2312–2315. [Google Scholar] [CrossRef]
- Bellono, N.W.; Kammel, L.G.; Zimmerman, A.L.; Oancea, E. UV light phototransduction activates transient receptor potential A1 ion channels in human melanocytes. Proc. Natl. Acad. Sci. USA 2013, 110, 2383–2388. [Google Scholar] [CrossRef]
- Larkin, C.; Chen, W.; Szabó, I.L.; Shan, C.; Dajnoki, Z.; Szegedi, A.; Buhl, T.; Fan, Y.; O’Neill, S.; Walls, D.; et al. Novel insights into the TRPV3-mediated itch in atopic dermatitis. J. Allergy Clin. Immunol. 2021, 147, 1110–1114.e5. [Google Scholar] [CrossRef]
- Choi, J.E.; Di Nardo, A. Skin neurogenic inflammation. Semin. Immunopathol. 2018, 40, 249–259. [Google Scholar] [CrossRef]
- Wilson, S.R.; Nelson, A.M.; Batia, L.; Morita, T.; Estandian, D.; Owens, D.M.; Lumpkin, E.A.; Bautista, D.M. The ion channel TRPA1 is required for chronic itch. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 9283–9294. [Google Scholar] [CrossRef]
- Andersen, H.H.; Elberling, J.; Sølvsten, H.; Yosipovitch, G.; Arendt-Nielsen, L. Nonhistaminergic and mechanical itch sensitization in atopic dermatitis. Pain 2017, 158, 1780–1791. [Google Scholar] [CrossRef]
- Oh, M.H.; Oh, S.Y.; Lu, J.; Lou, H.; Myers, A.C.; Zhu, Z.; Zheng, T. TRPA1-dependent pruritus in IL-13-induced chronic atopic dermatitis. J. Immunol. 2013, 191, 5371–5382. [Google Scholar] [CrossRef] [PubMed]
- Saarnilehto, M.; Chapman, H.; Savinko, T.; Lindstedt, K.; Lauerma, A.I.; Koivisto, A. Contact sensitizer 2,4-dinitrochlorobenzene is a highly potent human TRPA1 agonist. Allergy 2014, 69, 1424–1427. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Chen, C.; Zhou, W.; Ma, X.; Pu, X.; Zeng, Y.; Zhou, W.; Lv, F. TRPA1 deficiency alleviates inflammation of atopic dermatitis by reducing macrophage infiltration. Life Sci. 2021, 266, 118906. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Wheeler, J.J.; Pitake, S.; Ding, H.; Jiang, C.; Fukuyama, T.; Paps, J.S.; Ralph, P.; Coyne, J.; Parkington, M.; et al. Periostin Activation of Integrin Receptors on Sensory Neurons Induces Allergic Itch. Cell Rep. 2020, 31, 107472. [Google Scholar] [CrossRef] [PubMed]
- Lieu, T.; Jayaweera, G.; Zhao, P.; Poole, D.P.; Jensen, D.; Grace, M.; McIntyre, P.; Bron, R.; Wilson, Y.M.; Krappitz, M.; et al. The bile acid receptor TGR5 activates the TRPA1 channel to induce itch in mice. Gastroenterology 2014, 147, 1417–1428. [Google Scholar] [CrossRef] [PubMed]
- Kittaka, H.; Uchida, K.; Fukuta, N.; Tominaga, M. Lysophosphatidic acid-induced itch is mediated by signalling of LPA(5) receptor, phospholipase D and TRPA1/TRPV1. J. Physiol. 2017, 595, 2681–2698. [Google Scholar] [CrossRef]
- Morita, T.; McClain, S.P.; Batia, L.M.; Pellegrino, M.; Wilson, S.R.; Kienzler, M.A.; Lyman, K.; Olsen, A.S.; Wong, J.F.; Stucky, C.L.; et al. HTR7 Mediates Serotonergic Acute and Chronic Itch. Neuron 2015, 87, 124–138. [Google Scholar] [CrossRef]
- Afzal, R.; Shim, W.S. Activation of serotonin receptor 2 by glucosylsphingosine can be enhanced by TRPA1 but not TRPV1: Implication of a novel glucosylsphingosine-mediated itch pathway. Biochim. Et Biophys. Acta Biomembr. 2022, 1864, 184014. [Google Scholar] [CrossRef]
- Ruppenstein, A.; Limberg, M.M.; Loser, K.; Kremer, A.E.; Homey, B.; Raap, U. Involvement of Neuro-Immune Interactions in Pruritus with Special Focus on Receptor Expressions. Front. Med. 2021, 8, 627985. [Google Scholar] [CrossRef]
- Takahashi, S.; Ishida, A.; Kubo, A.; Kawasaki, H.; Ochiai, S.; Nakayama, M.; Koseki, H.; Amagai, M.; Okada, T. Homeostatic pruning and activity of epidermal nerves are dysregulated in barrier-impaired skin during chronic itch development. Sci. Rep. 2019, 9, 8625. [Google Scholar] [CrossRef]
- Denda, M.; Tsutsumi, M.; Goto, M.; Ikeyama, K.; Denda, S. Topical application of TRPA1 agonists and brief cold exposure accelerate skin permeability barrier recovery. J. Investig. Dermatol. 2010, 130, 1942–1945. [Google Scholar] [CrossRef] [PubMed]
- Lexchin, J.; Bero, L.A.; Djulbegovic, B.; Clark, O. Pharmaceutical industry sponsorship and research outcome and quality: Systematic review. BMJ (Clin. Res. Ed.) 2003, 326, 1167–1170. [Google Scholar] [CrossRef] [PubMed]
- Schulz, K.F.; Chalmers, I.; Hayes, R.J.; Altman, D.G. Empirical evidence of bias. Dimensions of methodological quality associated with estimates of treatment effects in controlled trials. JAMA 1995, 273, 408–412. [Google Scholar] [CrossRef] [PubMed]
- Augustin, M.; Misery, L.; Kobyletzski, L.V.; Mealing, S.; Redding, M.; Chuang, C.C.; Massey, R.; Cawkwell, M.; Bego Le-Bagousse, G.; Haddy, L.; et al. Systematic literature review assessing the overall costs and societal impacts of moderate-to-severe atopic dermatitis in Europe. J. Eur. Acad. Dermatol. Venereol. JEADV 2022, 36, 2316–2324. [Google Scholar] [CrossRef]
- Chatrath, S.; Lei, D.; Yousaf, M.; Chavda, R.; Gabriel, S.; Silverberg, J.I. Longitudinal course and predictors of depressive symptoms in atopic dermatitis. J. Am. Acad. Dermatol. 2022, 87, 582–591. [Google Scholar] [CrossRef]
- Iannone, M.; Janowska, A.; Panduri, S.; Morganti, R.; Davini, G.; Romanelli, M.; Dini, V. Impact of psychiatric comorbidities in psoriasis, hidradenitis suppurativa and atopic dermatitis: The importance of a psychodermatological approach. Exp. Dermatol. 2022, 31, 956–961. [Google Scholar] [CrossRef]
- Mann, C.; Dreher, M.; Weeß, H.G.; Staubach, P. Sleep Disturbance in Patients with Urticaria and Atopic Dermatitis: An Underestimated Burden. Acta Derm. Venereol. 2020, 100, adv00073. [Google Scholar] [CrossRef]
- Wan, J.; Shin, D.B.; Syed, M.N.; Abuabara, K.; Lemeshow, A.R.; Gelfand, J.M. Atopic dermatitis and risk of major neuropsychiatric disorders in children: A population-based cohort study. J. Eur. Acad. Dermatol. Venereol. JEADV 2022. [Google Scholar] [CrossRef]
- Schwartzman, G.; Lei, D.; Ahmed, A.; Chavda, R.; Gabriel, S.; Silverberg, J.I. Association of Adult Atopic Dermatitis Severity with Decreased Physical Activity: A Cross-sectional Study. Dermat. Contact Atopic Occup. Drug 2022. [Google Scholar] [CrossRef]
- Gilhooley, E.; O’Grady, C.; Roche, D.; Mahon, J.M.; Hambly, R.; Kelly, A.; Dhonncha, E.N.; Moriarty, B.; Connolly, M.; Kirby, B.; et al. High Levels of Psychological Distress, Sleep Disturbance, and Alcohol Use Disorder in Adults With Atopic Dermatitis. Dermat. Contact Atopic Occup. Drug 2021, 32, S33–S38. [Google Scholar] [CrossRef]
- McCall, M.N.; Murakami, P.N.; Lukk, M.; Huber, W.; Irizarry, R.A. Assessing affymetrix GeneChip microarray quality. BMC Bioinform. 2011, 12, 137. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Bolstad, B.M.; Collin, F.; Cope, L.M.; Hobbs, B.; Speed, T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003, 31, e15. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, D.; Cook, M. A statistical framework for consolidating “sibling” probe sets for Affymetrix GeneChip data. BMC Genom. 2008, 9, 188. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, Article3. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A powerful and practical approach to multiple testing. J. Roy. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swindell, W.R.; Bojanowski, K.; Chaudhuri, R.K. Isosorbide Fatty Acid Diesters Have Synergistic Anti-Inflammatory Effects in Cytokine-Induced Tissue Culture Models of Atopic Dermatitis. Int. J. Mol. Sci. 2022, 23, 14307. https://doi.org/10.3390/ijms232214307
Swindell WR, Bojanowski K, Chaudhuri RK. Isosorbide Fatty Acid Diesters Have Synergistic Anti-Inflammatory Effects in Cytokine-Induced Tissue Culture Models of Atopic Dermatitis. International Journal of Molecular Sciences. 2022; 23(22):14307. https://doi.org/10.3390/ijms232214307
Chicago/Turabian StyleSwindell, William R., Krzysztof Bojanowski, and Ratan K. Chaudhuri. 2022. "Isosorbide Fatty Acid Diesters Have Synergistic Anti-Inflammatory Effects in Cytokine-Induced Tissue Culture Models of Atopic Dermatitis" International Journal of Molecular Sciences 23, no. 22: 14307. https://doi.org/10.3390/ijms232214307
APA StyleSwindell, W. R., Bojanowski, K., & Chaudhuri, R. K. (2022). Isosorbide Fatty Acid Diesters Have Synergistic Anti-Inflammatory Effects in Cytokine-Induced Tissue Culture Models of Atopic Dermatitis. International Journal of Molecular Sciences, 23(22), 14307. https://doi.org/10.3390/ijms232214307





