1. Introduction
In the course of billions of evolution years, the mitochondria dramatically reduced the genome of their ancestors: almost all protomitochondrial genes were transferred to the nucleus or lost [
1]. Nowadays, most of the genes encoding mitochondria-specific proteins are localized and transcribed in the nucleus, their mRNAs are translated in the cytosol, and the proteins are imported into mitochondria via specific protein transport machinery [
2]. Nevertheless, most of modern mitochondria retained significantly reduced genomes containing limited set of genes along with the apparatus of genome maintenance and gene expression. In addition to the genes of ribosomal and transfer RNAs, mitochondrial genomes contain a set of protein-coding genes. The human mitochondrial genome contains 13 protein-coding genes, all of which encode subunits of electron transport chains [
3].
Mitochondrial protein synthesis was thought to be similar to bacterial translation for decades. However, recent advances in cryo-electron microscopy and mitochondrial molecular biology revealed profound differences between the mitochondrial and bacterial protein synthesis machineries observed in the structures of ribosomes, mechanisms of translation initiation, and other aspects [
3,
4,
5,
6,
7]. One of the peculiarities of mitochondrial translation worth noting is its membrane tethering, which facilitates the biosynthesis and insertion of OXPHOS subunits. As it was mentioned above, all proteins encoded in human mitochondrial DNA (mtDNA) are the subunits of electron transport chain complexes; however they altogether present a minor part of the complete set, whereas the major part is synthesized in cytosol. Therefore, cytosolic and mitochondrial translation processes must be tightly coordinated for the correct assembly of OXPHOS complexes. Such coordination is well described in baker’s yeast mitochondria, where it is modulated by a group of specific regulatory proteins—translational activators [
4]. The efficiency of translation initiation of every mitochondrial mRNA of
S. cerevisiae is governed by binding of specific translational activator to prolonged 5′-UTR and/or mitochondrial ribosome [
4]. Human mitochondrial mRNAs are leaderless and do not have any significant 5′-UTRs [
5]. At first glance, this fact makes translational activators unnecessary; however, the existence of analogous regulatory system cannot be excluded.
Pentatricopeptide-containing proteins (PPR proteins) are good candidates for the role of the regulators of mRNA translation in human mitochondria. These proteins belong to the α-solenoid RNA-binding proteins involved in almost all steps of the life cycle of mRNAs [
6]. All PPR proteins contain varying numbers of tandemly repeated sequence motifs of 35 amino acids (the PPR motifs) and are abundant in plant organelles but also found in all eukaryotic lineages [
7]. Proteins containing PPR motifs are known to have roles in transcription, RNA processing, splicing, stability, editing, and translation. Many yeast mitochondrial translational activators are PPR proteins [
4]. The human genome contains several genes encoding mitochondrial PPR proteins, e.g.,
POLMT,
LRPPRC,
PTCD1,
PTCD2,
PTCD3,
MRPS27, etc. [
8].
The focus of our research is PTCD2, a mysterious protein whose function was unknown despite the available data. Its mitochondrial localization and PPR family membership suggested its possible biological role in regulation of mitochondrial gene expression. A PTCD2 knock-out mouse model was studied previously. It demonstrated the reduction in activity of complex III in the heart, an increase in activity of complex IV in heart, muscle and liver along with defects in apocytochrome b mRNA maturation [
9]. However, the published model was not a true knock-out because truncated protein was still synthesized, and only exons 9 and 10 were missing. We made disruption of PTCD2 gene in human HeLa cells by introducing a frame-shift mutation close to the start codon and analyzed their mitochondrial translation, oxygen consumption, OXPHOS complex activity, and supercomplex formation. We show that PTCD2 deletion selectively decreased translation of COIII mRNA and the activity of the respiratory chain complex IV (CIV). As a consequence, it affects respiration and supercomplex formation. In addition, we obtained the data from sedimentation analysis and pull-down proteomics which allow us to suggest the interaction of PTCD2 with mitochondrial ribosomes.
3. Discussion
Regulation of human mitochondrial translation is far from being completely explained despite recent advances in structural biology providing insights in different stages of the life cycle of mitochondrial ribosomes. The molecular mechanisms governing the expression of each individual human mitochondrial mRNA and orchestrating its cross-talk with cytosolic translation are poorly studied. Such regulation in baker’s yeast is carried out via translational activators—special proteins which bind predominantly 5′-UTRs of mRNAs and increase its affinity to mitochondrial ribosome. Due to the absence of prolonged 5′-UTRs in human mitochondrial mRNAs, the existence of such system in human mitochondria has been questioned. Nevertheless, it was shown in 2009 that protein TACO1 selectively regulates the mitochondrial translation of COI mRNA [
11]. The mechanism of such regulation remains unclear but provides a proof of principle for the existence of translational activators for other mRNAs. The good candidates for being translational activators are pentatricopeptide-containing proteins (PPR proteins). In this work we probed PTCD2 PPR protein for its ability to affect mitochondrial translation in human HeLa cell line mitochondria.
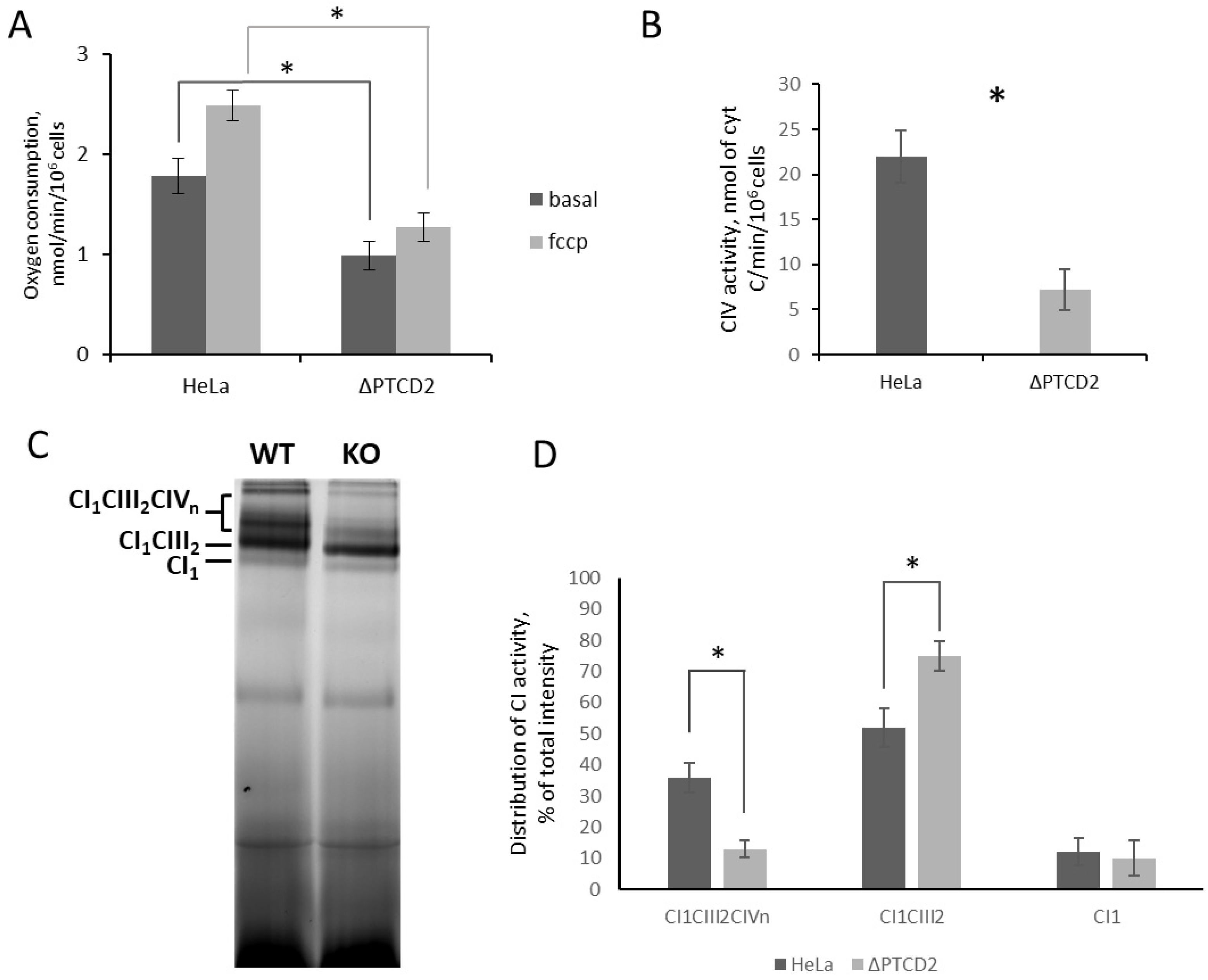
We found that functional deletion in the PTCD2 gene significantly decreased translation efficiency of COIII mRNA and led to reduced steady-state level of this mitochondrially encoded protein. The decrease in intensity observed in the band corresponding to COII/COIII in radio autography could not be attributed to any of these two proteins, but steady-state level of COIII assessed by Western blotting decreased much more significantly than COII. It suggests that a decrease in COII stationary level was caused by partial loss of COIII and incorrect assembly of cytochrome c oxidase.
The decrease in the amount of COIII, which is one of CIV subunits, should affect CIV activity and overall mitochondrial fitness. We found that the absence of PTCD2 in mitochondria of HeLa cells leads to significantly lower oxygen consumption rates in the presence and absence of uncoupler reagent compared to wild-type cells. Moreover, the OCRs of knock-out cells in both conditions were equal whereas in wild-type cells these parameters usually differ double to triple. This could be explained by the fact that normally CIV has an excess capacity necessary for fast adaptation of OXPHOS to changes in the environment [
12]. The amount of CIV is reduced in
PTCD2 gene knock-out cells, and therefore all CIV capacity is in use at coupled conditions.
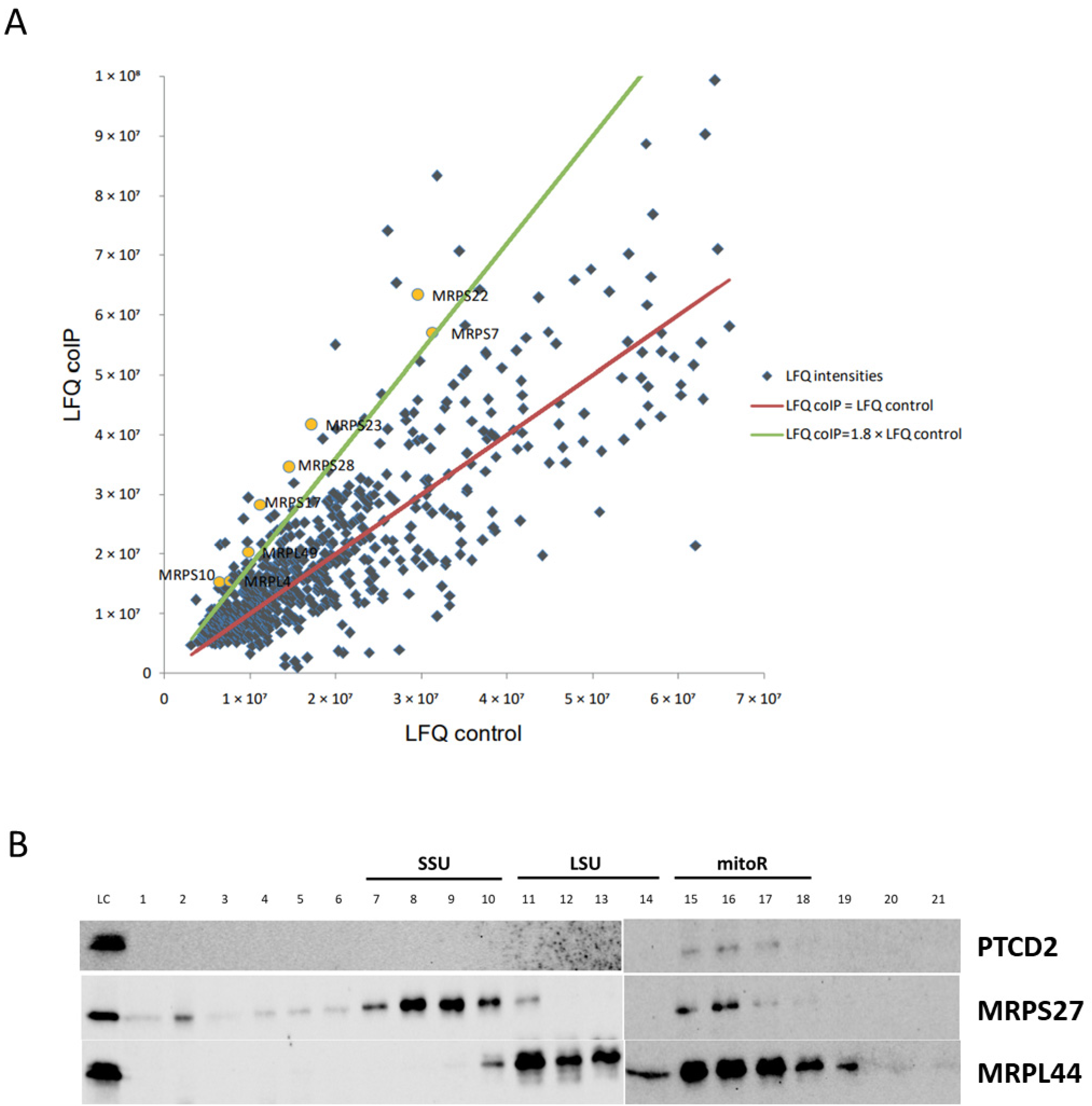
Almost all translational activators in baker’s yeast mitochondria interact with mitoribosome and/or mRNAs. Using coIP and proteomic approaches, we found that PTCD2 interacts with a portion of SSU and LSU proteins. This fact allows suggesting that PTCD2 is associated with mitochondrial translation apparatus. Moreover, PTCD2 was found co-sedimentated with associated mitoribosome but not with SSU or LSU alone. These observations suggest that PTCD2 could interact with a part of translating mitoribosomes. However, we cannot conclude the direct interaction between PTCD2 and mitochondrial ribosome as they both could interact with mRNA. We tried to detect any interactions of recombinant PTCD2 with mRNAs (COII, COIII, COI, CytB, and ND5) by several EMSA approaches, but in our experimental conditions such interactions were not found (Probably, PTCD2–mRNA interaction requires some additional factors, e.g., mitochondrial ribosome, CIV assembly factors, or other proteins. On the other hand, the molecular mechanism of translational regulation by PTCD2 does not necessarily involve its direct interaction with mRNA. It is possible that interaction of PTCD2 with associated mitoribosome is required for efficient elongation rather than COIII mRNA recognition. We also checked the assumption that decreased levels of COIII could be the result of PTCD2 being an assembly factor of CIV. In Blue-Native Western blots PTCD2 was not detected in association with any complexes.
Another explanation for PTCD2 role in COIII translation may be based on the presence of MRPS27 domain in its structure. It can be assumed that PTCD2 can weakly compete with MRPS27 for binding to mitochondrial ribosome, and mitoribosomes which contain PTCD2 translate COIII mRNA more efficiently. Nevertheless, the precise molecular mechanism of PTCD2 action should be studied during further research.
The latest published data based on mitochondrial ribosome profiling have shown that translation initiation of COIII in human mitochondria can be carried out from completely processed leaderless and unprocessed mRNA with prolonged 5′-UTR [
13]. The frequency of COIII mRNA translation initiation at processed or unprocessed transcripts varies across cell types. It could be assumed that PTCD2 is involved in initiation on one of these templates, which explains the reduction but not the complete arrest of COIII mRNA translation in our experiments. Moreover, the mode of PTCD2 action could be tissue-specific.
In summary, we have shown that deletion of PTCD2 leads to decrease in COIII translation in mitochondria, impairs the activity of CIV, OCR, and supercomplex composition in human HeLa cell mitochondria. Moreover, PTCD2 could be detected within the fraction of associated mitochondrial ribosomes and could bind mitoribosomal proteins in pull-down assays. Taken together, these facts allow us to suppose that PTCD2 can selectively regulate COIII translation in human HeLa cell mitochondria.
4. Materials and Methods
4.1. Strains, Media, Cell Lines
E. coli strains Top-10 One Shot (Thermo Scientific, Waltham, MA, USA) and BL21(DE3)/pLysS (Promega, Madison, WI, USA) were used for standard cloning and protein expression procedures. Human cell lines, HeLa (CCL-2 ATCC) and its derivatives, were cultured in DMEM (Corning, New York, NY, USA) with 4.5 g/L glucose, Corning @glutargo, sodium pyruvate, and 10% (v/v) FBS (Sigma, St. Louis, MO, USA), under a 5% CO2 humidified atmosphere at 37 °C. To prevent contamination DMEM was supplied with 1000 units/mL of penicillin, 100 µg/mL of streptomycin, and 2.5 µg/mL of Amphotericin B (all Sigma, St. Louis, MO, USA).
4.2. Generation of Knock-Out Cell Lines
The generation of knock-out cell lines with the CRISPR/Cas9 technology was performed as previously described in [
14]. sgRNA design was performed using CRISPOR online platform (
http://crispor.tefor.net (accessed on 2 October 2021)) [
15]. The selected sgRNA spacer cDNA sequences (5′-CACCGCAAAATCTGCAGCGCCTGCG-3′ and 5′-AAACGCAGGCGCTGCAGATTTTGCG-3′ for sgRNA1 and 5′-CACCGGCCCTCTCGGAGGTATCCG-3′ and 5′-AAACCGGATACCTCCGAGAGGGCC-3′ for sgRNA2) were synthesized (Evrogen, Moscow, Russia) and inserted between BbsI sites in the pU6-gRNA vector containing sgRNA scaffold (kindly provided by Dr. Boris Skryabin, University of Munster). HeLa cells were co-transfected with sgRNAs-pU6 vectors together with an Cas9/EGFP-containing plasmid [
16] using Lipofectamine 3000 reagent (Thermofisher Scientific, Waltham, MA, USA). Cells were sorted using FACS Aria SORP (Beckton Dickinson Biosciences, East Rutherford, NJ, USA). The gene editing was confirmed by PCR with specific primers (5′-GGACGAATCCCTTTTGTCGC-3′ and 5′-ATGCTAGCTGGGATCTAACAC-3′) followed by Sanger sequencing of resulting products.
4.3. Generation of Stable Cell Lines
Cell line expressing C-terminal c-Myc-tagged
PTCD2 under the control of a tetracycline-inducible promoter were generated in
PTCD2 gene knock-out HeLa cell line. Cells were co-transfected with the pSBtet-Neo vector (pSBtet-Neo was a gift from Eric Kowarz (Addgene plasmid #60509;
http://n2t.net/addgene:60509 (accessed on 1 November 2022); RRID:Addgene_60509)) containing the sequence of
PTCD2 gene with C-terminal c-Myc-tag, and the pCMV(CAT)T7-SB100 vector (pCMV(CAT)T7-SB100 was a gift from Zsuzsanna Izsvak (Addgene plasmid #34879;
http://n2t.net/addgene:34879 (accessed on 1 November 2022); RRID:Addgene_34879)) using Lipofectamin 3000 reagent (Thermofisher Scientific, Waltham, MA, USA). Selection of transfected cells was performed with G418 (1 mg/mL) for 14 days following cultivation in the media supplemented with 100 ng/mL of G418. The insertion of
PTCD2-c-myc was confirmed by PCR followed by detection of c-myc tagged protein after doxycycline induction by Western blotting.
4.4. Mitochondrial Translation Assay
Mitochondrial translation products were labeled as described in [
16]. Briefly, cells were pre-starved with methionine-free DMEM supplemented with 10% dialyzed FBS. Cytosolic translation was inactivated with 0.2 mg/mL of cycloheximide for 5 min. Then 0.5 mCi/mL [
35S] methionine was added. After incubation at 37 °C for 45 min the medium was replaced with fully supplemented DMEM containing cold methionine and the cells were harvested. Samples were further analyzed by 18% SDS-PAGE and autoradiography. Signals were recorded on Storage Phosphor screens using a STORM 865 scanner (GE Healthcare, Marlborough, MA, USA). ImageJ software (NIH, Bethesda, MD, USA) was used for quantification.
4.5. Western Blot
Western blot analyses were performed conventionally using following antibodies: anti-PTCD2 (NBP2-26176, Novusbio, Centennial, CO, USA), anti-MRPS27 (17280-1-AP, Proteintech, Manchester, UK), anti-MRPL44 (16394-1-AP, Proteintech, Manchester, UK), anti-Cox4 (11242-1-AP, Proteintech, Manchester, UK), anti-COII (55070-1-AP, Proteintech, Manchester, UK), anti-COIII (55082-1-AP, Proteintech, Manchester, UK), anti-goat IgG HRP conjugated (HAF017, Novusbio, Centennial, CO, USA), ECL Anti-rabbit IgG HRP linked (NA934V, GE Healthcare, Marlborough, MA, USA), according to manufacturers’ protocols. Blots were stained with SuperSignal West Pico ECL reagents (ThermoFisher Scientific, Waltham, MA, USA) and visualized with ChemiDoc imaging system (BioRad, Hercules, CA, USA).
4.6. Isolation of Mitochondria
The isolation of human mitochondria was performed according to [
17]. Cells were harvested, washed with PBS, and resuspended and swelled for 20 min in RSB Hypo buffer (10 mM NaCl, 10 mM MgCl
2, 1.5 mM Tris-HCl, pH 7.5). Cells were homogenized by tight pestle in a Dounce homogenizer. After addition of 2.5xMS Buffer (525 mM mannitol, 175 mM sucrose, 2.5 mM EDTA, 12.5 mM Tris-HCl, pH 7.5), cell debris was pelleted at 1300×
g for 5 min at 4 °C. The mitochondria were pelleted by centrifugation at 18,000×
g for 15 min at 4 °C. Mitochondria quantity was estimated by measurement of total mitochondria protein concentration by Bradford assay. Mitochondria pellets were stored at −80 °C until use.
4.7. Blue-Native Electrophoresis
BN-PAGE was performed as described in [
18]. Briefly, 100 μg of isolated mitochondria were resuspended in 10 μL of solubilization buffer A (50 mM NaCl, 2 mM 6-aminohexanoic acid, 1 mM EDTA, 50 mM imidazole-HCl, pH 7) and a proper amount of detergent was added (1.25 μL of dodecylmaltoside (20%) or 3 μL of digitonin (20%)). The mitochondria were solubilized 20 min on ice and debris was removed by 1 h centrifugation at 4 °C and 25,000×
g. Coomassie dye (5% Coomassie Blue G-250 dye stock solution, suspended 500 mM 6-aminohexanoic acid) was added to the supernatant to give a detergent/dye ratio of 8, and the samples were subjected to electrophoresis in a 4–14% gradient gel. After electrophoresis, either in-gel respiratory complex activities were measured, or Western blot analysis was performed. The measurement of complex I activity the gel was performed according to [
19]. Gels were incubated in solution containing 0.14 mM NADH, 1 mg/mL Nitro Blue tetrazolium (NBT) and 0.1 M Tris HCl, pH 7.4, until the appearance of violet bands. The reaction was stopped with 10% acetic acid solution.
4.8. CIV Activity
A reaction mixture containing 200 µL buffer (10 mM potassium phosphate buffer, 250 mM sucrose, 1 mg/mL BSA, pH 6.5), 10 µM reduced cytochrome c, and 2 µL dodecylmaltoside solution (20%) was added into a well of 96-well plate. The reaction was started by adding 2–10 × 104 cells, which were frozen/thawed two or three times in isotonic medium before using. To measure the activity of cytochrome c oxidase, the decrease in absorbance at 550 nm was monitored for 3 min using Infinite M200Pro plate reader (Tecan, Männedorf, Switzerland). The specificity of the activity of complex IV was checked by addition of 2 μL of 1M potassium cyanide (KCN). The resulting activity was calculated using formula “CIV activity (nmol cyt c/min/106 cells) = (ΔA(550 nm)/min)/(ε × cell number), where ε = extinction coefficient for reduced cytochrome c 18.5 mM−1cm−1.
4.9. Fractionation of Mitochondrial Ribosomes
Approximately 107 cells were collected, washed with PBS and lysed in 1 mL of 10 mM HEPES pH 7.4, 120 mM KCl, 20 mM magnesium acetate, 5 mM β-mercaptoethanol, 1.7% Triton X-100 for 30 min on ice. After clarification at 25,000× g for 30 min, lysates were applied to 10–40% linear sucrose gradient prepared on the same buffer without detergent. After ultracentrifugation at 80,000× g for 18 h (SW-40Ti rotor, Beckman Optima XE-90 centrifuge) gradients were fractionated with Gradient Station ip (Biocomp, Fredericton, NB, Canada). Proteins in fractions were sedimentated with methanol/chloroform and applied to Western blotting.
4.10. Recombinant PTCD2 Cloning, Expression and Purification
PTCD2 cDNA was amplified from total HeLa cDNA using 5′-ATCGAGGCCTCTGAGGCCATGGTCCGAGACAGTATGG-3′ and 5′-ATCGAGGCCTGACAGGCCTTAAAGATCTTCTTCGCTAATAAGTTTTTGTTCCTCAGCCAACAGGGACTGG. The fragment was cloned into pET30a vector (Novagen), and verified by Sanger sequencing. Resulting vector was transformed into BL21(DE3)/pLysS E. coli strain. 5 mL of LB medium supplemented with 50 ug/mL kanamycin and 20 ug/mL chloramphenicol were used as starting overnight culture grown at 37 °C with agitation; 1L of LB with appropriate antibiotics were inoculated with starting culture and incubated at 30 °C and agitation until OD600 reached 0.7–0.9. Expression was induced by adding IPTG up to 0.25 mM, cells were incubated for 4 h at 30 °C. Then cells were collected by centrifugation at 3000× g for 10 min, washed with PBS, resuspended in 40 mL of PBS, and lysed by sonication (10 × 15 s at 30% amplitude with 45 s intervals, Branson digital sonifier). Inclusion bodies were pelleted at 25,000× g for 20 min and dissolved in 7 mL of 25 mM Tris-HCl pH 7.5, 500 mM NaCl, 25 mM imidazole, 8M urea. After centrifugation (25,000× g, 20 min), dissolved inclusion bodies were applied to 1 mL Ni-NTA column (HisTrap, GE Healthcare, Marlborough, MA, USA) connected to AKTA Purifier chromatography system (GE Healthcare, Marlborough, MA, USA) and pre-equilibrated with the same buffer. After extensive washing with the same buffer (40 column volumes), recombinant PTCD2 was eluted with the same buffer supplemented with 300 mM of imidazole. After elution fractions contained PTCD2 were combined and dialyzed against 4 changes of 25 mM sodium phosphate buffer pH 7.5, 150 mM NaCl for 24 h. Protein concentration was measured by Coomassie Protein Assay (Thermo Fisher Scientific, Waltham, MA, USA).
4.11. Co-Immunoprecipitation
0.5 mg of HeLa mitochondria were resuspended in solubilizing buffer (50 mM imidazole-HCl (pH 7.0), 50 mM NaCl, 1 mM EDTA, 2 mM 6-aminohexanoic acid) supplemented with 2% dodecylmaltoside. After incubation for 40 min on ice, the insoluble material was removed by centrifugation at 18,000× g for 1 h at 4 °C. The supernatant was incubated first with 25 µg of recombinant 6xHis-tagged PTCD2 for 1 h at 4 °C with gentle agitation and then with Ni-NTA-Sepharose (Qiagene, Hilden, Germany). After washing the resin, the proteins were eluted according to the manufacturer’s standard protocol. The eluted proteins were precipitated by methanol/chloroform and subjected to proteomic analysis.
4.12. Comparative Proteomics
Proteome analysis was performed as described in [
20] with minor modifications. Eluates from the IP were loaded on to the 10% denaturing PAAG and when the bromophenol blue dye passed 5 mm of the separating gel current was stopped and the band containing total concentrated proteins was excised from the gel and sliced into pieces. Excised samples were washed twice with solution 1 (40% acetonitrile and 50 mM NH
4HCO
3). The pieces of gel were dried in acetonitrile, rehydrated in solution 2 (100 mM NH
4HCO
3 and 20 mM DTT) and incubated for 30 min at 56 °C. Next solution 2 was discarded and the samples were incubated in solution 3 (100 mM NH
4HCO
3 and 20 mM iodoacetamide) in the dark for 20 min, washed twice with water, three times with solution 1, dried in acetonitrile and trypsinized with Trypsin Gold (Promega, WI, USA) in 100 mM NH
4HCO
3 at 37 °C for 6 h. Peptides were desalted using Pierce C18 Tips, 100 ul according to manufacturer’s instructions. Peptides eluted from the tips were dried on vacuum manifold and solubilized in 0.1% formic acid.
One microgram of peptides in a volume of 1–4 uL was loaded onto the Acclaim μ-Precolumn (0.5 mm × 3 mm, 5 μm particle size, Thermo Scientific) at a flow rate of 10 uL/min for 4 min in an isocratic mode of Mobile Phase C (2% acetonitrile, 0.1% formic acid). Then the peptides were separated with high-performance liquid chromatography (HPLC, Ultimate 3000 Nano LC System, Thermo Scientific, Rockwell, IL, USA) in a 15 cm long C18 column (Acclaim® PepMap™ RSLC inner diameter of 75 μm, Thermo Fisher Scientific, Rockwell, IL, USA). The peptides were eluted with a gradient of buffer B (80% acetonitrile, 0.1% formic acid) at a flow rate of 0.3 μL/min. Total run time was 90 min, which included initial 4 min of column equilibration to buffer A (0.1% formic acid), then gradient from 5 to 35% of buffer B over 65 min, then 6 min to reach 99% of buffer B, flushing 10 min with 99% of buffer B and 5 min re-equilibration to buffer A.
MS analysis was performed at least in triplicate with a Q Exactive HF-X mass spectrometer (Q Exactive HF-X Hybrid Quadrupole-Orbitrap™ Mass spectrometer, Thermo Fisher Scientific, Rockwell, IL, USA). The temperature of capillary was 240 °C and the voltage at the emitter was 2.1 kV. Mass spectra were acquired at a resolution of 120,000 (MS) in a range of 300−1500 m/z. Tandem mass spectra of fragments were acquired at a resolution of 15,000 (MS/MS) in the range from 100 m/z to m/z value determined by a charge state of the precursor, but no >2000 m/z. The maximum integration time was 50 and 110 ms for precursor and fragment ions, correspondently. AGC target for precursor and fragment ions were set to 1 × 106 and 2 × 105, respectively. An isolation intensity threshold of 50,000 counts was determined for precursor’s selection and up to top 20 precursors were chosen for fragmentation with high-energy collisional dissociation (HCD) at 29 NCE. Precursors with a charge state of +1 and more than +5 were rejected and all measured precursors were dynamically excluded from triggering of a subsequent MS/MS for 20 s.
The obtained raw data were processed using the MaxQuant software [
21] with the built-in search engine Andromeda [
22]. All samples from one biological replicate were analyzed in one run. Protein sequences of the complete human proteome provided by Uniprot were used for protein identification with Andromeda. Carbamidomethylation of cysteines was set as fixed modification and protein N-terminal acetylation as well as oxidation of methionines was set as variable modification for the peptide search. A maximum mass deviation of 4.5 ppm was allowed for precursor’s identification and 20 ppm were set as match tolerance for fragment identification (acquisition in Orbitrap). Up to two missed cleavages were allowed for trypsin digestion. The false discovery rates (FDR) for peptide and protein identifications were set to 1%. Unique and razor peptides were used for label-free quantification (LFQ).