Robust Classification Using Posterior Probability Threshold Computation Followed by Voronoi Cell Based Class Assignment Circumventing Pitfalls of Bayesian Analysis of Biomedical Data
Abstract
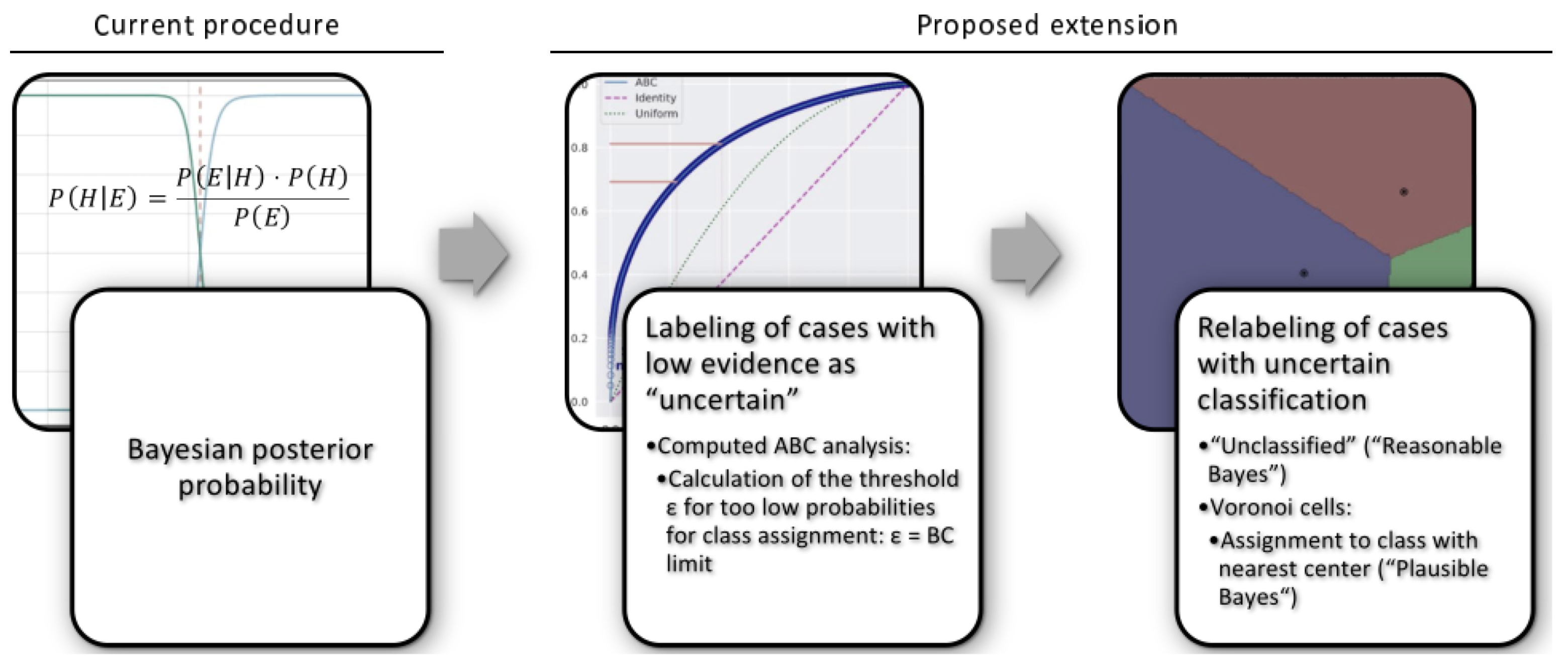
1. Introduction
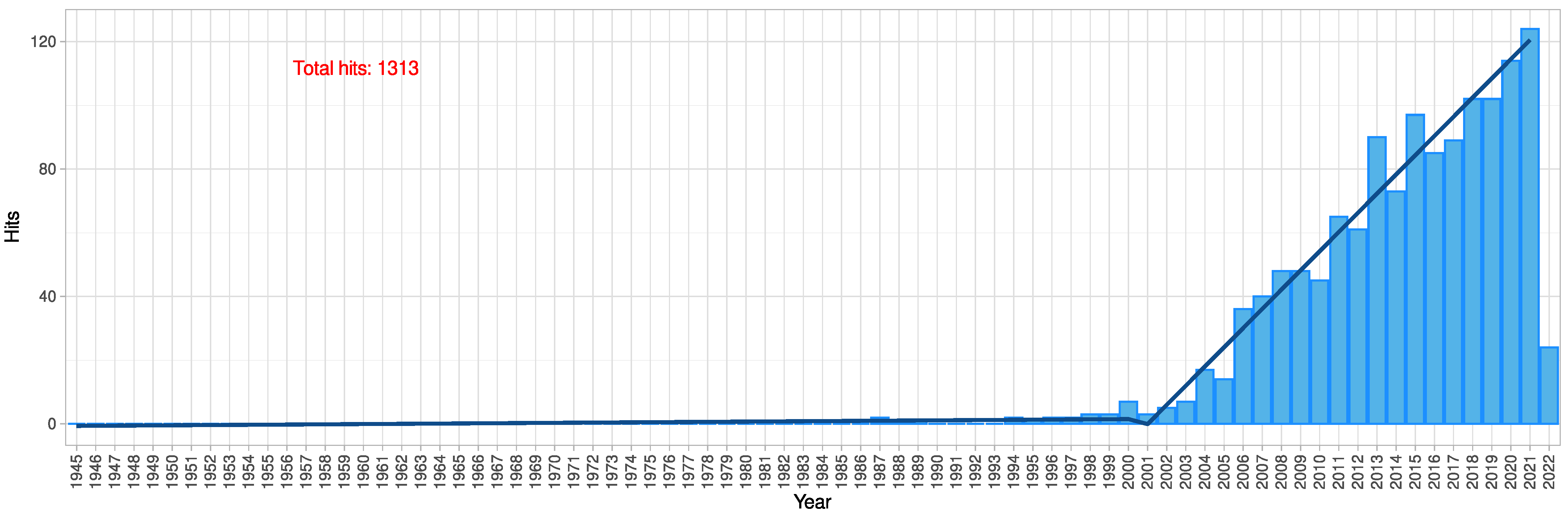
2. Results
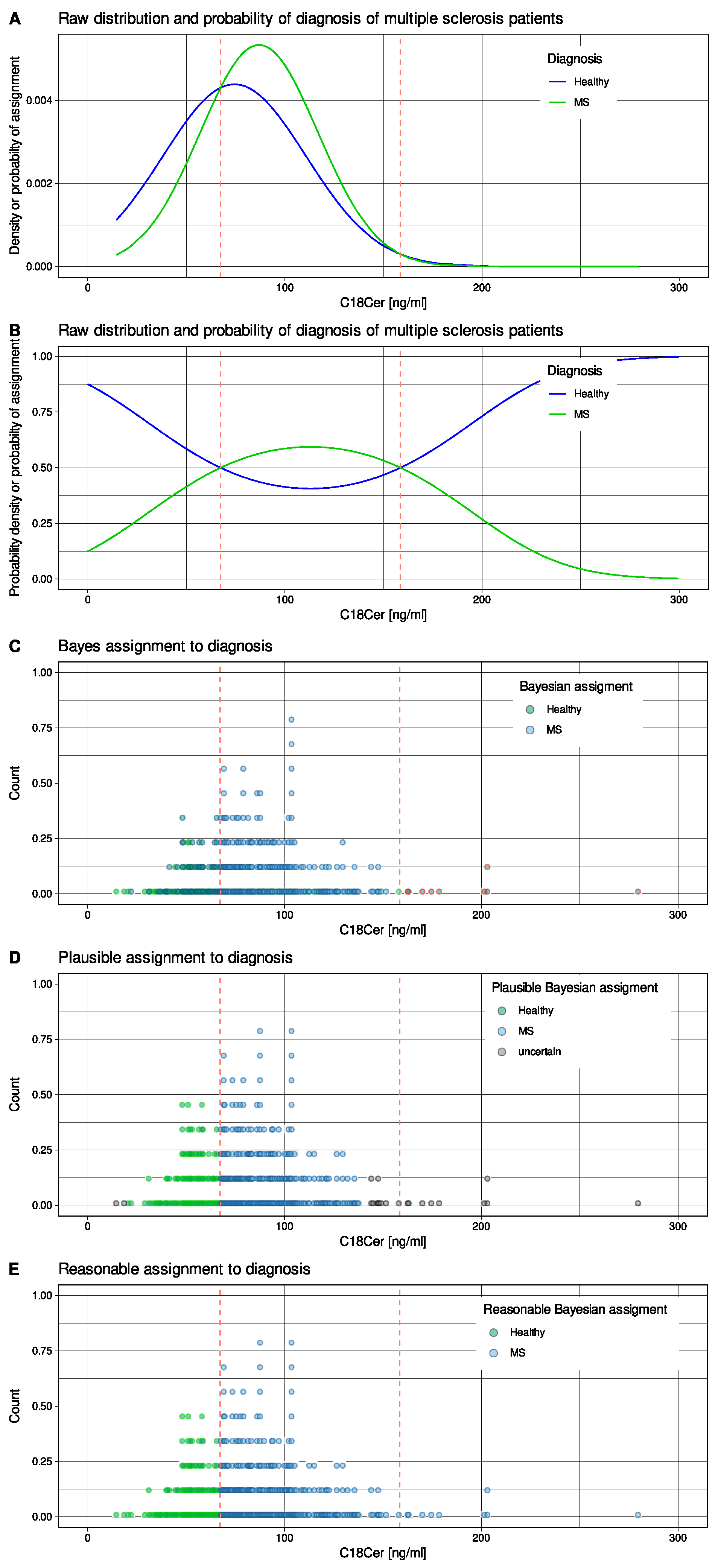
2.1. Multiple Sclerosis Lipidomics Data
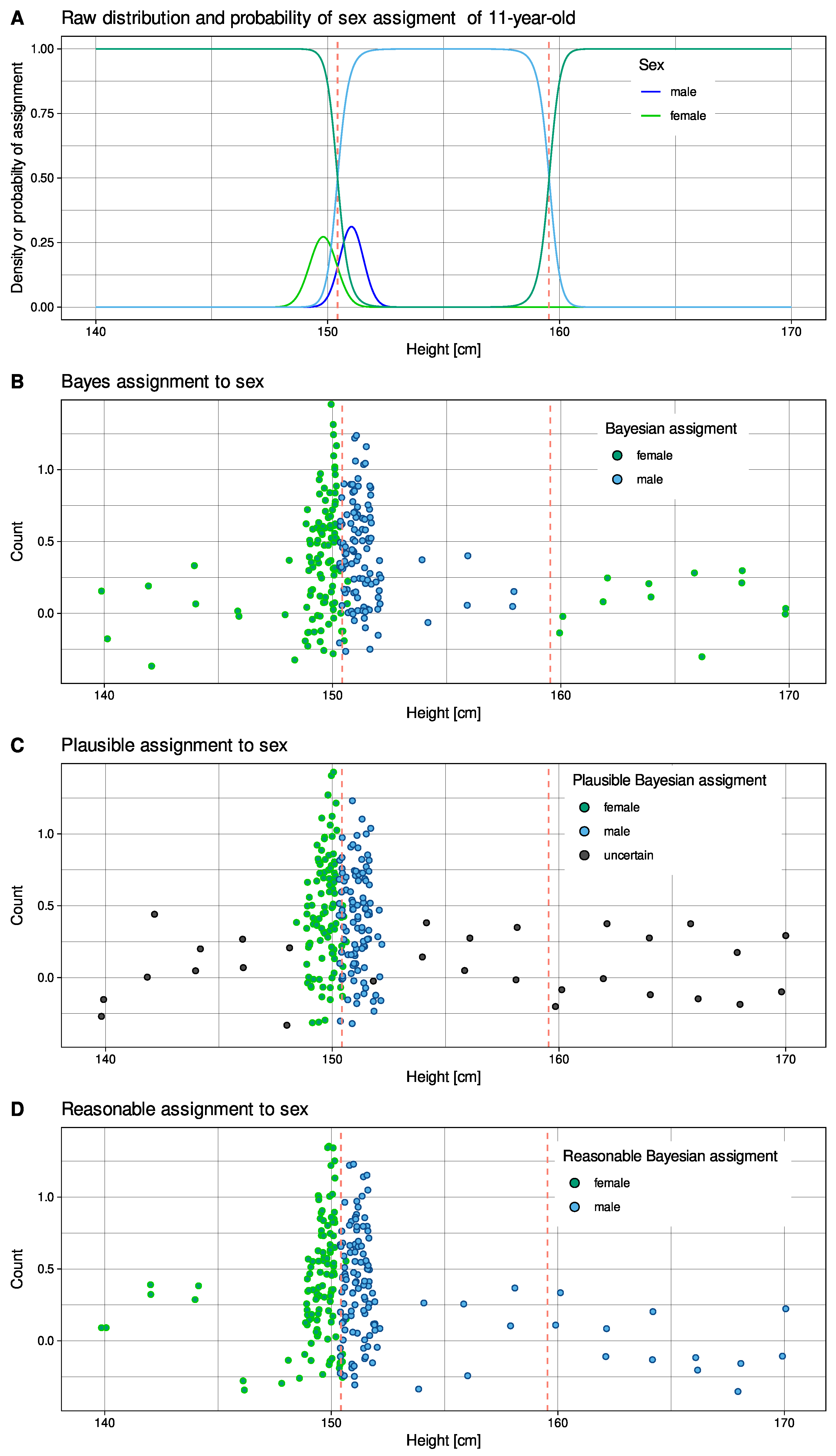
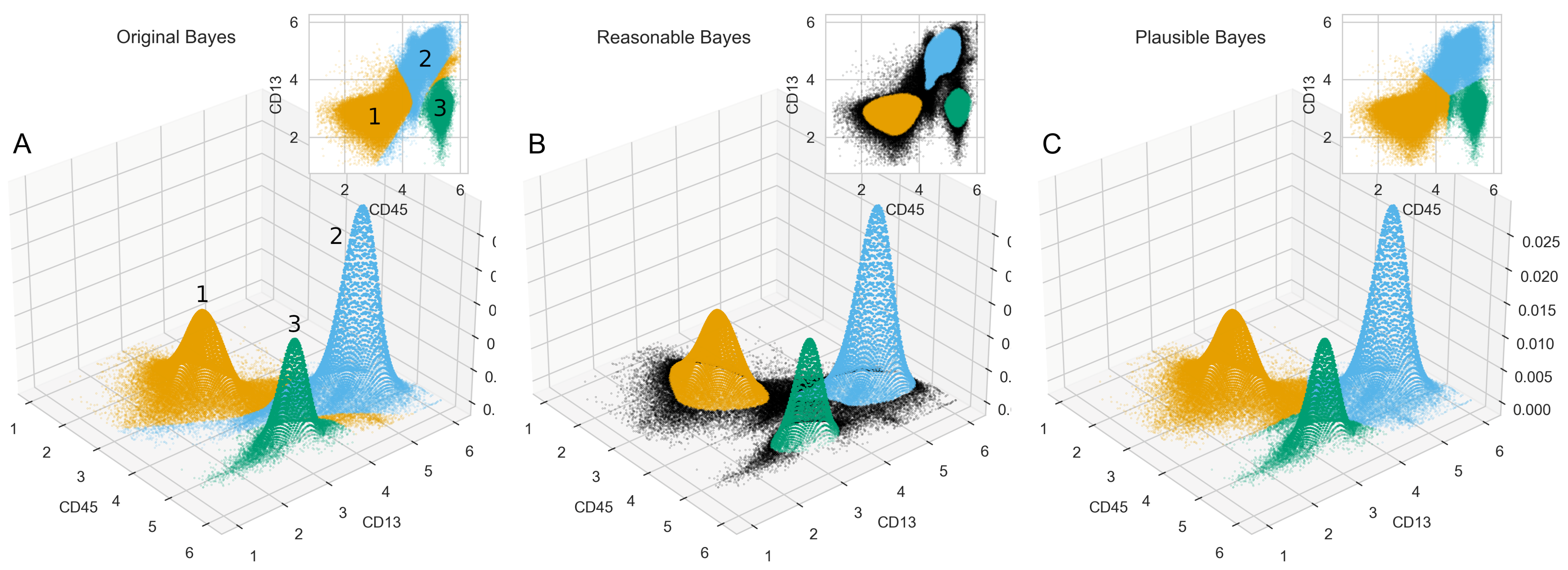
2.2. Flow Cytometric Data
3. Discussion
4. Materials and Methods
4.1. Bayesian Reasoning
4.2. Algorithm
4.2.1. Calculation of the Threshold for Low Probabilities
4.2.2. Corrected Assignments to Classes
Reasonable Bayes
Plausible Bayes
4.3. Experimentation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bayes, M.; Price, M. An Essay towards Solving a Problem in the Doctrine of Chances. By the Late Rev. Mr. Bayes, F. R. S. Communicated by Mr. Price, in a Letter to John Canton, A. M. F. R. S. Philos. Trans. 1763, 53, 370–418. [Google Scholar] [CrossRef]
- Tiberi, S.; Walsh, M.; Cavallaro, M.; Hebenstreit, D.; Finkenstädt, B. Bayesian inference on stochastic gene transcription from flow cytometry data. Bioinformatics 2018, 34, i647–i655. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.S.; Pertusi, D.A.; Adeniran, A.V.; Tyo, K.E.J. CellSort: A support vector machine tool for optimizing fluorescence-activated cell sorting and reducing experimental effort. Bioinformatics 2017, 33, 909–916. [Google Scholar] [CrossRef]
- Džunková, M.; Moya, A.; Vázquez-Castellanos, J.F.; Artacho, A.; Chen, X.; Kelly, C.; D’Auria, G. Active and Secretory IgA-Coated Bacterial Fractions Elucidate Dysbiosis in Clostridium difficile Infection. mSphere 2016, 1, e00101-16. [Google Scholar] [CrossRef] [PubMed]
- Comella, P.H.; Gonzalez-Kozlova, E.; Kosoy, R.; Charney, A.W.; Peradejordi, I.F.; Chandrasekar, S.; Tyler, S.R.; Wang, W.; Losic, B.; Zhu, J.; et al. A Molecular network approach reveals shared cellular and molecular signatures between chronic fatigue syndrome and other fatiguing illnesses. medRxiv 2021. [Google Scholar] [CrossRef]
- Kovalchik, S. RISmed: Download Content from NCBI Databases, 2020. Available online: https://CRAN.R-project.org/package=RISmed (accessed on 18 September 2022).
- Perfors, A.; Tenenbaum, J.B.; Griffiths, T.L.; Xu, F. A tutorial introduction to Bayesian models of cognitive development. Cognition 2011, 120, 302–321. [Google Scholar] [CrossRef]
- Gelman, A.; Yao, Y. Holes in Bayesian statistics. J. Phys. G Nucl. Part. Phys. 2020, 48, 014002. [Google Scholar] [CrossRef]
- Rodriguez-Martinez, A.; Zhou, B.; Sophiea, M.K.; Bentham, J.; Paciorek, C.J.; Iurilli, M.L.C.; Carrillo-Larco, R.M. Height and body-mass index trajectories of school-aged children and adolescents from 1985 to 2019 in 200 countries and territories: A pooled analysis of 2181 population-based studies with 65 million participants. Lancet 2020, 396, 1511–1524. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, Y.; Carroll, R.J. Variance estimation in the analysis of microarray data. J. R. Stat. Soc. Ser. B Stat. Methodol. 2009, 71, 425–445. [Google Scholar] [CrossRef]
- Archambeau, C.; Verleysen, M. Robust Bayesian clustering. Neural Netw. 2007, 20, 129–138. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Lotsch, J.; Schiffmann, S.; Schmitz, K.; Brunkhorst, R.; Lerch, F.; Ferreiros, N.; Wicker, S.; Tegeder, I.; Geisslinger, G.; Ultsch, A. Machine-learning based lipid mediator serum concentration patterns allow identification of multiple sclerosis patients with high accuracy. Sci. Rep. 2018, 8, 14884. [Google Scholar] [CrossRef] [PubMed]
- Sisignano, M.; Angioni, C.; Ferreiros, N.; Schuh, C.D.; Suo, J.; Schreiber, Y.; Dawes, J.M.; Antunes-Martins, A.; Bennett, D.L.; McMahon, S.B.; et al. Synthesis of lipid mediators during UVB-induced inflammatory hyperalgesia in rats and mice. PLoS ONE 2013, 8, e81228. [Google Scholar] [CrossRef] [PubMed]
- Zschiebsch, K.; Fischer, C.; Pickert, G.; Haeussler, A.; Radeke, H.; Grosch, S.; Ferreiros, N.; Geisslinger, G.; Werner, E.R.; Tegeder, I. Tetrahydrobiopterin attenuates DSS-evoked colitis in mice by rebalancing redox and lipid signaling. J. Crohns. Colitis. 2016, 10, 965–978. [Google Scholar] [CrossRef] [PubMed]
- Blachnio-Zabielska, A.U.; Chacinska, M.; Vendelbo, M.H.; Zabielski, P. The Crucial Role of C18-Cer in Fat-Induced Skeletal Muscle Insulin Resistance. Cell. Physiol. Biochem. 2016, 40, 1207–1220. [Google Scholar] [CrossRef]
- Rossi, C.; Cicalini, I.; Zucchelli, M.; di Ioia, M.; Onofrj, M.; Federici, L.; Del Boccio, P.; Pieragostino, D. Metabolomic Signature in Sera of Multiple Sclerosis Patients during Pregnancy. Int. J. Mol. Sci. 2018, 19, 3589. [Google Scholar] [CrossRef]
- Thrun, M.; Hoffmann, J.; Rohnert, M.; von Bonin, M.; Oelschlägel, U.; Brendel, C.; Ultsch, A. Flow Cytometry datasets consisting of peripheral blood and bone marrow samples for the evaluation of explainable artificial intelligence methods. Mendeley Data 2022. [Google Scholar] [CrossRef]
- Frearson, J.A.; Alexander, D.R. Protein tyrosine phosphatases in T-cell development, apoptosis and signalling. Immunol. Today 1996, 17, 385–391. [Google Scholar] [CrossRef]
- Woodhead, V.E.; Stonehouse, T.J.; Binks, M.H.; Speidel, K.; Fox, D.A.; Gaya, A.; Hardie, D.; Henniker, A.J.; Horejsi, V.; Sagawa, K.; et al. Novel molecular mechanisms of dendritic cell-induced T cell activation. Int. Immunol. 2000, 12, 1051–1061. [Google Scholar] [CrossRef]
- Horikoshi, A.; Sawada, S.; Endo, M.; Kawamura, M.; Murakami, J.; Iizuka, Y.; Takeuchi, J.; Ohshima, T.; Horie, T.; Motoyoshi, K. Relationship between responsiveness to colony stimulating factors (CSFs) and surface phenotype of leukemic blasts. Leuk. Res. 1995, 19, 195–201. [Google Scholar] [CrossRef]
- Rosenzwajg, M.; Tailleux, L.; Gluckman, J.C. CD13/N-aminopeptidase is involved in the development of dendritic cells and macrophages from cord blood CD34+ cells. Blood 2000, 95, 453–460. [Google Scholar] [CrossRef]
- Herzenberg, L.A.; Tung, J.; Moore, W.A.; Herzenberg, L.A.; Parks, D.R. Interpreting flow cytometry data: A guide for the perplexed. Nat. Immunol. 2006, 7, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Verschoor, C.P.; Lelic, A.; Bramson, J.L.; Bowdish, D.M. An Introduction to Automated Flow Cytometry Gating Tools and Their Implementation. Front. Immunol. 2015, 6, 380. [Google Scholar] [CrossRef] [PubMed]
- Moon, T.K. The expectation-maximization algorithm. IEEE Signal Process. Mag. 1996, 13, 47–60. [Google Scholar] [CrossRef]
- Waskom, M.L. seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Aut. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Devroye, L.; Gyorfi, L.; Lugosi, G. A Probabilistic Theory of Pattern Recognition; Stochastic Modelling and Applied Probability Series; Springer: New York, NY, USA, 1996; Volume 31, pp. 1–638. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: New York, NY, USA, 2009. [Google Scholar]
- Piryonesi, S.M.; El-Diraby Tamer, E. Role of Data Analytics in Infrastructure Asset Management: Overcoming Data Size and Quality Problems. J. Transp. Eng. Part B Pavements 2020, 146, 04020022. [Google Scholar] [CrossRef]
- Ziemski, M.; Wisanwanichthan, T.; Bokulich, N.A.; Kaehler, B.D. Beating Naive Bayes at Taxonomic Classification of 16S rRNA Gene Sequences. Front. Microbiol. 2021, 12, 644487. [Google Scholar] [CrossRef]
- Ontivero-Ortega, M.; Lage-Castellanos, A.; Valente, G.; Goebel, R.; Valdes-Sosa, M. Fast Gaussian Naïve Bayes for searchlight classification analysis. Neuroimage 2017, 163, 471–479. [Google Scholar] [CrossRef]
- Griffis, J.C.; Allendorfer, J.B.; Szaflarski, J.P. Voxel-based Gaussian naïve Bayes classification of ischemic stroke lesions in individual T1-weighted MRI scans. J. Neurosci. Methods 2016, 257, 97–108. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, Z.X.; Li, M.; Li, Y.Z.; Peng, C. Novel naïve Bayes classification models for predicting the carcinogenicity of chemicals. Food Chem. Toxicol. 2016, 97, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.S.; Shahjaman, M.; Rana, M.M.; Mollah, M.N.H. Robustification of Naïve Bayes Classifier and Its Application for Microarray Gene Expression Data Analysis. BioMed Res. Int. 2017, 2017, 3020627. [Google Scholar] [CrossRef]
- Zimek, A.; Schubert, E.; Kriegel, H.P. A survey on unsupervised outlier detection in high-dimensional numerical data. Stat. Anal. Data Min. ASA Data Sci. J. 2012, 5, 363–387. [Google Scholar] [CrossRef]
- Mor, B.; Garhwal, S.; Kumar, A. A Systematic Review of Hidden Markov Models and Their Applications. Arch. Comput. Methods Eng. 2020, 28, 1429–1448. [Google Scholar] [CrossRef]
- Freedman, D. Markov Chains; Springer: New York, NY, USA, 2012. [Google Scholar]
- Li, Q.; Li, R.; Ji, K.; Dai, W. Kalman Filter and Its Application. In Proceedings of the 2015 8th International Conference on Intelligent Networks and Intelligent Systems (ICINIS), Tianjin, China, 1–3 November 2015; pp. 74–77. [Google Scholar]
- Fischer, H. A History of the Central Limit Theorem: From Classical to Modern Probability Theory; Springer: New York, NY, USA, 2011; pp. 1–16. [Google Scholar] [CrossRef]
- Minker, J. On indefinite databases and the closed world assumption. In 6th Conference on Automated Deduction. CADE 1982; Loveland, D.W., Ed.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 1982; pp. 292–308. [Google Scholar]
- Ultsch, A.; Thrun, M.C.; Hansen-Goos, O.; Lotsch, J. Identification of Molecular Fingerprints in Human Heat Pain Thresholds by Use of an Interactive Mixture Model R Toolbox (AdaptGauss). Int. J. Mol. Sci. 2015, 16, 25897–25911. [Google Scholar] [CrossRef]
- Voronoi, G. Nouvelles applications des paramètres continus à la théorie des formes quadratiques. Premier mémoire. Sur quelques propriétés des formes quadratiques positives parfaites. J. FüR Die Reine Und Angew. Math. (Crelles J.) 1908, 1908, 97–102. [Google Scholar] [CrossRef]
- Ultsch, A.; Lotsch, J. Computed ABC Analysis for Rational Selection of Most Informative Variables in Multivariate Data. PLoS ONE 2015, 10, e0129767. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.C.; Wood, M.C. Joseph M. Juran: Critical Evaluations in Business and Management; Routledge: London, UK, 2005. [Google Scholar]
- Zhang, H. The Optimality of Naive Bayes. In Proceedings of the Seventeenth International Florida Artificial Intelligence Research Society Conference, Miami Beach, FL, USA, 12–14 May 2004; pp. 562–567. [Google Scholar]
- van der Meyden, R. Logical Approaches to Incomplete Information: A Survey. In Logics for Databases and Information Systems; Chomicki, J., Saake, G., Eds.; Springer: Boston, MA, USA, 1998; pp. 307–356. [Google Scholar] [CrossRef]
- Reosekar, R.S.; Pohekar, S.D. Six Sigma methodology: A structured review. Int. J. Lean Six Sigma 2014, 5, 392–422. [Google Scholar] [CrossRef]
- Habel, K.; Grasman, R.; Gramacy, R.B.; Mozharovskyi, P.; Sterratt, D.C. Geometry: Mesh Generation and Surface Tessellation, 2019. Available online: https://CRAN.R-project.org/package=geometry (accessed on 18 September 2022).
- Barber, C.B.; Dobkin, D.P.; Huhdanpaa, H. The quickhull algorithm for convex hulls. ACM Trans. Math. Softw. 1996, 22, 469–483. [Google Scholar] [CrossRef]
- Polianskii, V.; Pokorny, F.T. Voronoi Graph Traversal in High Dimensions with Applications to Topological Data Analysis and Piecewise Linear Interpolation. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery &Data Mining, Virtual, 6–10 July 2020; Association for Computing Machinery: New York, NY, USA, 2020; pp. 2154–2164. [Google Scholar] [CrossRef]
- Ihaka, R.; Gentleman, R. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Van Rossum, G.; Drake, F.L., Jr. Python Tutorial; Centrum voor Wiskunde en Informatica: Amsterdam, The Netherlands, 1995; Volume 620. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ultsch, A.; Lötsch, J. Robust Classification Using Posterior Probability Threshold Computation Followed by Voronoi Cell Based Class Assignment Circumventing Pitfalls of Bayesian Analysis of Biomedical Data. Int. J. Mol. Sci. 2022, 23, 14081. https://doi.org/10.3390/ijms232214081
Ultsch A, Lötsch J. Robust Classification Using Posterior Probability Threshold Computation Followed by Voronoi Cell Based Class Assignment Circumventing Pitfalls of Bayesian Analysis of Biomedical Data. International Journal of Molecular Sciences. 2022; 23(22):14081. https://doi.org/10.3390/ijms232214081
Chicago/Turabian StyleUltsch, Alfred, and Jörn Lötsch. 2022. "Robust Classification Using Posterior Probability Threshold Computation Followed by Voronoi Cell Based Class Assignment Circumventing Pitfalls of Bayesian Analysis of Biomedical Data" International Journal of Molecular Sciences 23, no. 22: 14081. https://doi.org/10.3390/ijms232214081
APA StyleUltsch, A., & Lötsch, J. (2022). Robust Classification Using Posterior Probability Threshold Computation Followed by Voronoi Cell Based Class Assignment Circumventing Pitfalls of Bayesian Analysis of Biomedical Data. International Journal of Molecular Sciences, 23(22), 14081. https://doi.org/10.3390/ijms232214081







