NB-LRR Lineage-Specific Equipment Is Sorted Out by Sequence Pattern Adaptation and Domain Segment Shuffling
Abstract
1. Introduction
2. Results
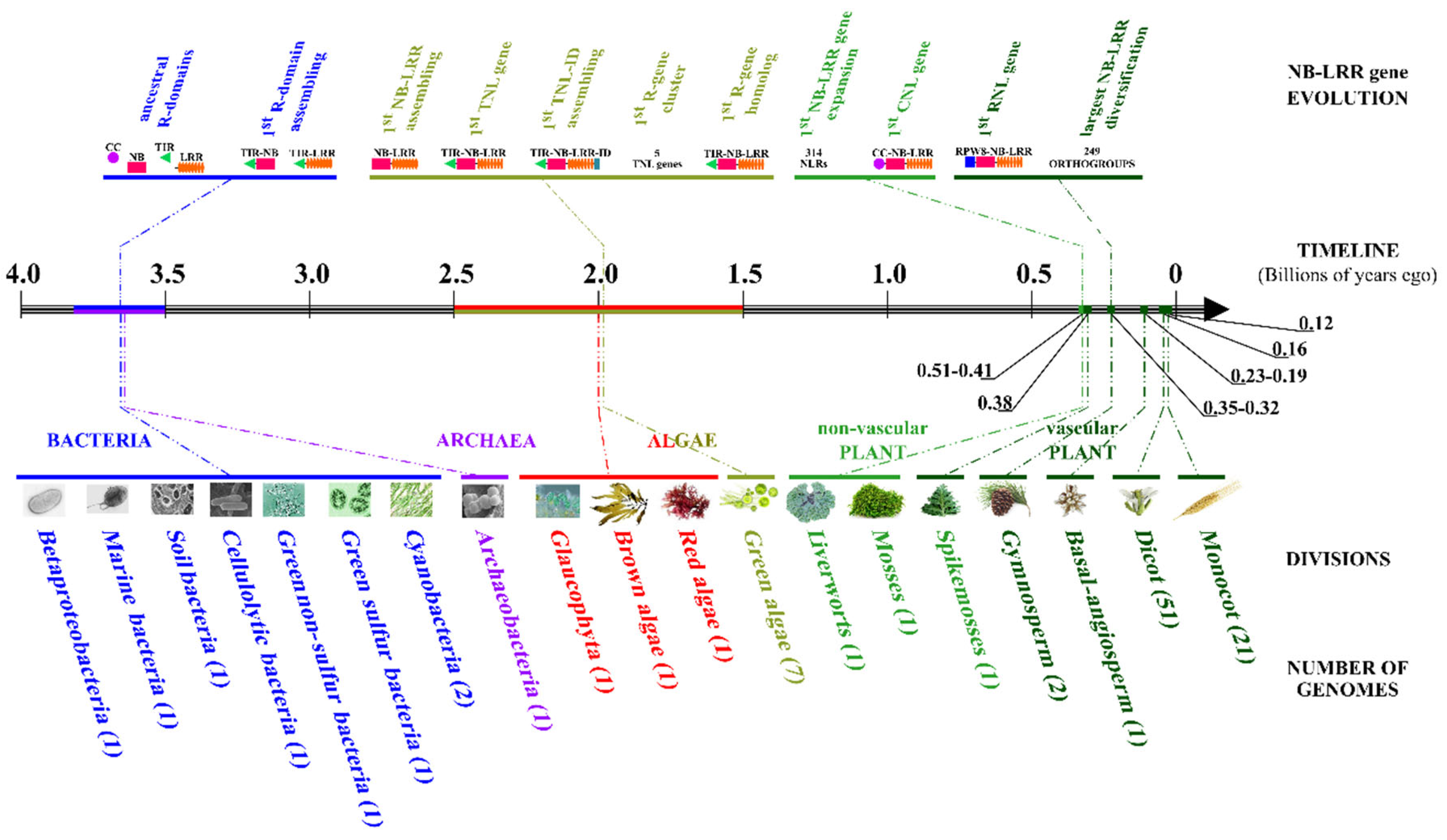
2.1. Diversification of NB-LRR Gene Receptors during Green Plant Evolution
2.2. Lineage-Specific R-gene Profiles in Major Crops
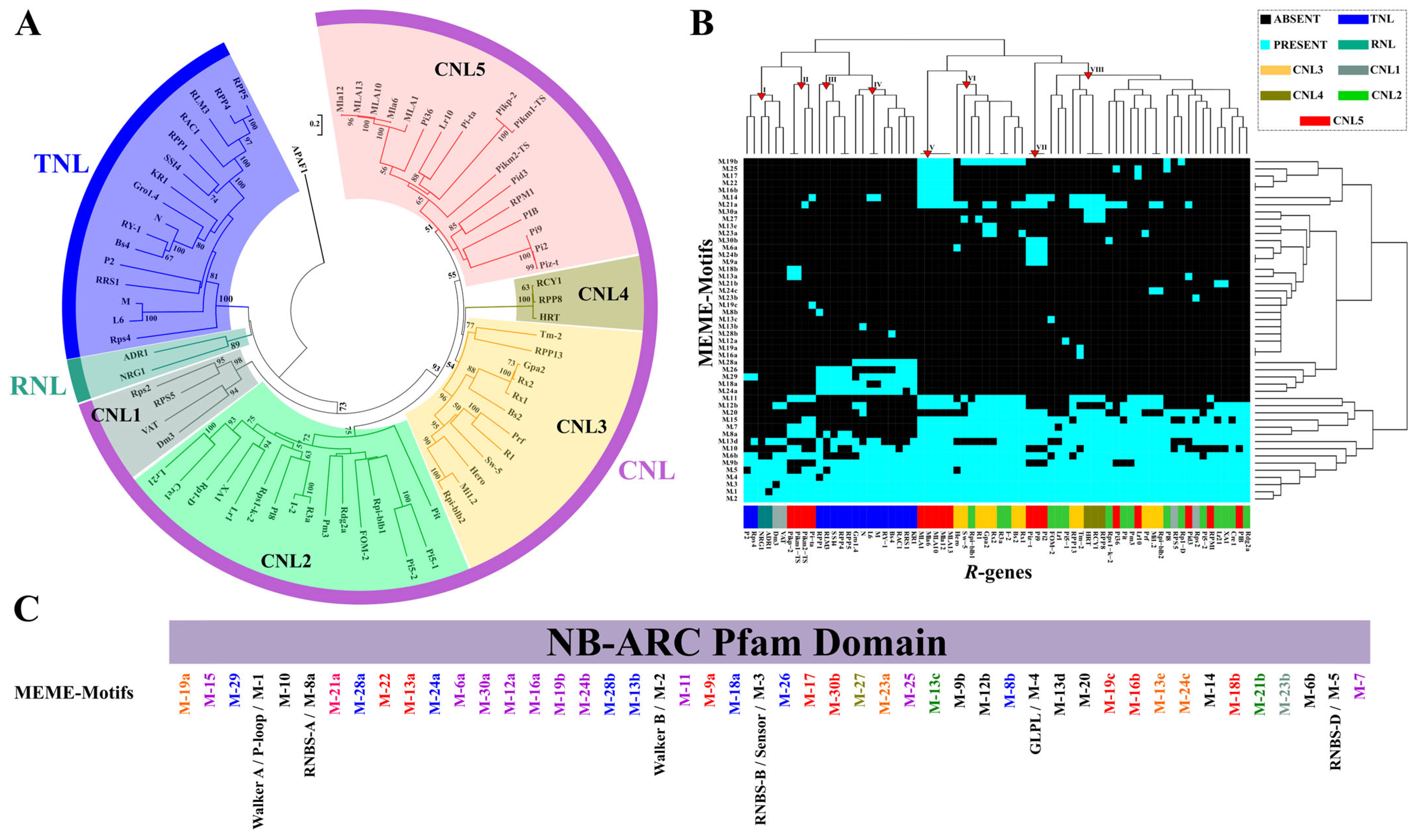
2.3. NB Domain Diversification in a R-genes Core Collection
3. Discussion
4. Materials and Methods
4.1. Taxa Dataset and NB-LRR Gene Annotation
4.2. Identification of Orthologous Groups and Physical R-Clusters
4.3. Maximum Likelihood Analysis
4.4. De Novo Prediction of NB-Encoding Genes Motifs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNL | CC-NB-LRR |
| LRR | Leucine-rich repeat |
| NB | Nucleotide-binding site |
| NB-LRR/NLR | Nucleotide-binding site and leucine-rich repeat |
| RNL | RPW8-NB-LRR |
| R-gene | Resistance gene |
| TNL | TIR-NB-LRR |
| PTI | pathogen-associated molecular pattern triggered immunity |
| ETI | effector-triggered immunity |
References
- Cappetta, E.; Andolfo, G.; Di Matteo, A.; Ercolano, M.R. Empowering Crop Resilience to Environmental Multiple Stress through the Modulation of Key Response Components. J. Plant Physiol. 2020, 246–247, 153134. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-H.; Derevnina, L.; Kamoun, S. Receptor Networks Underpin Plant Immunity. Science 2018, 360, 1300–1301. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The Plant Immune System. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Andolfo, G.; Ercolano, M.R. Plant Innate Immunity Multicomponent Model. Front. Plant Sci. 2015, 6, 987. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.M.; Iswanto, A.B.B.; Son, G.H.; Kim, S.H. Recent Advances in Effector-Triggered Immunity in Plants: New Pieces in the Puzzle Create a Different Paradigm. Int. J. Mol. Sci. 2021, 22, 4709. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-Wide Analysis of NBS-LRR-Encoding Genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef]
- Shao, Z.Q.; Xue, J.Y.; Wang, Q.; Wang, B.; Chen, J.Q. Revisiting the Origin of Plant NBS-LRR Genes. Trends Plant Sci. 2019, 24, 9–12. [Google Scholar] [CrossRef]
- Andolfo, G.; Di Donato, A.; Chiaiese, P.; De Natale, A.; Pollio, A.; Jones, J.D.G.; Frusciante, L.; Ercolano, M.R. Alien Domains Shaped the Modular Structure of Plant NLR Proteins. Genome Biol. Evol. 2019, 11, 3466–3477. [Google Scholar] [CrossRef]
- Ve, T.; Williams, S.J.; Kobe, B. Structure and Function of Toll/Interleukin-1 Receptor/Resistance Protein (TIR) Domains. Apoptosis 2015, 20, 250–261. [Google Scholar] [CrossRef]
- Bent, A.F.; Kunkel, B.N.; Dahlbeck, D.; Brown, K.L.; Schmidt, R.; Giraudat, J.; Leung, J.; Staskawicz, B.J. RPS2 of Arabidopsis thaliana: A Leucine-Rich Repeat Class of Plant Disease Resistance Genes. Science 1994, 265, 1856–1860. [Google Scholar] [CrossRef]
- Zhong, Y.; Cheng, Z.M. A Unique RPW8-Encoding Class of Genes That Originated in Early Land Plants and Evolved through Domain Fission, Fusion, and Duplication. Sci. Rep. 2016, 6, 32923. [Google Scholar] [CrossRef]
- Nieri, D.; Di Donato, A.; Ercolano, M.R. Analysis of Tomato Meiotic Recombination Profile Reveals Preferential Chromosome Positions for NB-LRR Genes. Euphytica 2017, 213, 206. [Google Scholar] [CrossRef]
- Barragan, A.C.; Weigel, D. Plant NLR Diversity: The Known Unknowns of Pan-NLRomes. Plant Cell 2021, 33, 814–831. [Google Scholar] [CrossRef]
- Sanseverino, W.; Ercolano, M.R. In Silico Approach to Predict Candidate R Proteins and to Define Their Domain Architecture. BMC Res. Notes 2012, 5, 678. [Google Scholar] [CrossRef]
- Sarris, P.F.; Cevik, V.; Dagdas, G.; Jones, J.D.G.; Krasileva, K.V. Comparative Analysis of Plant Immune Receptor Architectures Uncovers Host Proteins Likely Targeted by Pathogens. BMC Biol. 2016, 14, 8. [Google Scholar] [CrossRef]
- Bernoux, M.; Ve, T.; Williams, S.; Warren, C.; Valkov, E.; Zhang, X.; Ellis, J.G.; Kobe, B.; Dodds, P.N. TIR Domain Reveals Interfaces for Self-Association, Signaling, and Autoregulation. Cell Host Microbe 2011, 9, 200–211. [Google Scholar] [CrossRef]
- Maekawa, T.; Cheng, W.; Spiridon, L.N.; Töller, A.; Lukasik, E.; Saijo, Y.; Liu, P.; Shen, Q.H.; Micluta, M.A.; Somssich, I.E.; et al. Coiled-Coil Domain-Dependent Homodimerization of Intracellular Barley Immune Receptors Defines a Minimal Functional Module for Triggering Cell Death. Cell Host Microbe 2011, 9, 187–199. [Google Scholar] [CrossRef]
- Tameling, W.I.L.; Elzinga, S.D.J.; Darmin, P.S.; Vossen, J.H.; Takken, F.L.W.; Haring, M.A.; Cornelissen, B.J.C. The Tomato R Gene Products I-2 and Mi-1 Are Functional ATP Binding Proteins with ATPase Activity. Plant Cell 2002, 14, 2929–2939. [Google Scholar] [CrossRef]
- Moffett, P.; Farnham, G.; Peart, J.; Baulcombe, D.C. Interaction between Domains of a Plant NBS-LRR Protein in Disease Resistance-Related Cell Death. EMBO J. 2002, 21, 4511–4519. [Google Scholar] [CrossRef]
- Van Ooijen, G.; Mayr, G.; Kasiem, M.M.A.; Albrecht, M.; Cornelissen, B.J.C.; Takken, F.L.W. Structure-Function Analysis of the NB-ARC Domain of Plant Disease Resistance Proteins. J. Exp. Bot. 2008, 59, 1383–1397. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Dickerman, A.W.; Michelmore, R.W.; Sivaramakrishnan, S.; Sobral, B.W.; Young, N.D. Plant Disease Resistance Genes Encode Members of an Ancient and Diverse Protein Family within the Nucleotide-Binding Superfamily. Plant J. 1999, 20, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Wendel, J.; Fluhr, R. Divergent Evolution of Plant NBS-LRR Resistance Gene Homologues in Dicot and Cereal Genomes. J. Mol. Evol. 2000, 50, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Andolfo, G.; Villano, C.; Errico, A.; Frusciante, L.; Carputo, D.; Aversano, R.; Ercolano, M.R. Inferring RPW8-NLRs’s Evolution Patterns in Seed Plants: Case Study in Vitis vinifera. Planta 2020, 251, 32. [Google Scholar] [CrossRef] [PubMed]
- Bayer, P.E.; Edwards, D.; Batley, J. Bias in Resistance Gene Prediction Due to Repeat Masking. Nat. Plants 2018, 4, 762–765. [Google Scholar] [CrossRef]
- Andolfo, G.; Sanseverino, W.; Aversano, R.; Frusciante, L.; Ercolano, M.R. Genome-wide identification and analysis of candidate genes for disease resistance in tomato. Mol. Breed. 2014, 33, 227. [Google Scholar] [CrossRef]
- Andolfo, G.; Dohm, J.C.; Himmelbauer, H. Prediction of NB-LRR Resistance Genes Based on Full-Length Sequence Homology. Plant J. 2022, 110, 1592–1602. [Google Scholar] [CrossRef]
- Gu, L.; Si, W.; Zhao, L.; Yang, S.; Zhang, X. Dynamic Evolution of NBS–LRR Genes in Bread Wheat and Its Progenitors. Mol. Genet. Genom. 2015, 290, 727–738. [Google Scholar] [CrossRef]
- Di Donato, A.; Andolfo, G.; Ferrarini, A.; Delledonne, M.; Ercolano, M.R.; Crease, T. Investigation of Orthologous Pathogen Recognition Gene-Rich Regions in Solanaceous Species. Genome 2017, 60, 850–859. [Google Scholar] [CrossRef]
- Barchi, L.; Pietrella, M.; Venturini, L.; Minio, A.; Toppino, L.; Acquadro, A.; Andolfo, G.; Aprea, G.; Avanzato, C.; Bassolino, L.; et al. A Chromosome-Anchored Eggplant Genome Sequence Reveals Key Events in Solanaceae Evolution. Sci. Rep. 2019, 9, 11769. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, W.; Bolus, S.; Rouse, M.N.; Dubcovsky, J. Identification and Characterization of Wheat Stem Rust Resistance Gene Sr21 Effective against the Ug99 Race Group at High Temperature. PLoS Genet. 2018, 14, e1007287. [Google Scholar] [CrossRef]
- Wang, X.; Yan, X.; Hu, Y.; Qin, L.; Wang, D.; Jia, J.; Jiao, Y. A Recent Burst of Gene Duplications in Triticeae. Plant Commun. 2022, 3, 100268. [Google Scholar] [CrossRef]
- Feehan, J.M.; Castel, B.; Bentham, A.R.; Jones, J.D. Plant NLRs Get by with a Little Help from Their Friends. Curr. Opin. Plant Biol. 2020, 56, 99–108. [Google Scholar] [CrossRef]
- Ameline-Torregrosa, C.; Wang, B.B.; O’Bleness, M.S.; Deshpande, S.; Zhu, H.; Roe, B.; Young, N.D.; Cannon, S.B. Identification and Characterization of Nucleotide-Binding Site-Leucine-Rich Repeat Genes in the Model Plant Medicago Truncatula. Plant Physiol. 2008, 146, 5–21. [Google Scholar] [CrossRef]
- Yaish, M.W.F.; Sáenz De Miera, L.E.; Pérez De La Vega, M. Isolation of a Family of Resistance Gene Analogue Sequences of the Nucleotide Binding Site (NBS) Type from Lens Species. Genome 2004, 47, 650–659. [Google Scholar] [CrossRef]
- Palomino, C.; Satovic, Z.; Cubero, J.I.; Torres, A.M. Identification and Characterization of NBS-LRR Class Resistance Gene Analogs in Faba Bean (Vicia faba L.) and Chickpea (Cicer arietinum L.). Genome 2006, 49, 1227–1237. [Google Scholar] [CrossRef]
- Kanazin, V.; Marek, L.F.; Shoemaker, R.C. Resistance Gene Analogs Are Conserved and Clustered in Soybean. Proc. Natl. Acad. Sci. USA 1996, 93, 11746–11750. [Google Scholar] [CrossRef]
- Rairdan, G.J.; Moffett, P. Distinct Domains in the ARC Region of the Potato Resistance Protein Rx Mediate LRR Binding and Inhibition of Activation. Plant Cell 2006, 18, 2082–2093. [Google Scholar] [CrossRef]
- Takken, F.L.; Albrecht, M.; Tameling, W. IL Resistance Proteins: Molecular Switches of Plant Defence. Curr. Opin. Plant Biol. 2006, 9, 383–390. [Google Scholar] [CrossRef]
- Sueldo, D.J.; Shimels, M.; Spiridon, L.N.; Caldararu, O.; Petrescu, A.J.; Joosten, M.H.A.J.; Tameling, W.I.L. Random Mutagenesis of the Nucleotide-Binding Domain of NRC1 (NB-LRR Required for Hypersensitive Response-Associated Cell Death-1), a Downstream Signalling Nucleotide-Binding, Leucine-Rich Repeat (NB-LRR) Protein, Identifies Gain-of-Function Mutations in the Nucleotide-Binding Pocket. New Phytol. 2015, 208, 210–223. [Google Scholar] [CrossRef]
- Andolfo, G.; Iovieno, P.; Frusciante, L.; Ercolano, M.R. Genome-Editing Technologies for Enhancing Plant Disease Resistance. Front. Plant Sci. 2016, 7, 1813. [Google Scholar] [CrossRef] [PubMed]
- Hanson, P.I.; Whiteheart, S.W. AAA+ Proteins: Have Engine, Will Work. Nat. Rev. Mol. Cell Biol. 2005, 6, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Contreras, M.; Harant, A.; Wu, C.H.; Derevnina, L.; Sakai, T.; Duggan, C.; Moratto, E.; Bozkurt, T.O.; Maqbool, A.; et al. An N-Terminal Motif in NLR Immune Receptors Is Functionally Conserved across Distantly Related Plant Species. Elife 2019, 8, e49956. [Google Scholar] [CrossRef]
- Alexeyenko, A.; Tamas, I.; Liu, G.; Sonnhammer, E.L.L. Automatic Clustering of Orthologs and Inparalogs Shared by Multiple Proteomes. Bioinformatics 2006, 22, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Richly, E.; Kurth, J.; Leister, D. Mode of Amplification and Reorganization of Resistance Genes during Recent Arabidopsis Thaliana Evolution. Mol. Biol. Evol. 2002, 19, 76–84. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Warnes, A.G.R.; Bolker, B.; Bonebakker, L.; Huber, W.; Liaw, A.; Lumley, T.; Magnusson, A.; Moeller, S.; Schwartz, M. Package ‘ Gplots’. Available online: https://cran.r-project.org/web/packages/gplots/gplots.pdf (accessed on 6 October 2022).

| Plant Family | Average Number of Orthogroups | Average Number of Paralogs | Average Number of Gene Clusters |
|---|---|---|---|
| Brassicaceae | 52 (39–72) | 171 (105–322) | 128 (119–135) |
| Fabaceae | 70 (49–89) | 470 (295–803) | 392 (220–706) |
| Solanaceae | 96 (53–156) | 306 (157–645) | 338 (141–622) |
| Poaceae | 94.5 (41–149) | 384 (25–1033) | 172 (15–666) |
| Cucurbitaceae | 24 (20–27) | 54 (41–62) | 29 (-) |
| R-Gene of Group I | Protein Class | Orthogroup ID | Number of Orthologs | Number of Genomes |
|---|---|---|---|---|
| NRG1 | RNL | OG1028 | 227 | 44 |
| ADR1 | RNL | OG1033 | 193 | 77 |
| VAT | CNL | OG1169 | 17 | 6 |
| RPS4 | TNL | OG1043 | 128 | 10 |
| Dm3 | CNL | OG1093 | 44 | 1 |
| P2 | TNL | OG1106 | 34 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andolfo, G.; Di Donato, A.; Ercolano, M.R. NB-LRR Lineage-Specific Equipment Is Sorted Out by Sequence Pattern Adaptation and Domain Segment Shuffling. Int. J. Mol. Sci. 2022, 23, 14269. https://doi.org/10.3390/ijms232214269
Andolfo G, Di Donato A, Ercolano MR. NB-LRR Lineage-Specific Equipment Is Sorted Out by Sequence Pattern Adaptation and Domain Segment Shuffling. International Journal of Molecular Sciences. 2022; 23(22):14269. https://doi.org/10.3390/ijms232214269
Chicago/Turabian StyleAndolfo, Giuseppe, Antimo Di Donato, and Maria Raffaella Ercolano. 2022. "NB-LRR Lineage-Specific Equipment Is Sorted Out by Sequence Pattern Adaptation and Domain Segment Shuffling" International Journal of Molecular Sciences 23, no. 22: 14269. https://doi.org/10.3390/ijms232214269
APA StyleAndolfo, G., Di Donato, A., & Ercolano, M. R. (2022). NB-LRR Lineage-Specific Equipment Is Sorted Out by Sequence Pattern Adaptation and Domain Segment Shuffling. International Journal of Molecular Sciences, 23(22), 14269. https://doi.org/10.3390/ijms232214269






