Lactate Activates AMPK Remodeling of the Cellular Metabolic Profile and Promotes the Proliferation and Differentiation of C2C12 Myoblasts
Abstract
1. Introduction
2. Results
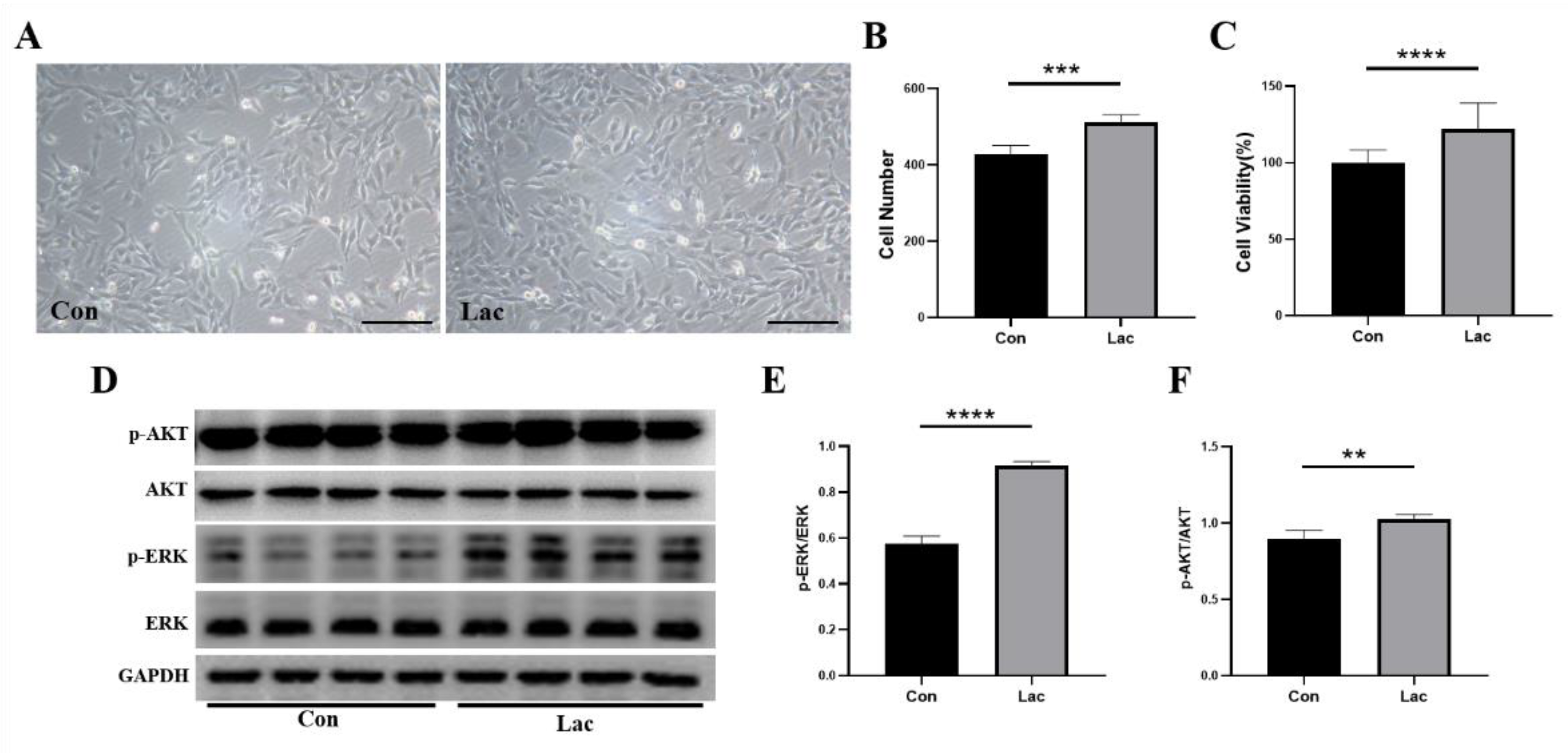
2.1. Lactate Treatment Promoted the Proliferation of Myoblasts
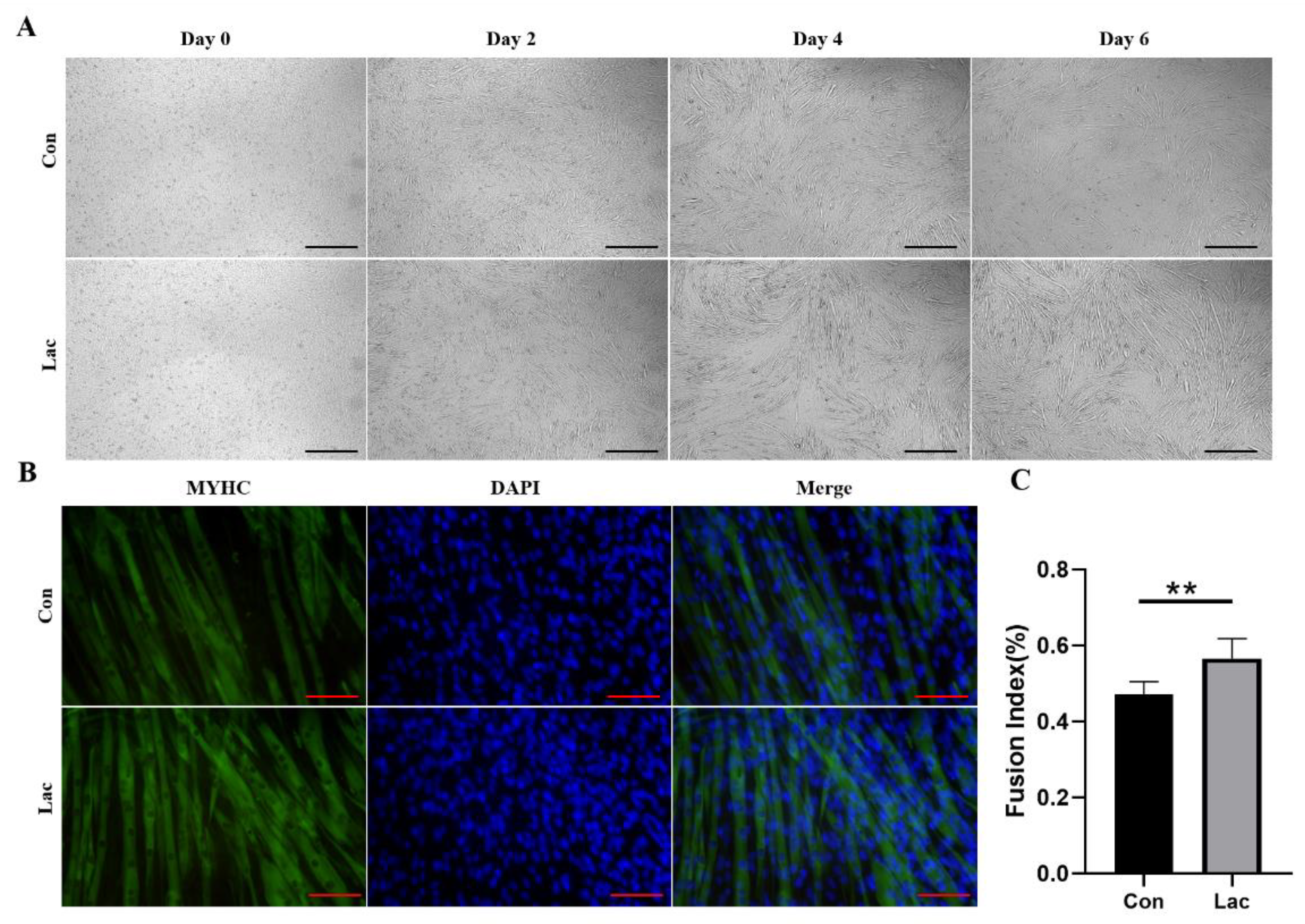
2.2. Lactate Treatment Promoted the Myogenic Differentiation of Myoblasts
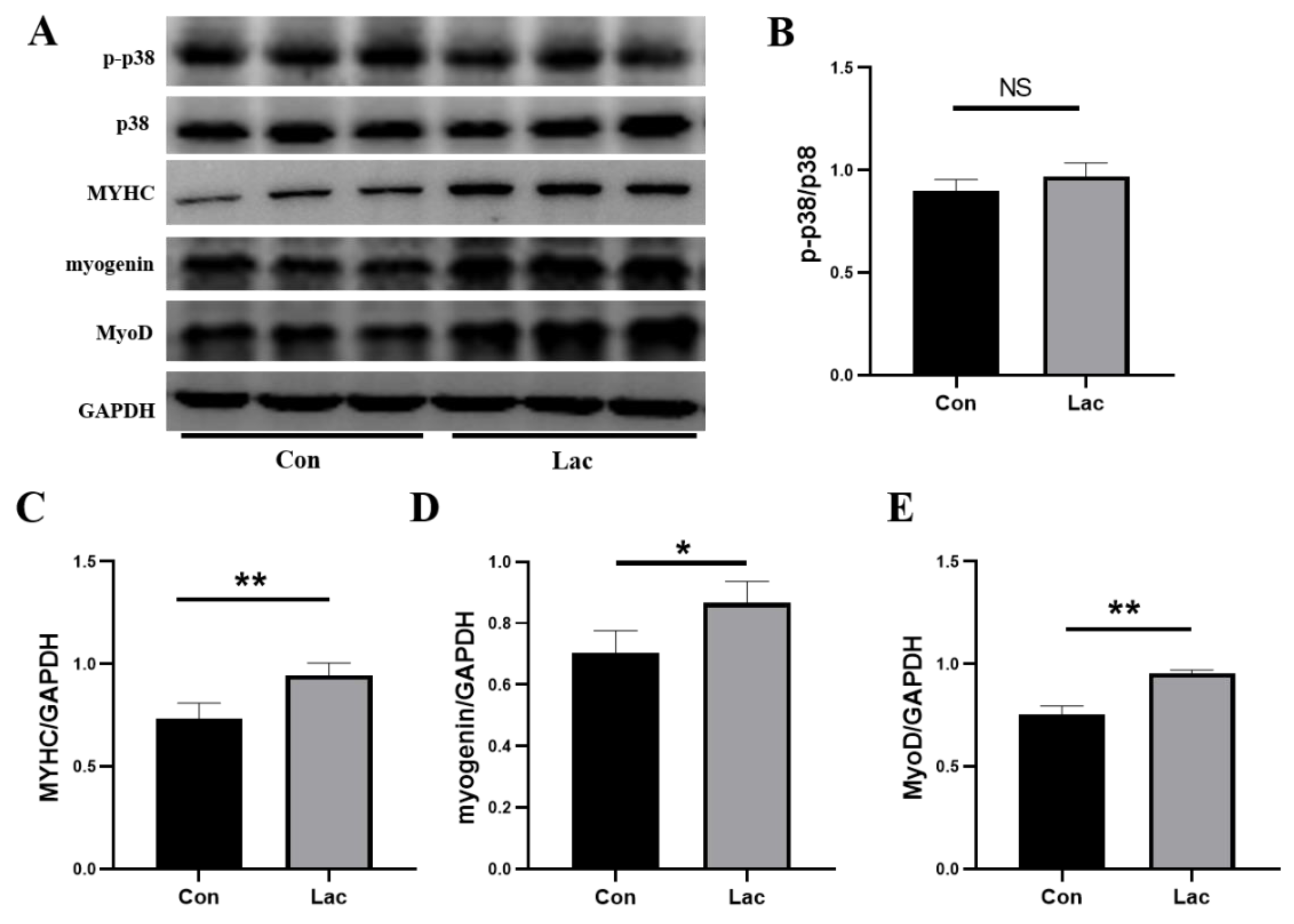
2.3. Lactate Treatment Promoted Expressions of Myogenic Regulatory Factors and Myosin Heavy Chain
2.4. Lactate Treatment Remodeled the Metabolic Profile of Myoblasts
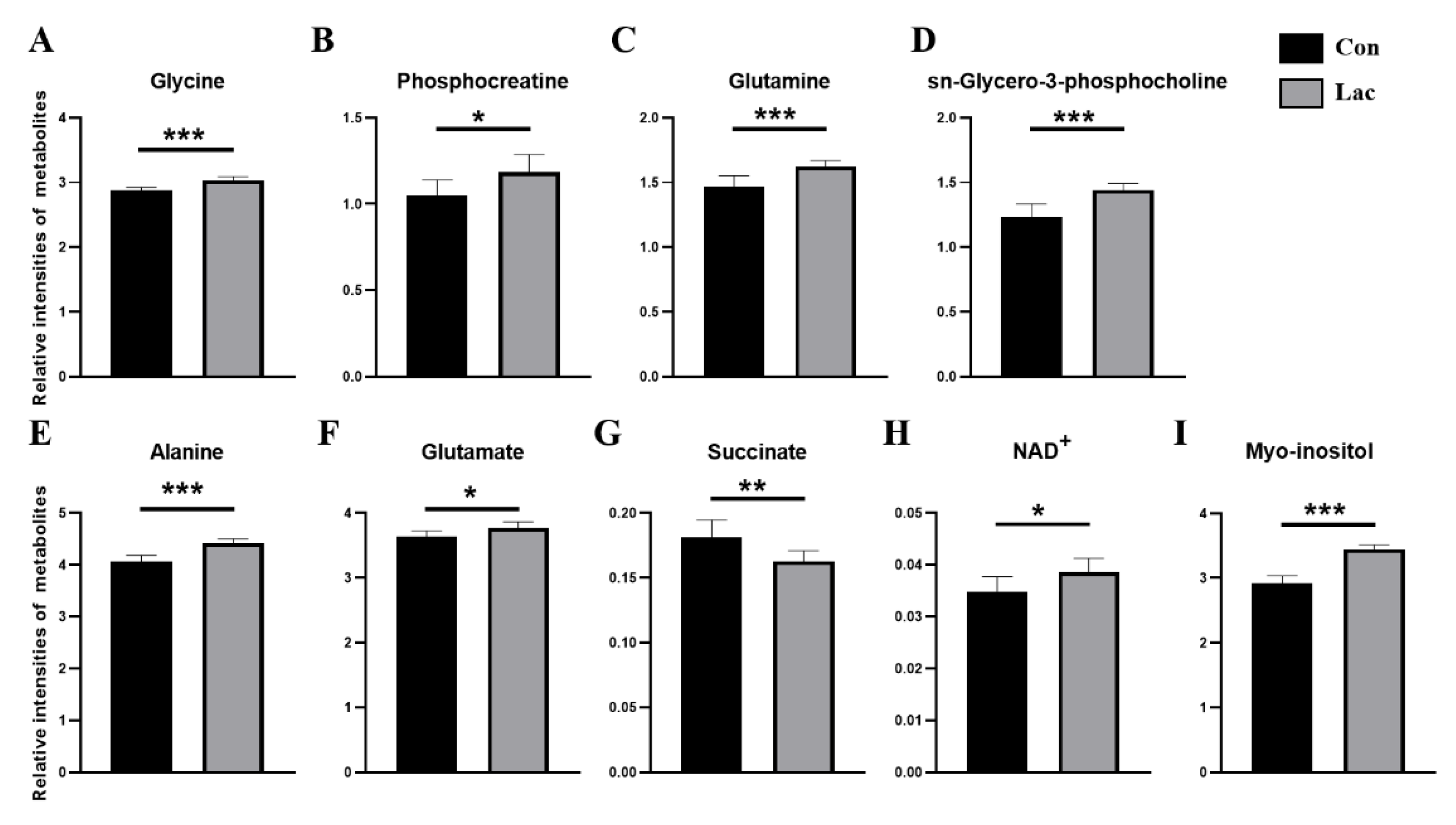
2.5. Lactate Treatment Changed the Levels of Metabolites in Myoblasts
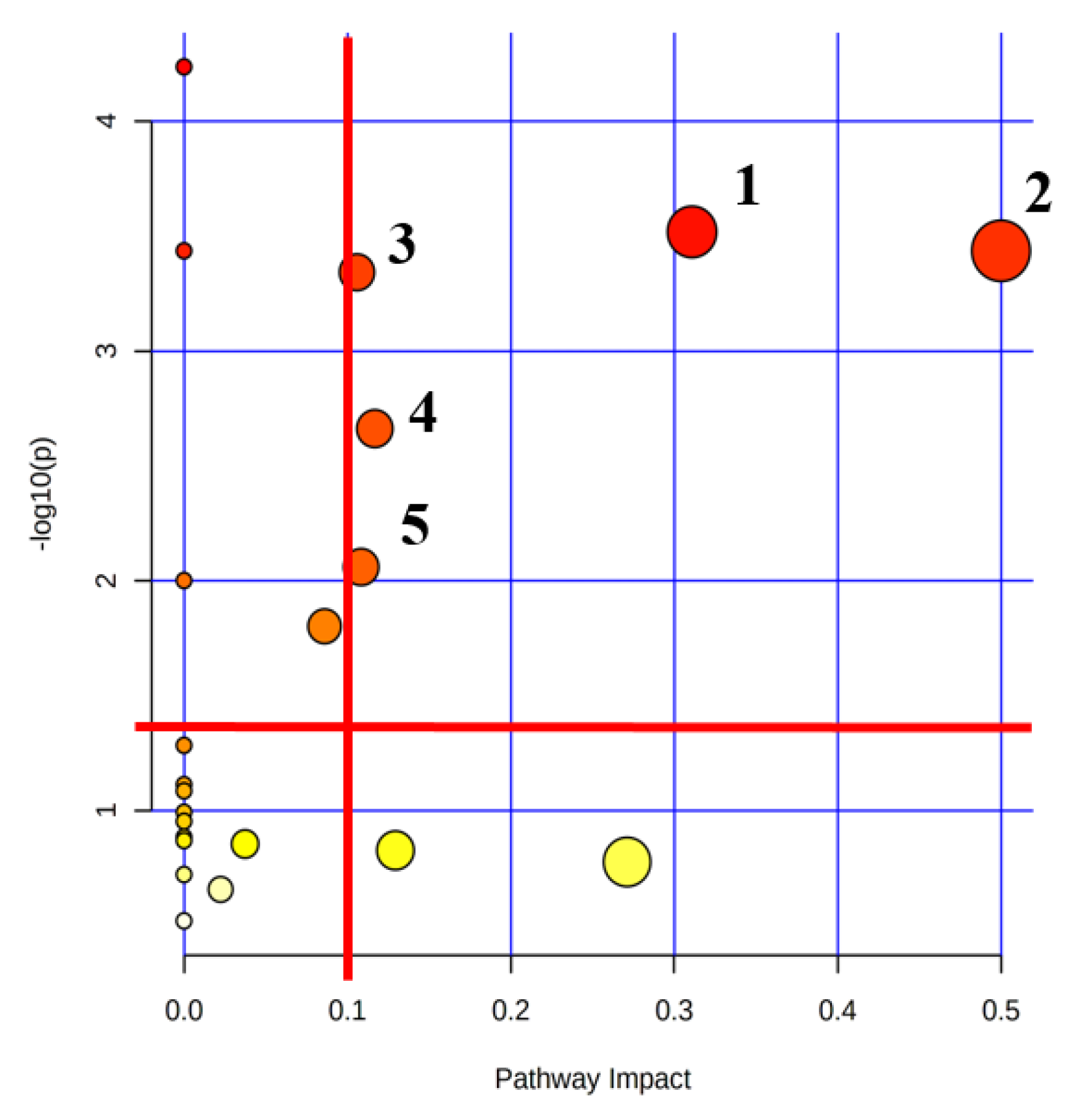
2.6. Lactate Treatment Altered the Metabolic Pathways in Myoblasts
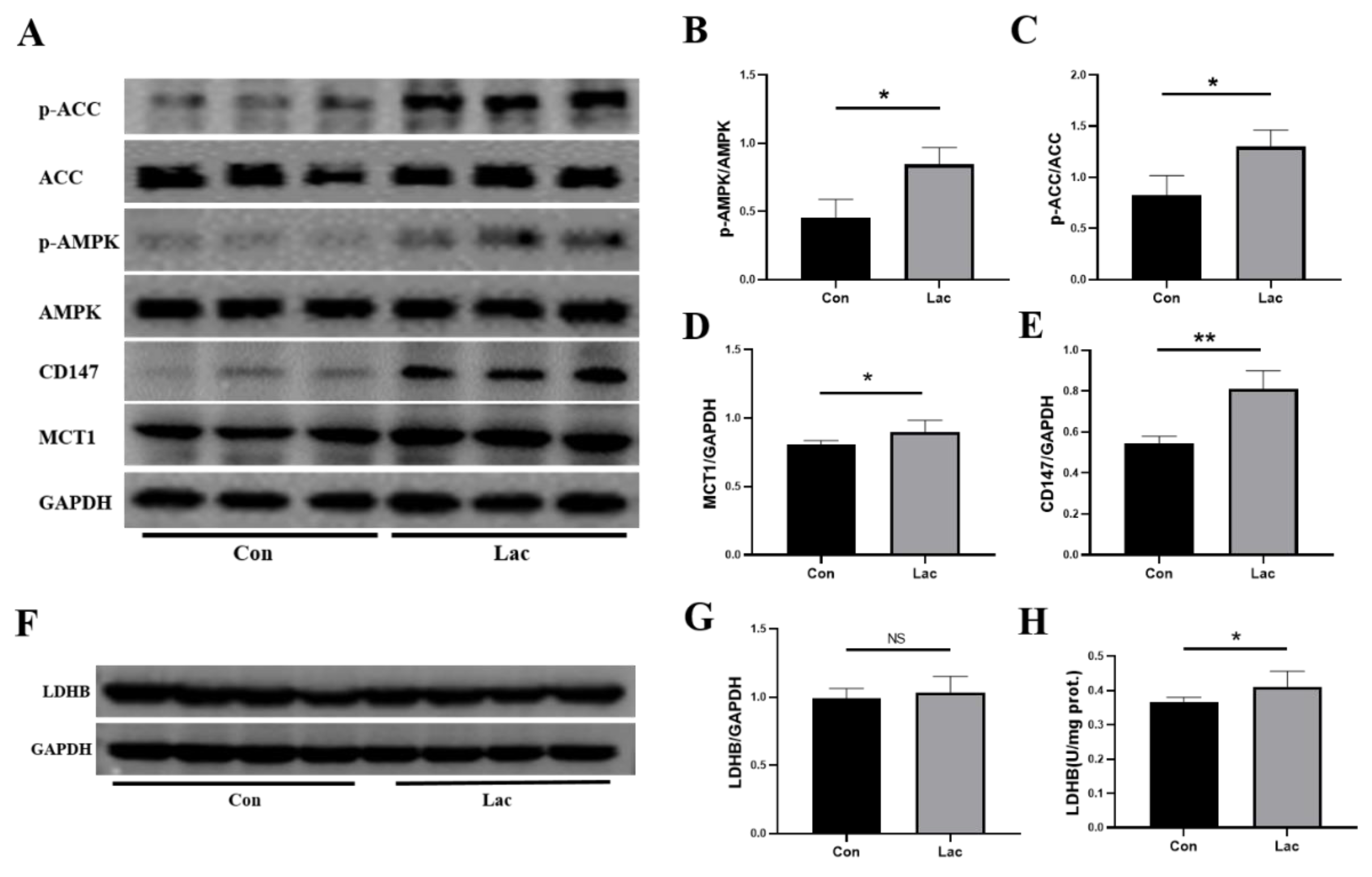
2.7. Lactate Treatment Upregulated the Energy Metabolism in Myoblasts
3. Discussion
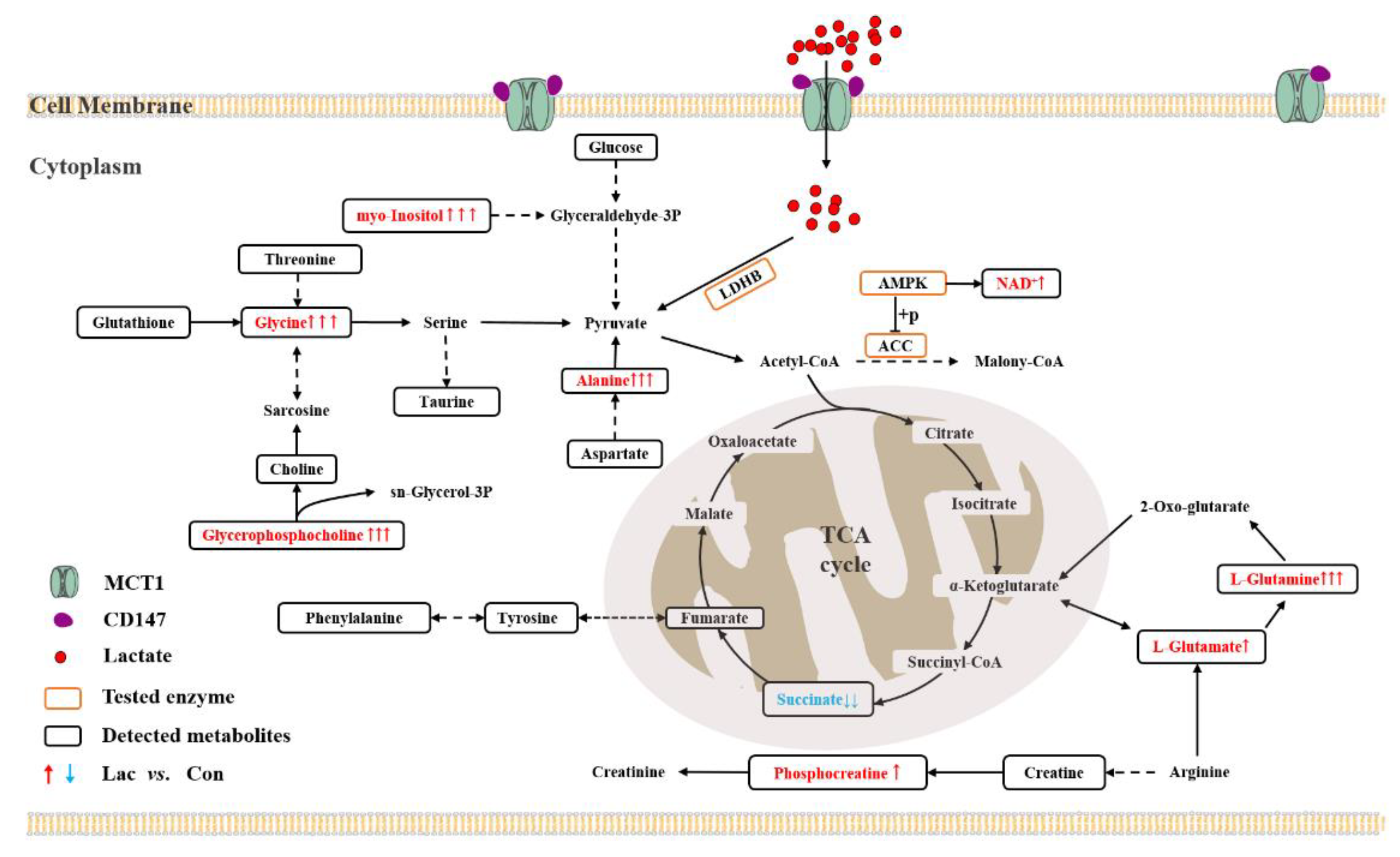
3.1. Lactate Treatment Promotes the Intake and Intracellular Utilization of Lactate
3.2. Lactate Treatment Activates TCA Cycle by Regulating Amino Acid Metabolism
3.3. Lactate Treatment Promotes Energy Production in Myoblasts
3.4. Lactate Treatment Activates AMPK Promoting the Proliferation and Differentiation of Myoblasts
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Viability Assay
4.3. Immunofluorescence Staining and Imaging
4.4. Enzyme Activities Assay
4.5. Western Blotting
4.6. Extraction of Aqueous Metabolites and NMR Sample Preparation
4.7. NMR Experiments
4.8. NMR Data Processing
4.9. NMR Resonance Assignments
4.10. Multivariate Statistics Analysis and Identification of Significant Metabolites
4.11. Univariate Statistics Analysis and Identification of Differential Metabolites
4.12. Metabolic Pathway Analysis and Identification of Crucial Metabolic Pathways
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Frontera, W.R.; Ochala, J. Skeletal muscle: A brief review of structure and function. Calcif. Tissue Int. 2015, 96, 183–195. [Google Scholar] [CrossRef]
- Zierath, J.R.; Hawley, J.A. Skeletal muscle fiber type: Influence on contractile and metabolic properties. PLoS Biol. 2004, 2, e348. [Google Scholar] [CrossRef]
- Mauro, A. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961, 9, 493–495. [Google Scholar] [CrossRef]
- Verdijk, L.B.; Koopman, R.; Schaart, G.; Meijer, K.; Savelberg, H.H.; van Loon, L.J. Satellite cell content is specifically reduced in type II skeletal muscle fibers in the elderly. Am. J. Physiol. Endocrinol. Metab. 2007, 292, E151–E157. [Google Scholar] [CrossRef]
- Verma, M.; Asakura, Y.; Murakonda, B.S.; Pengo, T.; Latroche, C.; Chazaud, B.; McLoon, L.K.; Asakura, A. Muscle Satellite Cell Cross-Talk with a Vascular Niche Maintains Quiescence via VEGF and Notch Signaling. Cell Stem Cell 2018, 23, 530–543.e9. [Google Scholar] [CrossRef]
- Alameddine, H.S.; Dehaupas, M.; Fardeau, M. Regeneration of skeletal muscle fibers from autologous satellite cells multiplied in vitro. An experimental model for testing cultured cell myogenicity. Muscle Nerve 1989, 12, 544–555. [Google Scholar] [CrossRef]
- Arentson-Lantz, E.J.; English, K.L.; Paddon-Jones, D.; Fry, C.S. Fourteen days of bed rest induces a decline in satellite cell content and robust atrophy of skeletal muscle fibers in middle-aged adults. J. Appl. Physiol. 2016, 120, 965–975. [Google Scholar] [CrossRef]
- Zammit, P.S. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Dev. Biol. 2017, 72, 19–32. [Google Scholar] [CrossRef]
- Zanou, N.; Gailly, P. Skeletal muscle hypertrophy and regeneration: Interplay between the myogenic regulatory factors (MRFs) and insulin-like growth factors (IGFs) pathways. Cell. Mol. Life Sci. 2013, 70, 4117–4130. [Google Scholar] [CrossRef]
- Kjellgren, D.; Thornell, L.E.; Andersen, J.; Pedrosa-Domellof, F. Myosin heavy chain isoforms in human extraocular muscles. Investig. Ophthalmol. Vis. Sci. 2003, 44, 1419–1425. [Google Scholar] [CrossRef][Green Version]
- Sokoloff, A.J.; Daugherty, M.; Li, H. Myosin heavy-chain composition of the human hyoglossus muscle. Dysphagia 2010, 25, 81–93. [Google Scholar] [CrossRef]
- Pedrosa-Domellof, F.; Kjellgren, D.; Thornell, L.E. Myosin heavy chain isoforms in adult human extraocular muscles. FASEB J. 2001, 15, A386. [Google Scholar]
- Wu, Z.; Woodring, P.J.; Bhakta, K.S.; Tamura, K.; Wen, F.; Feramisco, J.R.; Karin, M.; Wang, J.Y.; Puri, P.L. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol. Cell. Biol. 2000, 20, 3951–3964. [Google Scholar] [CrossRef]
- Li, J.; Johnson, S.E. ERK2 is required for efficient terminal differentiation of skeletal myoblasts. Biochem. Biophys. Res. Commun. 2006, 345, 1425–1433. [Google Scholar] [CrossRef]
- Lluis, F.; Perdiguero, E.; Nebreda, A.R.; Munoz-Canoves, P. Regulation of skeletal muscle gene expression by p38 MAP kinases. Trends Cell Biol. 2006, 16, 36–44. [Google Scholar] [CrossRef]
- Elia, D.; Madhala, D.; Ardon, E.; Reshef, R.; Halevy, O. Sonic hedgehog promotes proliferation and differentiation of adult muscle cells: Involvement of MAPK/ERK and PI3K/Akt pathways. Biochim. Biophys. Acta 2007, 1773, 1438–1446. [Google Scholar] [CrossRef]
- Bodine, S.C.; Stitt, T.N.; Gonzalez, M.; Kline, W.O.; Stover, G.L.; Bauerlein, R.; Zlotchenko, E.; Scrimgeour, A.; Lawrence, J.C.; Glass, D.J.; et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001, 3, 1014–1019. [Google Scholar] [CrossRef]
- Ryall, J.G. Metabolic reprogramming as a novel regulator of skeletal muscle development and regeneration. FEBS J. 2013, 280, 4004–4013. [Google Scholar] [CrossRef]
- Dell’Orso, S.; Juan, A.H.; Ko, K.D.; Naz, F.; Perovanovic, J.; Gutierrez-Cruz, G.; Feng, X.; Sartorelli, V. Single cell analysis of adult mouse skeletal muscle stem cells in homeostatic and regenerative conditions. Development 2019, 146, dev174177. [Google Scholar] [CrossRef]
- Theret, M.; Gsaier, L.; Schaffer, B.; Juban, G.; Ben Larbi, S.; Weiss-Gayet, M.; Bultot, L.; Collodet, C.; Foretz, M.; Desplanches, D.; et al. AMPK alpha 1-LDH pathway regulates muscle stem cell self-renewal by controlling metabolic homeostasis. EMBO J. 2017, 36, 1946–1962. [Google Scholar] [CrossRef]
- Fu, X.; Zhu, M.J.; Dodson, M.V.; Du, M. AMP-activated Protein Kinase Stimulates Warburg-like Glycolysis and Activation of Satellite Cells during Muscle Regeneration. J. Biol. Chem. 2015, 290, 26445–26456. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Cappellesso, F.; Amorim, R.; Serneels, J.; Virga, F.; Eelen, G.; Carobbio, S.; Rincon, M.Y.; Maechler, P.; De Bock, K.; et al. Macrophage-derived glutamine boosts satellite cells and muscle regeneration. Nature 2020, 587, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Rabinowitz, J.D.; Enerback, S. Lactate: The ugly duckling of energy metabolism. Nat. Metab. 2020, 2, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, J.L.; Tietz, E.; Two-Feathers, T.; Paull, J.; Chapman, K. Lactate, fructose and glucose oxidation profiles in sports drinks and the effect on exercise performance. PLoS ONE 2007, 2, e927. [Google Scholar] [CrossRef]
- Sun, S.R.; Li, H.; Chen, J.H.; Qian, Q. Lactic Acid: No Longer an Inert and End-Product of Glycolysis. Physiology 2017, 32, 453–463. [Google Scholar] [CrossRef]
- Brooks, G.A. The Science and Translation of Lactate Shuttle Theory. Cell Metab. 2018, 27, 757–785. [Google Scholar] [CrossRef]
- Brooks, G.A. Lactate as a fulcrum of metabolism. Redox Biol. 2020, 35, 101454. [Google Scholar] [CrossRef]
- Sekine, N.; Cirulli, V.; Regazzi, R.; Brown, L.J.; Gine, E.; Tamarit-Rodriguez, J.; Girotti, M.; Marie, S.; MacDonald, M.J.; Wollheim, C.B. Low Lactate-Dehydrogenase and High Mitochondrial Glycerol Phosphate Dehydrogenase in Pancreatic Beta-Cells. Potential Role in Nutrient Sensing. J. Biol. Chem. 1994, 269, 4895–4902. [Google Scholar] [CrossRef]
- Halestrap, A.P. Monocarboxylic Acid Transport. Compr. Physiol. 2013, 3, 1611–1643. [Google Scholar] [CrossRef]
- Juel, C.; Halestrap, A.P. Lactate transport in skeletal muscle—Role and regulation of the monocarboxylate transporter. J. Physiol. 1999, 517, 633–642. [Google Scholar] [CrossRef]
- Zhao, C.; Wilson, M.C.; Schuit, F.; Halestrap, A.P.; Rutter, G.A. Expression and distribution of lactate/monocarboxylate transporter isoforms in pancreatic islets and the exocrine pancreas. Diabetes 2001, 50, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Muri, J.; Fitzgerald, G.; Gorski, T.; Gianni-Barrera, R.; Masschelein, E.; D’Hulst, G.; Gilardoni, P.; Turiel, G.; Fan, Z.; et al. Endothelial Lactate Controls Muscle Regeneration from Ischemia by Inducing M2-like Macrophage Polarization. Cell Metab. 2020, 31, 1136–1153. [Google Scholar] [CrossRef] [PubMed]
- Willkomm, L.; Schubert, S.; Jung, R.; Elsen, M.; Borde, J.; Gehlert, S.; Suhr, F.; Bloch, W. Lactate regulates myogenesis in C2C12 myoblasts in vitro. Stem Cell Res. 2014, 12, 742–753. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.S.; Kim, S.; Moon, S.; Park, D.H.; Kang, J.H. Lactate Overload Inhibits Myogenic Activity in C2C12 Myotubes. Open Life Sci. 2019, 14, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Ohno, Y.; Oyama, A.; Kaneko, H.; Egawa, T.; Yokoyama, S.; Sugiura, T.; Ohira, Y.; Yoshioka, T.; Goto, K. Lactate increases myotube diameter via activation of MEK/ERK pathway in C2C12 cells. Acta Physiol. 2018, 223, e13042. [Google Scholar] [CrossRef]
- Ohno, Y.; Ando, K.; Ito, T.; Suda, Y.; Matsui, Y.; Oyama, A.; Kaneko, H.; Yokoyama, S.; Egawa, T.; Goto, K. Lactate Stimulates a Potential for Hypertrophy and Regeneration of Mouse Skeletal Muscle. Nutrients 2019, 11, 869. [Google Scholar] [CrossRef]
- Philp, A.; Macdonald, A.L.; Watt, P.W. Lactate—A signal coordinating cell and systemic function. J. Exp. Biol. 2005, 208, 4561–4575. [Google Scholar] [CrossRef]
- Yun, K.; Wold, B. Skeletal muscle determination and differentiation: Story of a core regulatory network and its context. Curr. Opin. Cell Biol. 1996, 8, 877–889. [Google Scholar] [CrossRef]
- Le Grand, F.; Rudnicki, M.A. Skeletal muscle satellite cells and adult myogenesis. Curr. Opin. Cell Biol. 2007, 19, 628–633. [Google Scholar] [CrossRef]
- Soukup, T.; Jirmanova, I.; Mrackova, K.; Zacharova, G.; Thornell, L.E. Expression of myosin heavy chain (MyHC) isoforms in rat intrafusal muscle fibres after neonatal deefferentation and subsequent denervation. Gen. Physiol. Biophys. 1999, 18 (Suppl. 1), 81–83. [Google Scholar]
- Lee, D.C.; Sohn, H.A.; Park, Z.Y.; Oh, S.; Kang, Y.K.; Lee, K.M.; Kang, M.; Jang, Y.J.; Yang, S.J.; Hong, Y.K.; et al. A Lactate-Induced Response to Hypoxia. Cell 2015, 161, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Nalbandian, M.; Takeda, M. Lactate as a Signaling Molecule That Regulates Exercise-Induced Adaptations. Biology 2016, 5, 38. [Google Scholar] [CrossRef] [PubMed]
- Feldman, J.L.; Stockdale, F.E. Skeletal muscle satellite cell diversity: Satellite cells form fibers of different types in cell culture. Dev. Biol. 1991, 143, 320–334. [Google Scholar] [CrossRef]
- Cerda-Kohler, H.; Henríquez-Olguín, C.; Casas, M.; Jensen, T.E.; Llanos, P.; Jaimovich, E. Lactate administration activates the ERK1/2, mTORC1, and AMPK pathways differentially according to skeletal muscle type in mouse. Physiol. Rep. 2018, 6, e13800. [Google Scholar] [CrossRef]
- Hardie, D.G. AMPK: A key regulator of energy balance in the single cell and the whole organism. Int. J. Obes. 2008, 32, S7–S12. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Gao, Y.; Huang, C.; Lin, D. NMR-Based Metabolomic Analysis for the Effects of alpha-Ketoglutarate Supplementation on C2C12 Myoblasts in Different Energy States. Molecules 2021, 26, 1841. [Google Scholar] [CrossRef] [PubMed]
- Stainsby, W.N.; Brechue, W.F.; O’Drobinak, D.M. Regulation of muscle lactate production. Med. Sci. Sports Exerc. 1991, 23, 907–911. [Google Scholar] [CrossRef]
- Hui, S.; Ghergurovich, J.M.; Morscher, R.J.; Jang, C.; Teng, X.; Lu, W.; Esparza, L.A.; Reya, T.; Zhan, L.; Yanxiang Guo, J.; et al. Glucose feeds the TCA cycle via circulating lactate. Nature 2017, 551, 115–118. [Google Scholar] [CrossRef]
- Van Hall, G.; Stømstad, M.; Rasmussen, P.; Jans, Ø.; Zaar, M.; Gam, C.; Quistorff, B.; Secher, N.H.; Nielsen, H.B. Blood lactate is an important energy source for the human brain. J. Cereb. Blood Flow Metab. 2009, 29, 1121–1129. [Google Scholar] [CrossRef]
- Halestrap, A.P. The SLC16 gene family—Structure, role and regulation in health and disease. Mol. Asp. Med. 2013, 34, 337–349. [Google Scholar] [CrossRef]
- Liu, Z.Q.; Huang, C.H.; Liu, Y.; Lin, D.H.; Zhao, Y.F. NMR-based metabolomic analysis of the effects of alanyl-glutamine supplementation on C2C12 myoblasts injured by energy deprivation. RSC Adv. 2018, 8, 16114–16125. [Google Scholar] [CrossRef] [PubMed]
- Matrka, M.C.; Watanabe, M.; Muraleedharan, R.; Lambert, P.F.; Lane, A.N.; Romick-Rosendale, L.E.; Wells, S.I. Overexpression of the human DEK oncogene reprograms cellular metabolism and promotes glycolysis. PLoS ONE 2017, 12, e0177952. [Google Scholar] [CrossRef] [PubMed]
- Dohl, J.; Passos, M.E.; Foldi, J.; Chen, Y.; Pithon-Curi, T.; Curi, R.; Gorjao, R.; Deuster, P.A.; Yu, T. Glutamine depletion disrupts mitochondrial integrity and impairs C2C12 myoblast proliferation, differentiation, and the heat-shock response. Nutr. Res. 2020, 84, 42–52. [Google Scholar] [CrossRef]
- Westerblad, H.; Bruton, J.D.; Katz, A. Skeletal muscle: Energy metabolism, fiber types, fatigue and adaptability. Exp. Cell Res. 2010, 316, 3093–3099. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Hussien, R.; Oommen, S.; Gohil, K.; Brooks, G.A. Lactate sensitive transcription factor network in L6 cells: Activation of MCT1 and mitochondrial biogenesis. FASEB J. 2007, 21, 2602–2612. [Google Scholar] [CrossRef]
- Kitaoka, Y.; Takeda, K.; Tamura, Y.; Hatta, H. Lactate administration increases mRNA expression of PGC-1 alpha and UCP3 in mouse skeletal muscle. Appl. Physiol. Nutr. Metab. 2016, 41, 695–698. [Google Scholar] [CrossRef]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef]
- Zhou, L.; Lu, R.; Huang, C.; Lin, D. Taurine Protects C2C12 Myoblasts From Impaired Cell Proliferation and Myotube Differentiation Under Cisplatin-Induced ROS Exposure. Front. Mol. Biosci. 2021, 8, 685362. [Google Scholar] [CrossRef]
- Xu, W.; Lin, D.; Huang, C. NMR-based metabolomic analysis for the effects of creatine supplementation on mouse myoblast cell line C2C12. Acta Biochim. Biophys. Sin. 2017, 49, 617–627. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Liu, X.; Huang, C.; Lin, D. Lactate Activates AMPK Remodeling of the Cellular Metabolic Profile and Promotes the Proliferation and Differentiation of C2C12 Myoblasts. Int. J. Mol. Sci. 2022, 23, 13996. https://doi.org/10.3390/ijms232213996
Zhou Y, Liu X, Huang C, Lin D. Lactate Activates AMPK Remodeling of the Cellular Metabolic Profile and Promotes the Proliferation and Differentiation of C2C12 Myoblasts. International Journal of Molecular Sciences. 2022; 23(22):13996. https://doi.org/10.3390/ijms232213996
Chicago/Turabian StyleZhou, Yu, Xi Liu, Caihua Huang, and Donghai Lin. 2022. "Lactate Activates AMPK Remodeling of the Cellular Metabolic Profile and Promotes the Proliferation and Differentiation of C2C12 Myoblasts" International Journal of Molecular Sciences 23, no. 22: 13996. https://doi.org/10.3390/ijms232213996
APA StyleZhou, Y., Liu, X., Huang, C., & Lin, D. (2022). Lactate Activates AMPK Remodeling of the Cellular Metabolic Profile and Promotes the Proliferation and Differentiation of C2C12 Myoblasts. International Journal of Molecular Sciences, 23(22), 13996. https://doi.org/10.3390/ijms232213996





