GRP94 Inhabits the Immortalized Porcine Hepatic Stellate Cells Apoptosis under Endoplasmic Reticulum Stress through Modulating the Expression of IGF-1 and Ubiquitin
Abstract
1. Introduction
2. Results
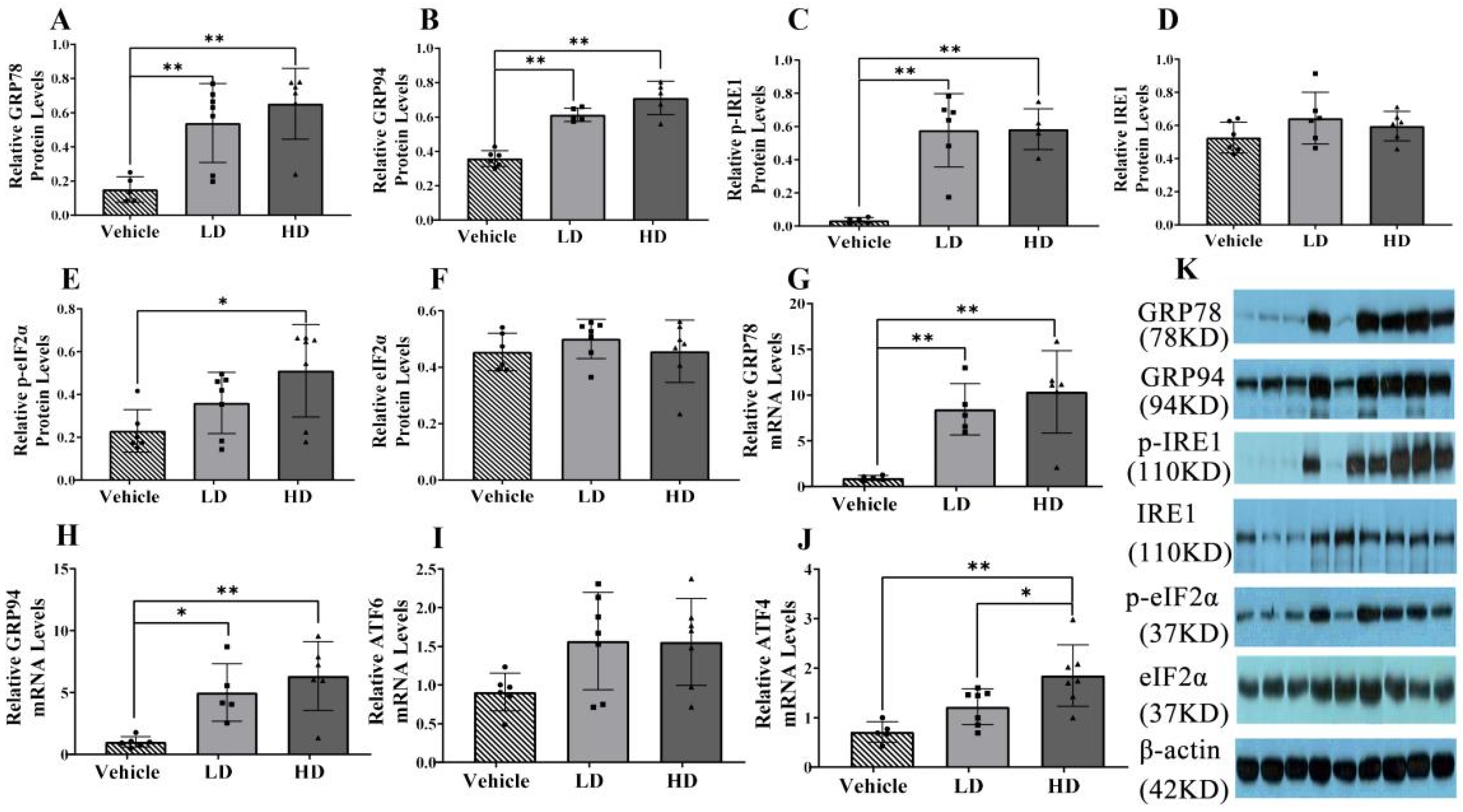
2.1. Establishment of an ERS Model in Piglets
2.2. ERS-Dependent Changes in IGF-1 Efficiency of Piglet Livers and Plasmas
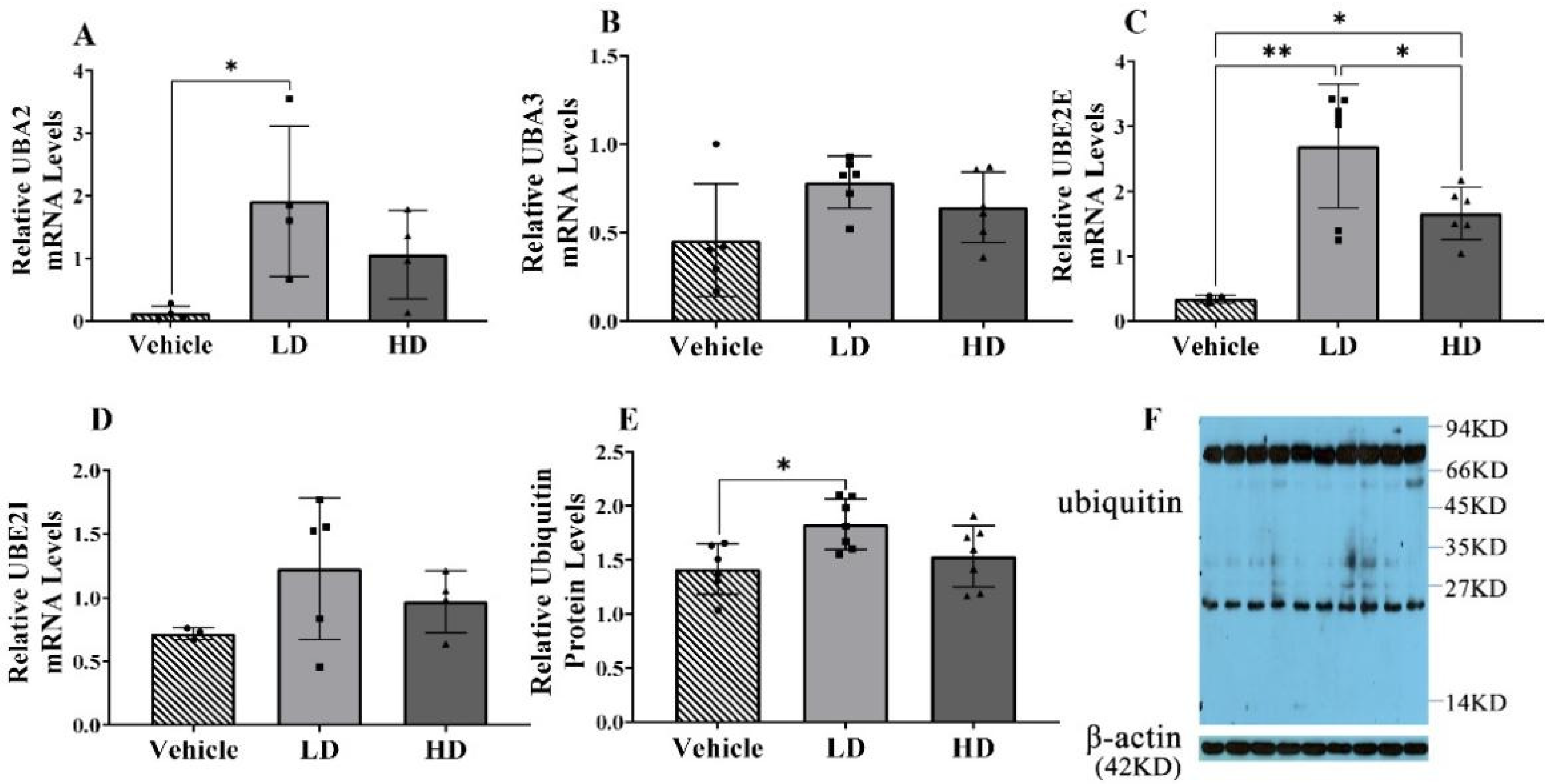
2.3. ERS-Dependent Changes in Ubiquitination of Piglet Livers
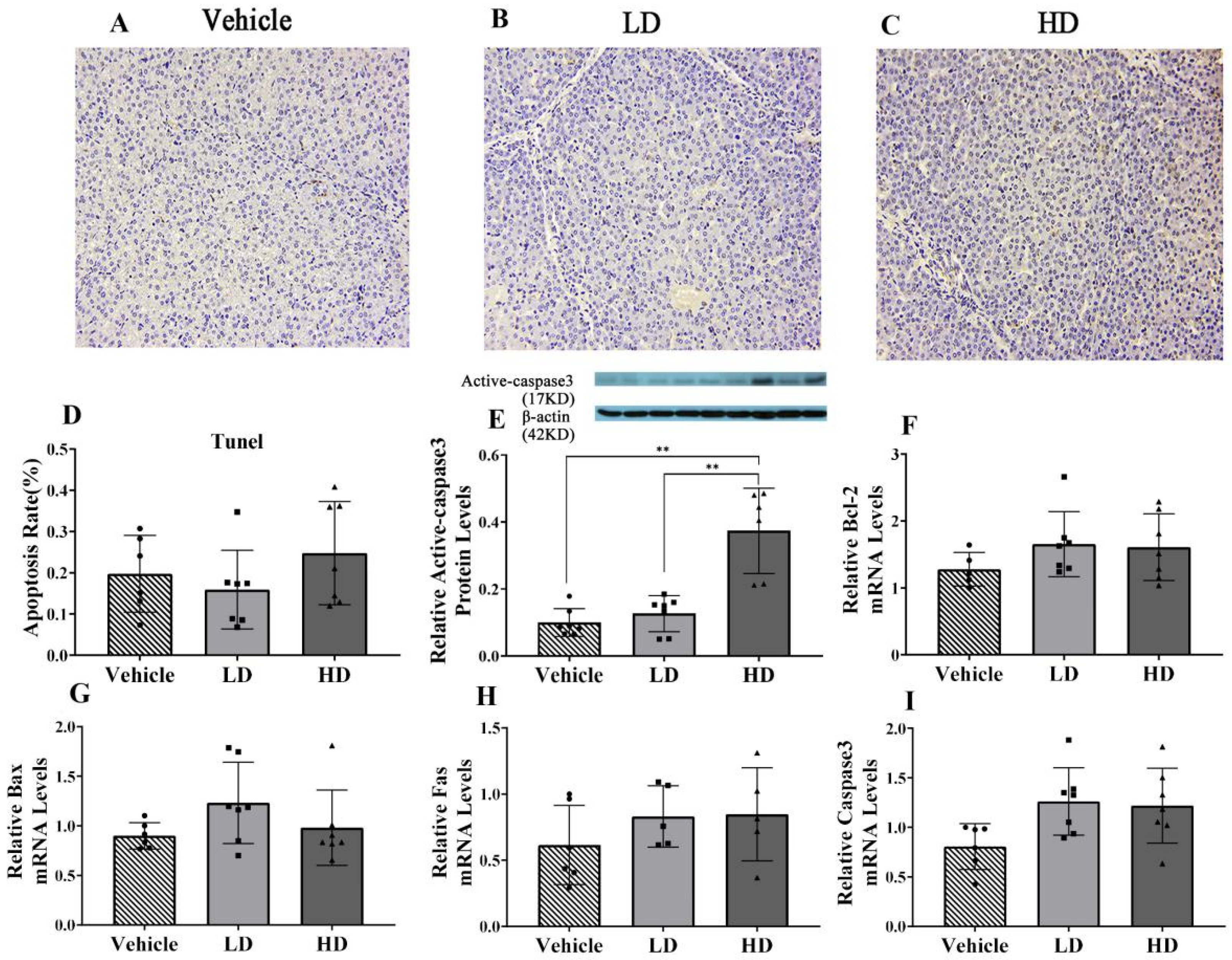
2.4. ERS-Dependent Changes in Hepatocellular Apoptosis of Piglet Livers
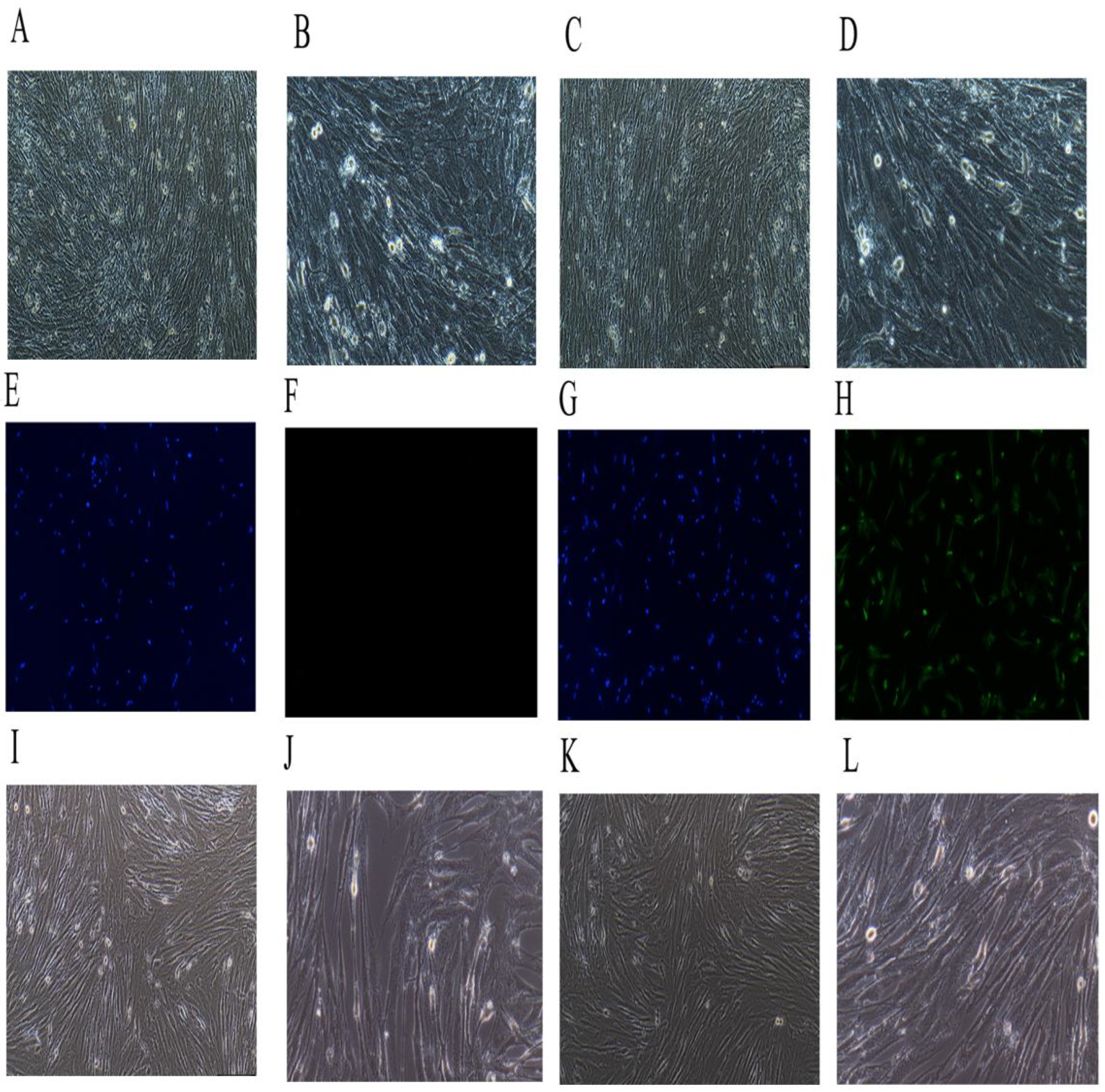
2.5. Immortalized the Porcine HSCs
2.6. Inhibition of GRP94 by GRP94-shRNA Lentivirus Transfection
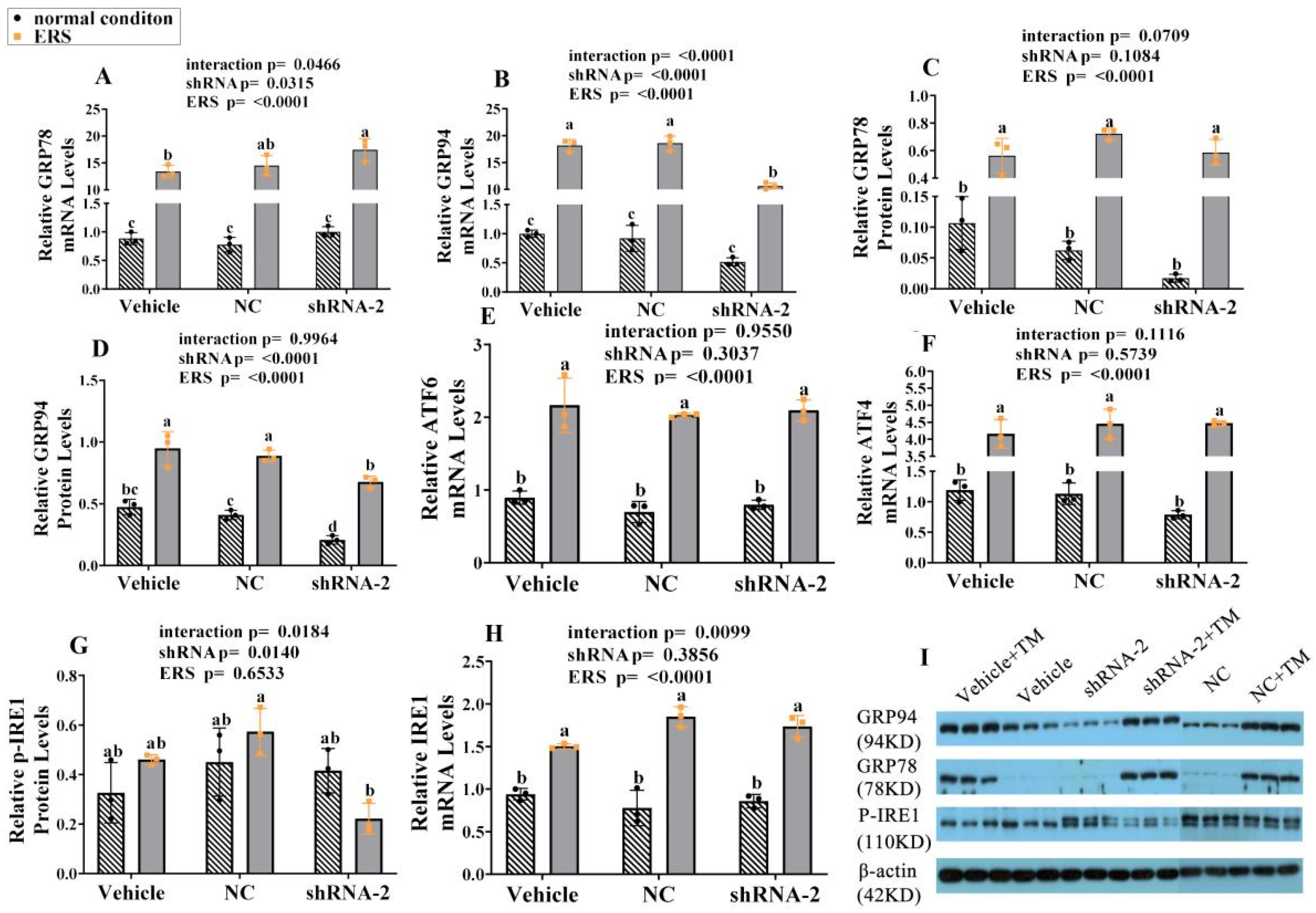
2.7. The Effect of GRP94 Knockdown and ERS on ERS Mark Genes and Proteins in HSCs
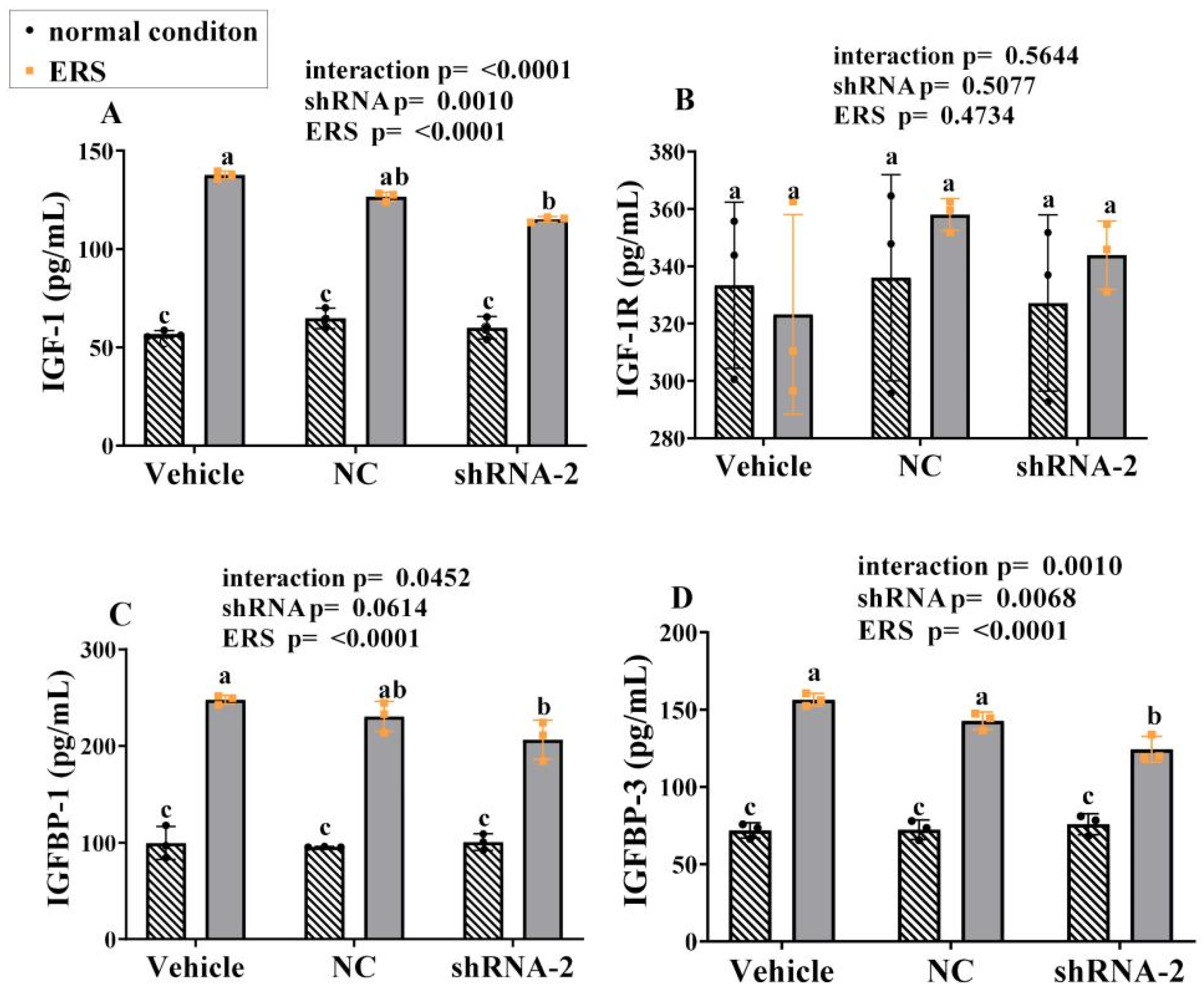
2.8. The Effect of GRP94 Knockdown and ERS on IGF-1 in HSCs
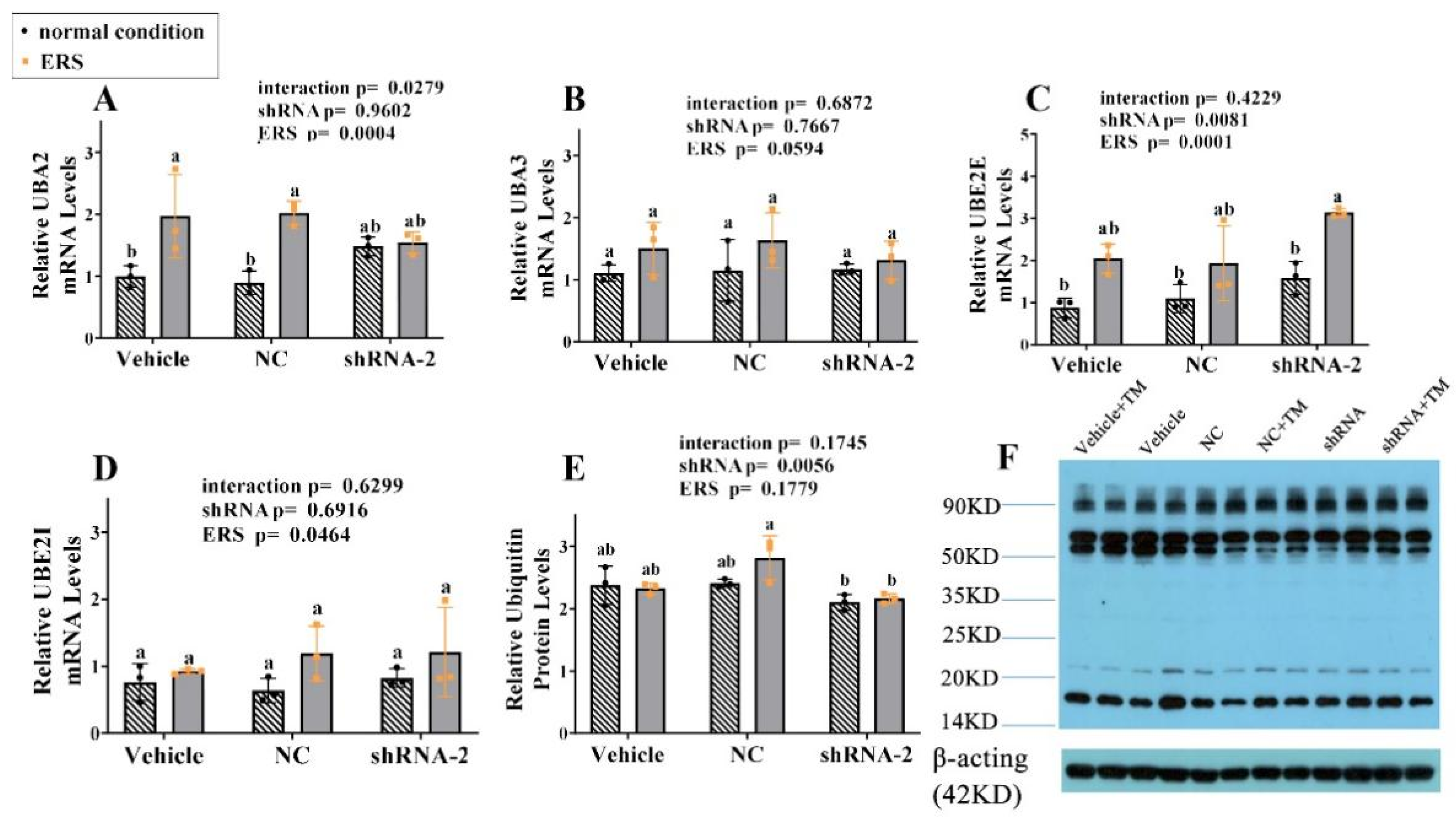
2.9. The Effect of GRP94 Knockdown and ERS on Ubiquitination in HSCs
2.10. The Effect of GRP94 Knockdown and ERS on Apoptosis in HSCs
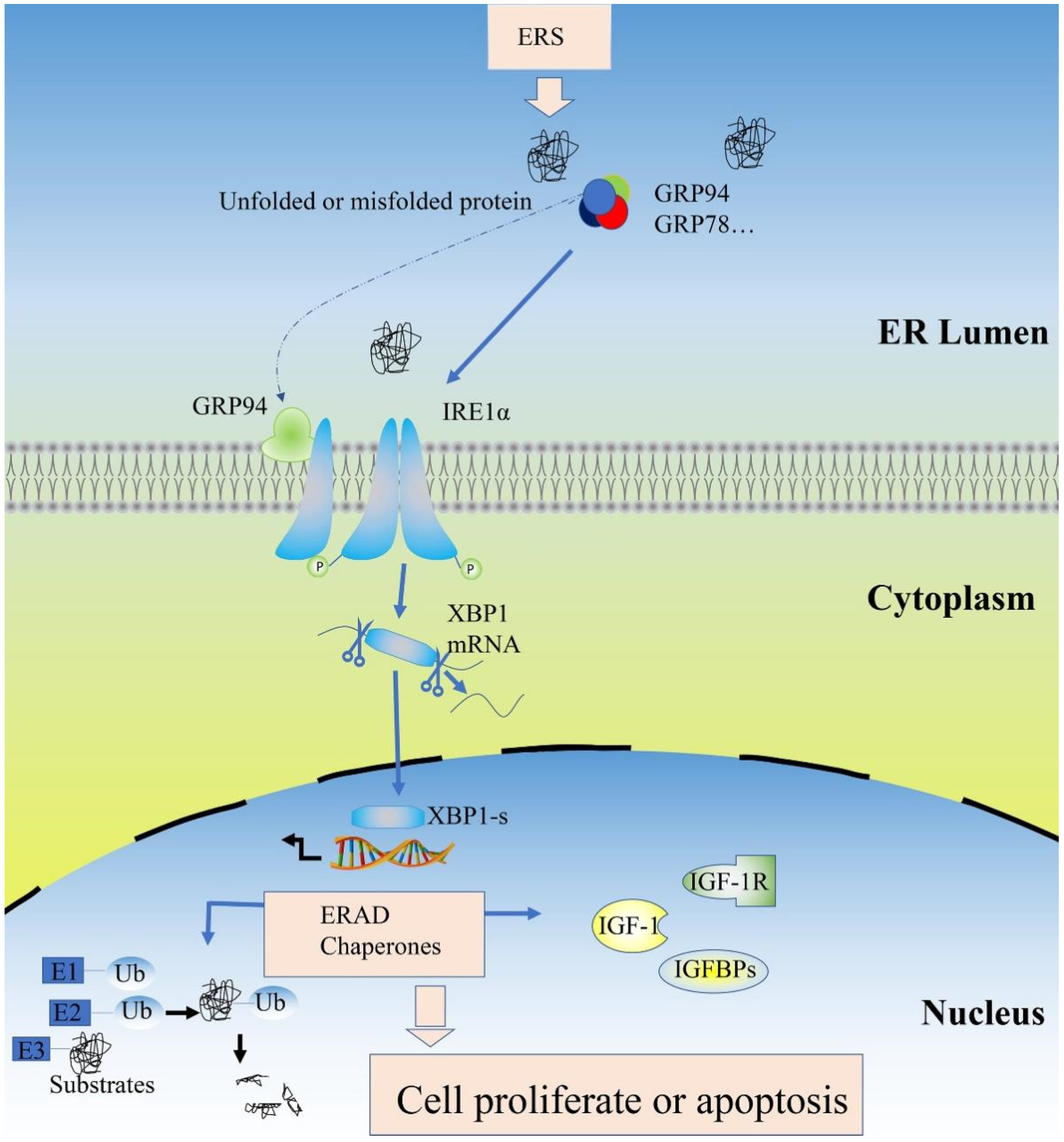
3. Discussion
4. Materials and Methods
4.1. Animals and Establishment of ERS Model
4.2. Preparation of Porcine Immortalized HSCs
4.2.1. Separation and Culture of Porcine Hepatic Stellate Cells
4.2.2. Immunofluorescence Identification
4.2.3. Lentivirus Transfection
4.2.4. Screening of Porcine HSCs
4.2.5. The Morphology of Immortalized Porcine HSCs
4.3. Plasmid Construction and Transfection
4.4. TUNEL Assay
4.5. Immunofluorescence Identification
4.6. RT-qPCR Analysis
4.7. IGF-1, IGF-1R, IGFBP-1, and IGFBP-3 Measurement in the Liver, Plasma and Cell Supernatant
4.8. Cell Viability Measurement
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Wu, J.; Kaufman, R.J. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006, 13, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Amen, O.M.; Sarker, S.D.; Ghildyal, R.; Arya, A. Endoplasmic Reticulum Stress Activates Unfolded Protein Response Signaling and Mediates Inflammation, Obesity, and Cardiac Dysfunction: Therapeutic and Molecular Approach. Front. Pharmacol. 2019, 10, 977. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.; Oxford, A.E.; Tawara, K.; Jorcyk, C.L.; Oxford, J.T. Endoplasmic Reticulum Stress and Unfolded Protein Response in Cartilage Pathophysiology; Contributing Factors to Apoptosis and Osteoarthritis. Int. J. Mol. Sci. 2017, 18, 665. [Google Scholar] [CrossRef]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef]
- Lu, X.; Huang, H.; Fu, X.; Chen, C.; Liu, H.; Wang, H.; Wu, D. The Role of Endoplasmic Reticulum Stress and NLRP3 Inflammasome in Liver Disorders. Int. J. Mol. Sci. 2022, 23, 3528. [Google Scholar] [CrossRef]
- Chen, Q.; Fang, W.; Cui, K.; Chen, Q.; Xiang, X.; Zhang, J.; Zhang, Y.; Mai, K.; Ai, Q. Endoplasmic reticulum stress induces hepatic steatosis by transcriptional upregulating lipid droplet protein perilipin2. FASEB J. 2021, 35, e21900. [Google Scholar] [CrossRef]
- Chen, Q.; Fang, W.; Shen, Y.; Xu, D.; Chen, Q.; Cui, K.; Mai, K.; Ai, Q. Suppression of cideb under endoplasmic reticulum stress exacerbated hepatic inflammation by inducing hepatic steatosis and oxidative stress. Free Radic. Biol. Med. 2022, 185, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zheng, S.; Zhang, J.; Xu, S.; Miao, Z. Cadmium induces endoplasmic reticulum stress-mediated apoptosis in pig pancreas via the increase of Th1 cells. Toxicology 2021, 457, 152790. [Google Scholar] [CrossRef]
- Li, T.; Chen, Y.; Tan, P.; Shi, H.; Huang, Z.; Cai, T.; Cheng, Y.; Du, Y.; Fu, W. Dihydroartemisinin alleviates steatosis and inflammation in nonalcoholic steatohepatitis by decreasing endoplasmic reticulum stress and oxidative stress. Bioorg. Chem. 2022, 122, 105737. [Google Scholar] [CrossRef]
- Liu, Y.; Pan, X.; Li, S.; Yu, Y.; Chen, J.; Yin, J.; Li, G. Endoplasmic reticulum stress restrains hepatocyte growth factor expression in hepatic stellate cells and rat acute liver failure model. Chem. Biol. Interact. 2017, 277, 43–54. [Google Scholar] [CrossRef]
- Takeichi, Y.; Miyazawa, T.; Sakamoto, S.; Hanada, Y.; Wang, L.; Gotoh, K.; Uchida, K.; Katsuhara, S.; Sakamoto, R.; Ishihara, T.; et al. Non-alcoholic fatty liver disease in mice with hepatocyte-specific deletion of mitochondrial fission factor. Diabetologia 2021, 64, 2092–2107. [Google Scholar] [CrossRef] [PubMed]
- Wang, K. Autophagy and apoptosis in liver injury. Cell Cycle 2015, 14, 1631–1642. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, X.; Wang, Y.; Wang, H.; Huang, C.; Li, J. Endoplasmic reticulum stress-induced hepatic stellate cell apoptosis through calcium-mediated JNK/P38 MAPK and Calpain/Caspase-12 pathways. Mol. Cell Biochem. 2014, 394, 1–12. [Google Scholar] [CrossRef] [PubMed]
- De Minicis, S.; Candelaresi, C.; Agostinelli, L.; Taffetani, S.; Saccomanno, S.; Rychlicki, C.; Trozzi, L.; Marzioni, M.; Benedetti, A.; Svegliati-Baroni, G. Endoplasmic Reticulum stress induces hepatic stellate cell apoptosis and contributes to fibrosis resolution. Liver Int. 2012, 32, 1574–1584. [Google Scholar] [CrossRef]
- Torres, S.; Baulies, A.; Insausti-Urkia, N.; Alarcón-Vila, C.; Fucho, R.; Solsona-Vilarrasa, E.; Núñez, S.; Robles, D.; Ribas, V.; Wakefield, L.; et al. Endoplasmic Reticulum Stress-Induced Upregulation of STARD1 Promotes Acetaminophen-Induced Acute Liver Failure. Gastroenterology 2019, 157, 552–568. [Google Scholar] [CrossRef]
- Wang, K. Molecular mechanisms of hepatic apoptosis. Cell Death Dis. 2014, 5, e996. [Google Scholar] [CrossRef]
- Keating, M.F.; Drew, B.G.; Calkin, A.C. Antisense Oligonucleotide Technologies to Combat Obesity and Fatty Liver Disease. Front. Physiol. 2022, 13, 839471. [Google Scholar] [CrossRef]
- Chen, W.T.; Ha, D.; Kanel, G.; Lee, A.S. Targeted deletion of ER chaperone GRP94 in the liver results in injury, repopulation of GRP94-positive hepatocytes, and spontaneous hepatocellular carcinoma development in aged mice. Neoplasia 2014, 16, 617–626. [Google Scholar] [CrossRef]
- Zhu, X.; Xiao, Z.; Chen, X.; Li, Y.; Zhang, X.; Xu, Y.; Feng, X.; Wang, J. Parenteral nutrition-associated liver injury and increased GRP94 expression prevented by omega-3 fish oil-based lipid emulsion supplementation. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 708–713. [Google Scholar] [CrossRef]
- Lebeau, P.F.; Wassef, H.; Byun, J.H.; Platko, K.; Ason, B.; Jackson, S.; Dobroff, J.; Shetterly, S.; Richards, W.G.; Al-Hashimi, A.A.; et al. The loss-of-function PCSK9Q152H variant increases ER chaperones GRP78 and GRP94 and protects against liver injury. J. Clin. Investig. 2021, 131, e128650. [Google Scholar] [CrossRef]
- Fornasiero, F.; Scapin, C.; Vitadello, M.; Pizzo, P.; Gorza, L. Active nNOS Is Required for Grp94-Induced Antioxidant Cytoprotection: A Lesson from Myogenic to Cancer Cells. Int. J. Mol. Sci. 2022, 23, 2915. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Anversa, P. The insulin-like growth factor I system: Physiological and pathophysiological implication in cardiovascular diseases associated with metabolic syndrome. Biochem. Pharmacol. 2015, 93, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Carlson, S.W.; Saatman, K.E. Central Infusion of Insulin-Like Growth Factor-1 Increases Hippocampal Neurogenesis and Improves Neurobehavioral Function after Traumatic Brain Injury. J. Neurotrauma 2018, 35, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Chen, C.; Chen, J.; Zhan, R.; Zhang, Q.; Xu, X.; Li, D.; Li, M. Cell surface GRP78 facilitates hepatoma cells proliferation and migration by activating IGF-IR. Cell. Signal. 2017, 35, 154–162. [Google Scholar] [CrossRef]
- Edwards, S.L.; Erdenebat, P.; Morphis, A.C.; Kumar, L.; Wang, L.; Chamera, T.; Georgescu, C.; Wren, J.D.; Li, J. Insulin/IGF-1 signaling and heat stress differentially regulate HSF1 activities in germline development. Cell Rep. 2021, 36, 109623. [Google Scholar] [CrossRef]
- Guan, C.P.; Li, Q.T.; Jiang, H.; Geng, Q.W.; Xu, W.; Li, L.Y.; Xu, A.E. IGF-1 resist oxidative damage to HaCaT and depigmentation in mice treated with H2O2. Biochem. Biophys. Res. Commun. 2018, 503, 2485–2492. [Google Scholar] [CrossRef]
- Wang, D.; Zheng, T.; Ge, X.; Xu, J.; Feng, L.; Jiang, C.; Tao, J.; Chen, Y.; Liu, X.; Yu, B.; et al. Unfolded protein response-induced expression of long noncoding RNA Ngrl1 supports peripheral axon regeneration by activating the PI3K-Akt pathway. Exp. Neurol. 2022, 352, 114025. [Google Scholar] [CrossRef]
- Cui, Y.; Hao, Y.; Li, J.; Bao, W.; Li, G.; Gao, Y.; Gu, X. Chronic Heat Stress Induces Immune Response, Oxidative Stress Response, and Apoptosis of Finishing Pig Liver: A Proteomic Approach. Int. J. Mol. Sci. 2016, 17, 393. [Google Scholar] [CrossRef]
- Xia, W.; Wang, Y.; Zhang, Y.; Ge, X.; Lv, P.; Cheng, J.; Wei, J. Endoplasmic reticulum stress induces growth retardation by inhibiting growth hormone IGF-I axis. Growth Horm. IGF Res. 2020, 55, 101341. [Google Scholar] [CrossRef]
- Bilginer, Y.; Topaloglu, R.; Alikasifoglu, A.; Kara, N.; Besbas, N.; Ozen, S.; Bakkaloglu, A. Low cortisol levels in active juvenile idiopathic arthritis. Clin. Rheumatol. 2010, 29, 309–314. [Google Scholar] [CrossRef]
- Qu, J.; Zou, T.; Lin, Z. The Roles of the Ubiquitin-Proteasome System in the Endoplasmic Reticulum Stress Pathway. Int. J. Mol. Sci. 2021, 22, 1526. [Google Scholar] [CrossRef] [PubMed]
- Cybulsky, A.V. The intersecting roles of endoplasmic reticulum stress, ubiquitin- proteasome system, and autophagy in the pathogenesis of proteinuric kidney disease. Kidney Int. 2013, 84, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Sullivan, D.K.; Azizian, N.G.; Liu, Y.; Li, Y. Inhibition of ERAD synergizes with FTS to eradicate pancreatic cancer cells. BMC Cancer 2021, 21, 237. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Stone, S.; Yue, Y.; Lin, W. Endoplasmic reticulum associated degradation is required for maintaining endoplasmic reticulum homeostasis and viability of mature Schwann cells in adults. Glia 2021, 69, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.H.; Lee, J.S.; Kim, H.G.; Lee, H.W.; Fang, Z.; Kwon, H.H.; Kim, D.W.; Lee, C.M.; Jeong, J.W. Ethyl Acetate Fraction of Amomum villosum var. xanthioides Attenuates Hepatic Endoplasmic Reticulum Stress-Induced Non-Alcoholic Steatohepatitis via Improvement of Antioxidant Capacities. Antioxidants 2021, 10, 998. [Google Scholar] [CrossRef]
- Kim, S.H.; Seo, H.; Kwon, D.; Yuk, D.Y.; Jung, Y.S. Taurine Ameliorates Tunicamycin-Induced Liver Injury by Disrupting the Vicious Cycle between Oxidative Stress and Endoplasmic Reticulum Stress. Life 2022, 12, 354. [Google Scholar] [CrossRef]
- Jeong, P.S.; Yoon, S.B.; Lee, M.H.; Son, H.C.; Lee, H.Y.; Lee, S.; Koo, B.S.; Jeong, K.J.; Lee, J.H.; Jin, Y.B.; et al. Embryo aggregation regulates in vitro stress conditions to promote developmental competence in pigs. PeerJ 2019, 7, e8143. [Google Scholar] [CrossRef]
- Saga, K.; Iwashita, Y.; Hidano, S.; Aso, Y.; Isaka, K.; Kido, Y.; Tada, K.; Takayama, H.; Masuda, T.; Hirashita, T.; et al. Secondary Unconjugated Bile Acids Induce Hepatic Stellate Cell Activation. Int. J. Mol. Sci. 2018, 19, 3043. [Google Scholar] [CrossRef]
- Wang, Z.J.; Yu, H.; Hao, J.J.; Peng, Y.; Yin, T.T.; Qiu, Y.N. PM2.5 promotes Drp1-mediated mitophagy to induce hepatic stellate cell activation and hepatic fibrosis via regulating miR-411. Exp. Cell Res. 2021, 407, 112828. [Google Scholar] [CrossRef]
- Huang, Y.H.; Chen, M.H.; Guo, Q.L.; Chen, Z.X.; Chen, Q.D.; Wang, X.Z. Interleukin-10 induces senescence of activated hepatic stellate cells via STAT3-p53 pathway to attenuate liver fibrosis. Cell. Signal. 2020, 66, 109445. [Google Scholar] [CrossRef]
- Mannaerts, I.; Thoen, L.F.R.; Eysackers, N.; Cubero, F.J.; Batista Leite, S.; Coldham, I.; Colle, I.; Trautwein, C.; van Grunsven, L.A. Unfolded protein response is an early, non-critical event during hepatic stellate cell activation. Cell Death Dis. 2019, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Dai, L.; Wang, M.; Feng, F.; Xiao, Y. Tunicamycin Induces Hepatic Stellate Cell Apoptosis Through Calpain-2/Ca(2 +)-Dependent Endoplasmic Reticulum Stress Pathway. Front. Cell Dev. Biol. 2021, 9, 684857. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, L.; Han, X.; Li, C.; Liu, R.; Ma, Z.; Han, B.; Xie, R.; Yang, Q. Endoplasmic reticulum stress in hepatic stellate cells induced by tunicamycin promotes apoptosis and cell cycle arrest. Chin. J. Cell. Mol. Immunol. 2021, 37, 794–800. [Google Scholar]
- Lepenies, J.; Wu, Z.; Stewart, P.M.; Strasburger, C.J.; Quinkler, M. IGF-1, IGFBP-3 and ALS in adult patients with chronic kidney disease. Growth Horm. IGF Res. 2010, 20, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Maiers, J.L.; Malhi, H. Endoplasmic Reticulum Stress in Metabolic Liver Diseases and Hepatic Fibrosis. Semin. Liver Dis. 2019, 39, 235–248. [Google Scholar] [CrossRef]
- Fernandes-da-Silva, A.; Miranda, C.S.; Santana-Oliveira, D.A.; Oliveira-Cordeiro, B.; Rangel-Azevedo, C.; Silva-Veiga, F.M.; Martins, F.F.; Souza-Mello, V. Endoplasmic reticulum stress as the basis of obesity and metabolic diseases: Focus on adipose tissue, liver, and pancreas. Eur. J. Nutr. 2021, 6, 2949–2960. [Google Scholar] [CrossRef]
- Chen, C.; Wu, H.; Ye, H.; Tortajada, A.; Rodriguez-Perales, S.; Torres-Ruiz, R.; Vidal, A.; Peligros, M.I.; Reissing, J.; Bruns, T.; et al. Activation of the Unfolded Protein Response (UPR) Is Associated with Cholangiocellular Injury, Fibrosis and Carcinogenesis in an Experimental Model of Fibropolycystic Liver Disease. Cancers 2021, 14, 78. [Google Scholar] [CrossRef]
- Marzec, M.; Eletto, D.; Argon, Y. GRP94: An HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2012, 1823, 774–787. [Google Scholar] [CrossRef]
- Zhu, G.; Lee, A.S. Role of the unfolded protein response, GRP78 and GRP94 in organ homeostasis. J. Cell. Physiol. 2015, 230, 1413–1420. [Google Scholar] [CrossRef]
- Poirier, S.; Mamarbachi, M.; Chen, W.T.; Lee, A.S.; Mayer, G. GRP94 Regulates Circulating Cholesterol Levels through Blockade of PCSK9-Induced LDLR Degradation. Cell Rep. 2015, 13, 2064–2071. [Google Scholar] [CrossRef]
- Randow, F.; Seed, B. Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat. Cell Biol. 2001, 3, 891–896. [Google Scholar] [CrossRef]
- Lu, T.; Wang, Y.; Xu, K.; Zhou, Z.; Gong, J.; Zhang, Y.; Gong, H.; Dai, Q.; Yang, J.; Xiong, B.; et al. Co-downregulation of GRP78 and GRP94 Induces Apoptosis and Inhibits Migration in Prostate Cancer Cells. Open Life Sci. 2019, 14, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Ogura, K.; Goshima, Y. IRE-1/XBP-1 pathway of the unfolded protein response is required for properly localizing neuronal UNC-6/Netrin for axon guidance in C. elegans. Genes Cells 2015, 20, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Changhui, M.; Miao, W.; Biquan, L.; Shiuan, W.; Dezheng, D.; Robin, W.; Stephen, R.; Lee, A.S. Targeted mutation of the mouse Grp94 gene disrupts development and perturbs endoplasmic reticulum stress signaling. PLoS ONE 2010, 5, e10852. [Google Scholar]
- Lin, Y.W.; Weng, X.F.; Huang, B.L.; Guo, H.P.; Xu, Y.W.; Peng, Y.H. IGFBP-1 in cancer: Expression, molecular mechanisms, and potential clinical implications. Am. J. Transl. Res. 2021, 13, 813–832. [Google Scholar]
- Petäjä, E.M.; Zhou, Y.; Havana, M.; Hakkarainen, A.; Lundbom, N.; Ihalainen, J.; Yki-Järvinen, H. Phosphorylated IGFBP-1 as a non-invasive predictor of liver fat in NAFLD. Sci. Rep. 2016, 6, 24740. [Google Scholar] [CrossRef]
- Yan, X.D.; Yao, M.; Wang, L.; Zhang, H.J.; Yan, M.J.; Gu, X.; Shi, Y.; Chen, J.; Dong, Z.Z.; Yao, D.F. Overexpression of insulin-like growth factor-I receptor as a pertinent biomarker for hepatocytes malignant transformation. World J. Gastroenterol. 2013, 19, 6084–6092. [Google Scholar] [CrossRef] [PubMed]
- Marchand, A.; Tomkiewicz, C.; Magne, L.; Barouki, R.; Garlatti, M. Endoplasmic reticulum stress induction of insulin-like growth factor-binding protein-1 involves ATF4. J. Biol. Chem. 2006, 281, 19124–19133. [Google Scholar] [CrossRef]
- Leu, J.I.; Crissey, M.A.; Taub, R. Massive hepatic apoptosis associated with TGF-beta1 activation after Fas ligand treatment of IGF binding protein-1-deficient mice. J. Clin. Investig. 2003, 111, 129–139. [Google Scholar] [CrossRef]
- Gatti, R.; De Palo, E.F.; Antonelli, G.; Spinella, P. IGF-I/IGFBP system: Metabolism outline and physical exercise. J. Endocrinol. Investig. 2012, 35, 699–707. [Google Scholar] [CrossRef]
- Xin, H.; Zhang, X.; Sun, D.; Zhang, C.; Hao, Y.; Gu, X. Chronic heat stress increases insulin-like growth factor-1(IGF-1) but does not affect IGF-binding proteins in growing pigs. J. Therm. Biol. 2018, 77, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S. Role of insulin-like growth factor binding protein-3 in glucose and lipid metabolism. Ann. Pediatr. Endocrinol. Metab. 2013, 18, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Argon, Y.; Bresson, S.E.; Marzec, M.T.; Grimberg, A. Glucose-Regulated Protein 94 (GRP94): A Novel Regulator of Insulin-Like Growth Factor Production. Cells 2020, 9, 1844. [Google Scholar] [CrossRef] [PubMed]
- Barton, E.R.; Park, S.; James, J.K.; Makarewich, C.A.; Philippou, A.; Eletto, D.; Lei, H.; Brisson, B.; Ostrovsky, O.; Li, Z.; et al. Deletion of muscle GRP94 impairs both muscle and body growth by inhibiting local IGF production. FASEB J. 2012, 26, 3691–3702. [Google Scholar] [CrossRef]
- Xia, S.W.; Wang, Z.M.; Sun, S.M.; Su, Y.; Li, Z.H.; Shao, J.J.; Tan, S.Z.; Chen, A.P.; Wang, S.J.; Zhang, Z.L.; et al. Endoplasmic reticulum stress and protein degradation in chronic liver disease. Pharmacol. Res. 2020, 161, 105218. [Google Scholar] [CrossRef]
- Wang, T.; Yang, L.; Li, C.; Wang, J.; Zhang, J.; Zhou, Y.; Sun, F.; Wang, H.; Ma, F.; Qian, H. Comprehensive analysis reveals GRP94 is associated with worse prognosis of breast cancer. Transl. Cancer Res. 2021, 10, 298–309. [Google Scholar] [CrossRef]
- Di, X.J.; Wang, Y.J.; Han, D.Y.; Fu, Y.L.; Duerfeldt, A.S.; Blagg, B.S.; Mu, T.W. Grp94 Protein Delivers γ-Aminobutyric Acid Type A (GABAA) Receptors to Hrd1 Protein-mediated Endoplasmic Reticulum-associated Degradation. J. Biol. Chem. 2016, 291, 9526–9539. [Google Scholar] [CrossRef]
- Christianson, J.C.; Shaler, T.A.; Tyler, R.E.; Kopito, R.R. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 2008, 10, 272–282. [Google Scholar] [CrossRef]
- Le Thomas, A.; Ferri, E.; Marsters, S.; Harnoss, J.M.; Lawrence, D.A.; Zuazo-Gaztelu, I.; Modrusan, Z.; Chan, S.; Solon, M.; Chalouni, C.; et al. Decoding non-canonical mRNA decay by the endoplasmic-reticulum stress sensor IRE1alpha. Nat. Commun. 2021, 12, 7310. [Google Scholar] [CrossRef]
- Hetz, C.; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef]
- Lerner, A.G.; Upton, J.P.; Praveen, P.V.; Ghosh, R.; Nakagawa, Y.; Igbaria, A.; Shen, S.; Nguyen, V.; Backes, B.J.; Heiman, M.; et al. IRE1α induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 2012, 16, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Upton, J.P.; Wang, L.; Han, D.; Wang, E.S.; Huskey, N.E.; Lim, L.; Truitt, M.; McManus, M.T.; Ruggero, D.; Goga, A.; et al. IRE1α cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science 2012, 338, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Lei, K.; Davis, R.J. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc. Natl. Acad. Sci. USA 2003, 100, 2432–2437. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.L.; Guo, W.L.; Chen, X.M. Overexpressing microRNA-150 attenuates hypoxia-induced human cardiomyocyte cell apoptosis by targeting glucose-regulated protein-94. Mol. Med. Rep. 2018, 17, 4181–4186. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Ren, H.; Zhang, J.; Yao, W.; Zhao, B.; Miao, J. An Inhibitor of Grp94 Inhibits OxLDL-Induced Autophagy and Apoptosis in VECs and Stabilized Atherosclerotic Plaques. Front. Cardiovasc. Med. 2021, 8, 757591. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Erkan, M.; Streit, S.; Friess, H.; Kleeff, J. Silencing of GRP94 expression promotes apoptosis in pancreatic cancer cells. Int. J. Oncol. 2009, 35, 823–828. [Google Scholar] [CrossRef][Green Version]
- Ho, K.Y.; Yeh, T.S.; Huang, H.H.; Hung, K.F.; Chai, C.Y.; Chen, W.T.; Tsai, S.M.; Chang, N.C.; Chien, C.Y.; Wang, H.M.; et al. Upregulation of phosphorylated HSP27, PRDX2, GRP75, GRP78 and GRP94 in acquired middle ear cholesteatoma growth. Int. J. Mol. Sci. 2013, 14, 14439–14459. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, Y.; Bai, B.; Shang, C.; Fang, J.; Cong, J.; Li, W.; Li, S.; Song, G.; Liu, Z.; et al. Effects of Apoptin-Induced Endoplasmic Reticulum Stress on Lipid Metabolism, Migration, and Invasion of HepG-2 Cells. Front. Oncol. 2021, 11, 614082. [Google Scholar] [CrossRef]
- Bian, Z.M.; Elner, S.G.; Elner, V.M. Regulated expression of caspase-12 gene in human retinal pigment epithelial cells suggests its immunomodulating role. Investig. Ophthalmol. Vis. Sci. 2008, 49, 5593–5601. [Google Scholar] [CrossRef]
- Eletto, D.; Dersh, D.; Argon, Y. GRP94 in ER quality control and stress responses. Semin. Cell Dev. Biol. 2010, 21, 479–485. [Google Scholar] [CrossRef]
- Lee, S.H.; Song, R.; Lee, M.N.; Kim, C.S.; Lee, H.; Kong, Y.Y.; Kim, H.; Jang, S.K. A molecular chaperone glucose-regulated protein 94 blocks apoptosis induced by virus infection. Hepatology 2008, 47, 854–866. [Google Scholar] [CrossRef] [PubMed]



| Carrier No. | Insert Content | shRNA Sequence |
|---|---|---|
| pHS-AVC-1 | GRP94-shRNA1 | 5′-GCTGGAAATGAGGAGTTAACG CGAACGTTAACTCCTCATTTCCAGC-3′ |
| pHS-AVC-2 | GRP94-shRNA2 | 5′-GCAGCAACATAAACTCCTTAA CGAATTAAGGAGTTTATGTTGCTGC-3′ |
| pHS-AVC-3 | GRP94-shRNA3 | 5′-GGAAAGAATTTGGTACCAACA CGAATGTTGGTACCAAATTCTTTCC-3′ |
| Gene Name | Primer | Product (bp) |
|---|---|---|
| GRP94 | F: ACACTGCGGTCAGGGTAT R: TTCTTCGTCGTCGTCTTG | 180 |
| GRP78 | F: AATGGCCGTGTGGAGATCA R: GAGCTGGTTCTTGGCTGCAT | 114 |
| ATF6 | F: CCAGTCCTTGCTGTCACT R: CACACTTCTCATGGTACTCTG | 141 |
| ATF4 | F: GCCCCCGCAGATAGTGAA R: TGGGGAAAGGGGAAGAGTTTG | 103 |
| UBA2 | F: CAGTGCAAAAAGGTCACGCA R: GGCCTAATTCGGACAAGGGT | 105 |
| UBA3 | F: GCGGAGAACAATATGGCGGA R: ACCTTCCCAGTCTCCAGTGT | 120 |
| UBE2E | F: CATGCTCACCAAGTGCATCG R: GTGAAGGGTTGGGAAGGGAG | 136 |
| UBE2I | F: ATTCCACCCGAACGTGTACC R: ATCGTGTAGGCCTCTGCTTG | 165 |
| Bcl2 | F: TGGAGAGCGTAGACAAGGAGA R: CATCGGTTGAAGCGTTCCTG | 184 |
| Caspase-3 | F: AGAATTGGACTGTGGGATTGAGACG R: GCCAGGAATAGTAACCAGGTGCTG | 122 |
| Bax | F: GCTGACGGCAACTTCAACTG R: CCGATCTCGAAGGAAGTCCA | 141 |
| Fas | F: TGATGCCCAAGTGACTGACC R: GCAGAATTGACCCTCACGAT | 161 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Xin, H.; Zhang, C.; Gu, X.; Hao, Y. GRP94 Inhabits the Immortalized Porcine Hepatic Stellate Cells Apoptosis under Endoplasmic Reticulum Stress through Modulating the Expression of IGF-1 and Ubiquitin. Int. J. Mol. Sci. 2022, 23, 14059. https://doi.org/10.3390/ijms232214059
Wang X, Xin H, Zhang C, Gu X, Hao Y. GRP94 Inhabits the Immortalized Porcine Hepatic Stellate Cells Apoptosis under Endoplasmic Reticulum Stress through Modulating the Expression of IGF-1 and Ubiquitin. International Journal of Molecular Sciences. 2022; 23(22):14059. https://doi.org/10.3390/ijms232214059
Chicago/Turabian StyleWang, Xiaohong, Hairui Xin, Chuang Zhang, Xianhong Gu, and Yue Hao. 2022. "GRP94 Inhabits the Immortalized Porcine Hepatic Stellate Cells Apoptosis under Endoplasmic Reticulum Stress through Modulating the Expression of IGF-1 and Ubiquitin" International Journal of Molecular Sciences 23, no. 22: 14059. https://doi.org/10.3390/ijms232214059
APA StyleWang, X., Xin, H., Zhang, C., Gu, X., & Hao, Y. (2022). GRP94 Inhabits the Immortalized Porcine Hepatic Stellate Cells Apoptosis under Endoplasmic Reticulum Stress through Modulating the Expression of IGF-1 and Ubiquitin. International Journal of Molecular Sciences, 23(22), 14059. https://doi.org/10.3390/ijms232214059









