Metabolomic Profiling of Plasma, Urine, and Saliva of Kidney Transplantation Recipients
Abstract
1. Introduction
2. Results
2.1. Comparison of Plasma Metabolomics among Donors and Kidney Transplant Recipients
2.2. Comparison of Plasma Metabolomics between Patients with Kidney Transplantations with Stable (S) and Impaired Graft (I) Kidney Functions
2.3. Comparison of Plasma, Urine, and Saliva Metabolomics of Group I
2.4. Comparison of Metabolome Profiles in Patients with Kidney Transplantation Grouped by Banff Classification
2.4.1. Comparison of (i + t)
2.4.2. Comparison of c4d
2.4.3. Comparison of aah
3. Discussion
4. Material and Methods
4.1. Study Design
4.2. Sample Processing for Metabolomic Analysis
4.3. Instrument of Metabolomic Analysis
4.4. CE-TOFMS Conditions for Cationic Metabolite Analysis
4.5. CE-TOFMS Conditions for Anionic Metabolite Analysis
4.6. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Item | i + t (−) | i + t (+) | p-Value |
|---|---|---|---|
| N | 27 | 19 | |
| Sex, Male/Female | 20/7 | 10/9 | 0.13 a |
| Age | 48 (23–74) | 54 (34–74) | 0.14 b |
| Donor’s age | 53 (20–71) | 54 (5–72) | 0.98 b |
| BMI | 22.4 ± 3.47 | 23.4 ± 3.31 | 0.65 b |
| Creatinine | 1.53 ± 0.72 | 1.73 ± 0.58 | 0.20 b |
| Item | c4d (−) | c4d (+) | p-Value |
|---|---|---|---|
| n | 28 | 18 | |
| Sex, Male/Female | 18/10 | 12/6 | 0.87 a |
| Age | 49.5 (26–74) | 51 (23–68) | 0.83 b |
| Donor’s age | 48 (5–72) | 55.5 (26–67) | 0.44 b |
| BMI | 22.7 ± 3.62 | 22.9 ± 3.13 | 0.97 b |
| Creatinine | 1.50 ± 0.74 | 1.75 ± 0.51 | 0.027 *b |
| Item | aah (−) | aah (+) | p-Value |
|---|---|---|---|
| n | 41 | 5 | |
| Sex, Male/Female | 25/5 | 16/0 | 0.084 a |
| Age | 50 (23–74) | 42 (30–68) | 0.78 b |
| Donor’s age | 53 (5–72) | 54 (36–71) | 0.76 b |
| BMI | 22.9 (±3.45) | 22.1 (±3.21) | 0.75 b |
| Creatinine | 1.54 ± 0.64 | 2.1 ± 0.71 | 0.13 b |
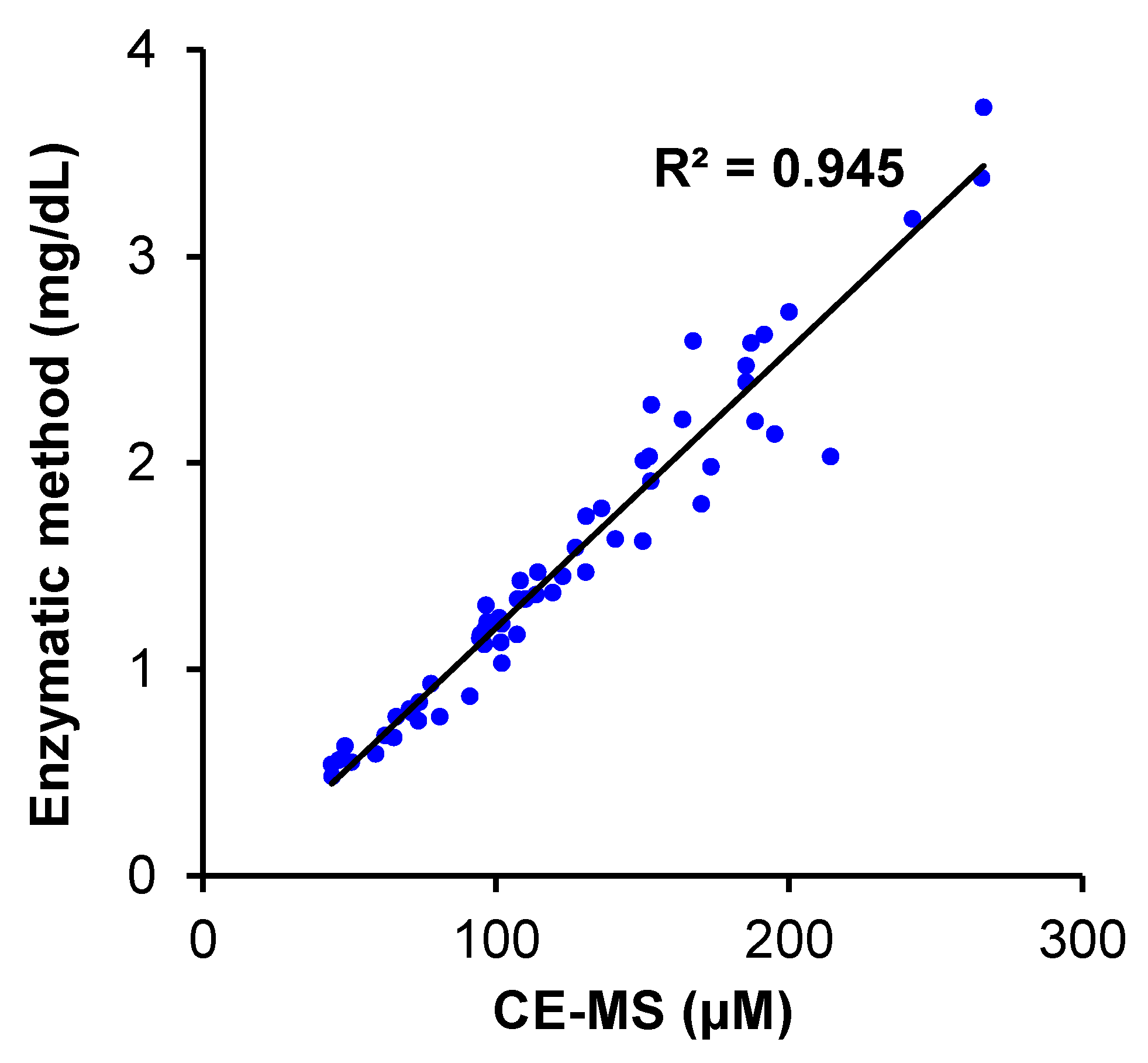
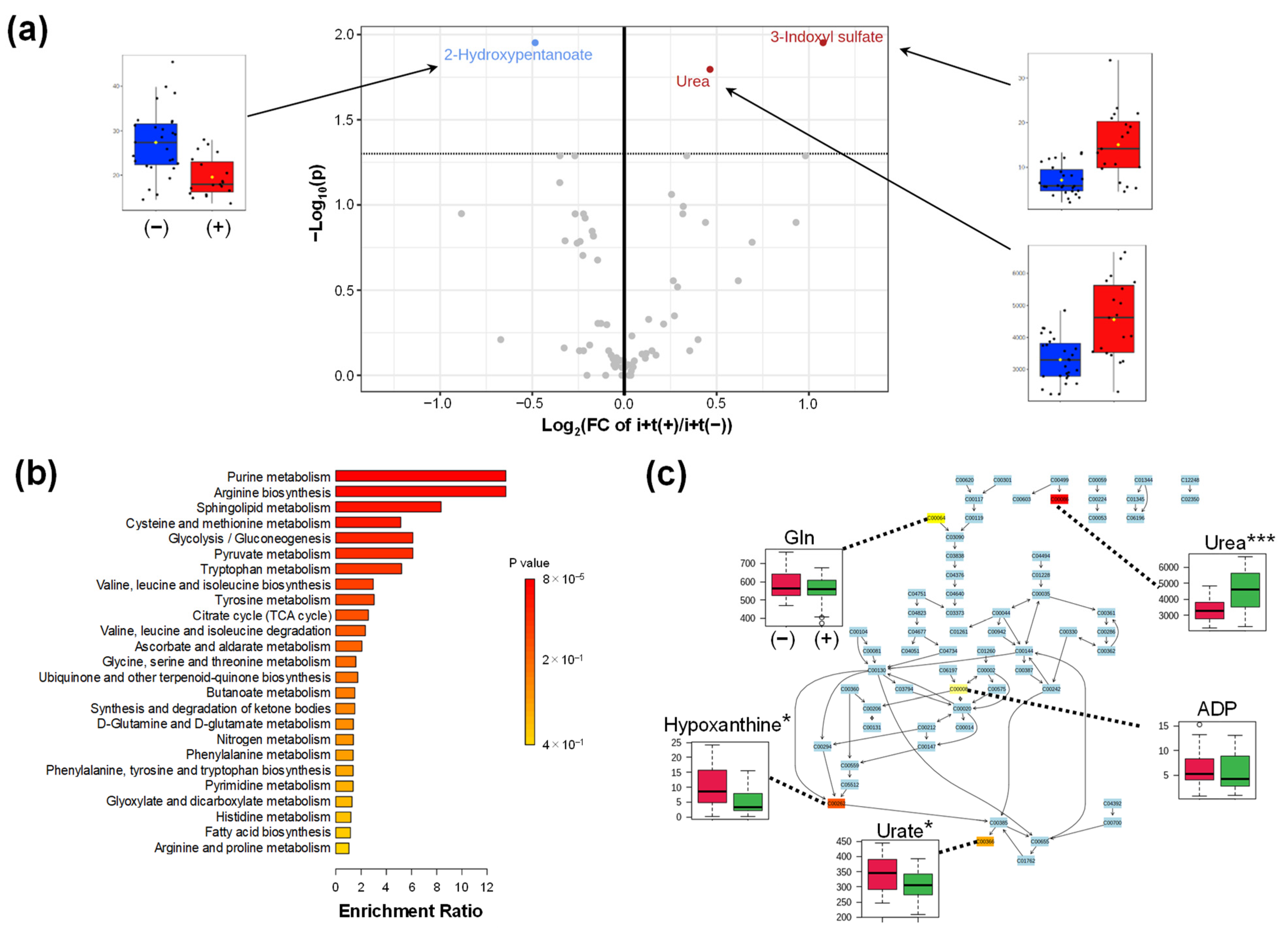
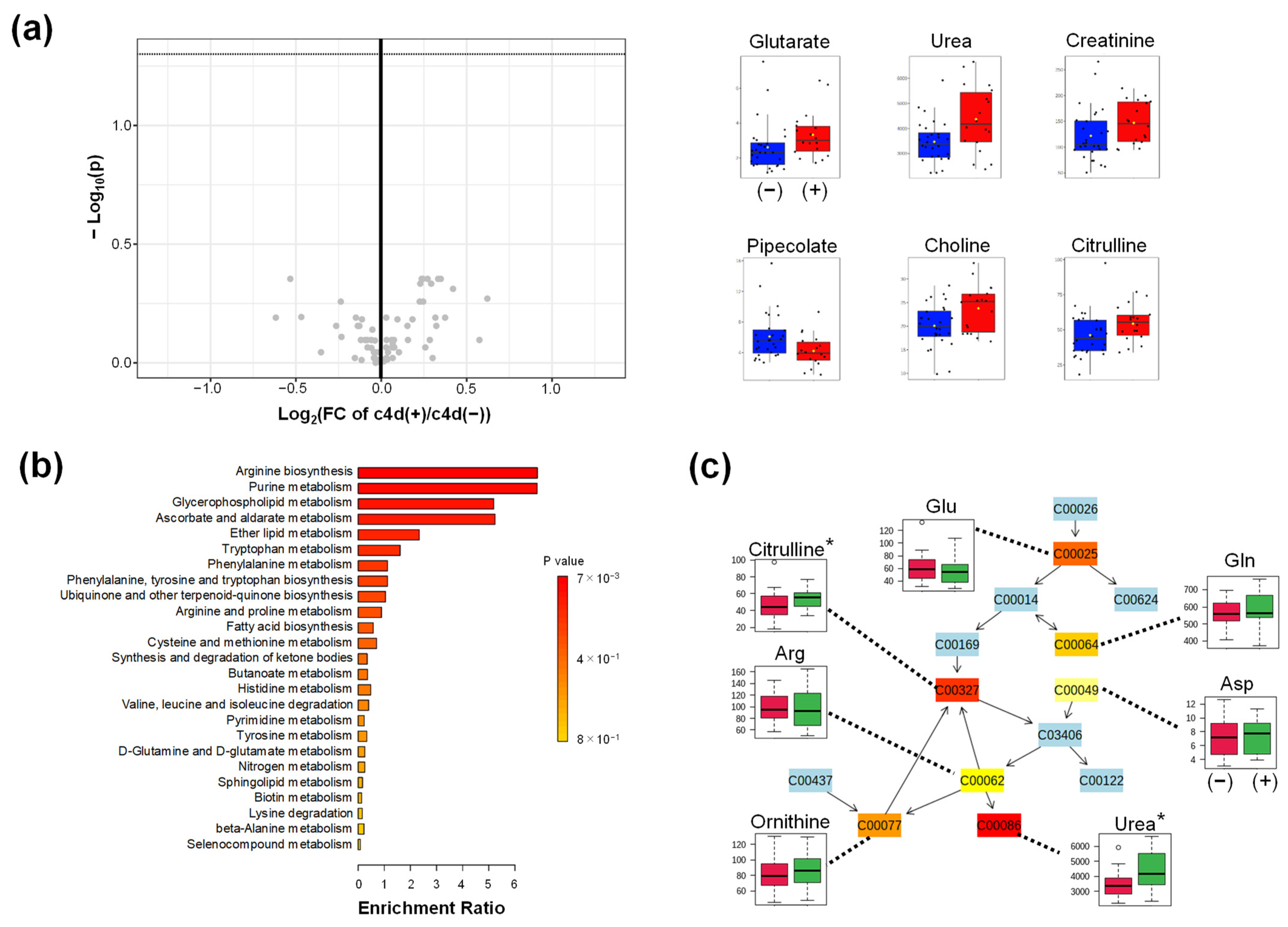
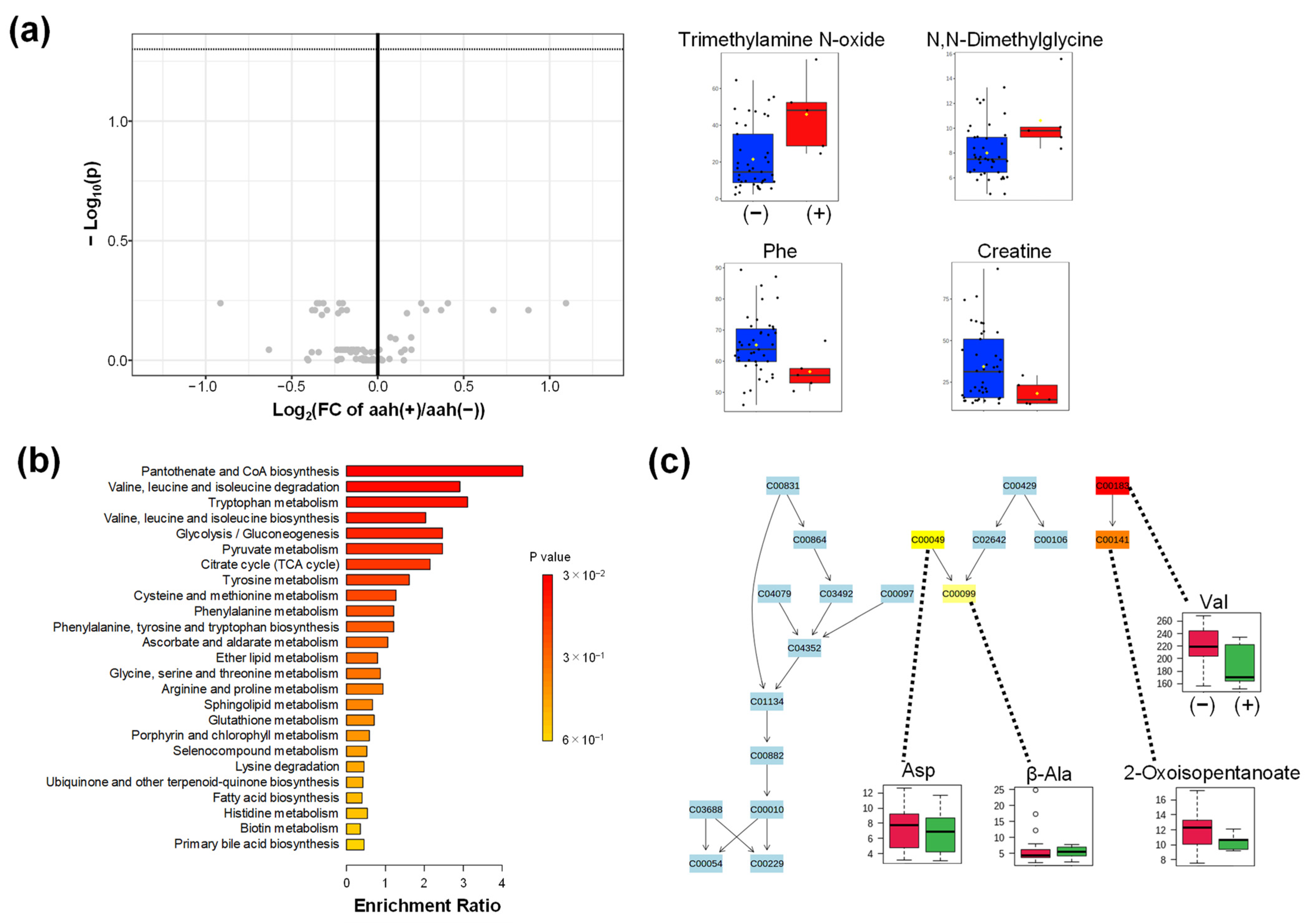
Appendix B
- The MetaboAnalyst database was accessed from https://www.metaboanalyst.ca/ (accessed on 1 June 2022) and started by pressing “Click here to start” and selecting “Statistical Analysis [one factor]”;
- The data matrix of quantified concentrations (metabolite × sample) was uploaded. (The first line and the second line include sample and group names. The first line includes metabolite names. The other cells include the quantified values);
- The “None” option was selected for normalization, data transformation, and data scaling options. Then, “Normalize” and “Proceed” to go to the statistics page were clicked;
- Afterward, the “heatmap” menu was clicked, and the following options were selected: Data source: Normalized data, Standardization: Autoscale features, Distance measure: Euclidean; clustering methods: Ward; Color contrast: Default. The “use top 25” option with the T-test/ANOVA method was checked, and the option to “Submit” was clicked to show the heatmap;
- Then, the “volcano plot” menu was clicked. The following options—Analysis: Unpaired. Non-parametric test: Yes, p-value threshold: 0.05 with FDR, Group variance: Equal, were selected. Then “Submit” was pressed to show a volcano plot.
- Then, the “PLS-DA” menu was clicked. The “Cross-Validation” tab was opened. We selected the following options: Maximum components to search: 5, Cross-validation method: 10-fold CV: Performance measure: Q2. Then, “Update” to measure R2 and Q2 was clicked. Score plots and VIP scores were obtained using the “2D Scores Plots” and “Imp. Features” tab;
- Then, “Home” menu and “Click here to start” were clicked, and “Enrichment analysis” was selected;
- Afterward, we opened the “Quantitative Enrichment Analysis” tab, uploaded the data matrix, and performed the data processing as described in step 3;
- Finally, the “KEGG” option was selected in the “pathway-based analysis” menu, and the “Submit” option was clicked to conduct an enrichment analysis.
References
- Jalalzadeh, M.; Mousavinasab, N.; Peyrovi, S.; Ghadiani, M.H. The impact of acute rejection in kidney transplantation on long-term allograft and patient outcome. Nephro-Urol. Mon. 2015, 7, e24439. [Google Scholar] [CrossRef] [PubMed]
- Koo, E.H.; Jang, H.R.; Lee, J.E.; Park, J.B.; Kim, S.J.; Kim, D.J.; Kim, Y.G.; Oh, H.Y.; Huh, W. The impact of early and late acute rejection on graft survival in renal transplantation. Kidney Res. Clin. Pract. 2015, 34, 160–164. [Google Scholar] [CrossRef]
- Keown, P.A. Predicting long-term outcome in renal transplantation. Kidney Int. 2013, 84, 650–652. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meier-Kriesche, H.U.; Schold, J.D.; Srinivas, T.R.; Kaplan, B. Lack of improvement in renal allograft survival despite a marked decrease in acute rejection rates over the most recent era. Am. J. Transplant. 2004, 4, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Humar, A.; Matas, A.J. Surgical complications after kidney transplantation. In Seminars in Dialysis; Blackwell Science Inc.: Malden, MA, USA, 2005; pp. 505–510. [Google Scholar]
- Furness, P.N.; Philpott, C.M.; Chorbadjian, M.T.; Nicholson, M.L.; Bosmans, J.L.; Corthouts, B.L.; Bogers, J.J.; Schwarz, A.; Gwinner, W.; Haller, H.; et al. Protocol biopsy of the stable renal transplant: A multicenter study of methods and complication rates. Transplantation 2003, 76, 969–973. [Google Scholar] [CrossRef]
- Schwarz, A.; Gwinner, W.; Hiss, M.; Radermacher, J.; Mengel, M.; Haller, H. Safety and adequacy of renal transplant protocol biopsies. Am. J. Transplant. 2005, 5, 1992–1996. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics: The principles and potential applications to transplantation. Am. J. Transplant. 2005, 5, 2814–2820. [Google Scholar] [CrossRef]
- Bonneau, E.; Tetreault, N.; Robitaille, R.; Boucher, A.; De Guire, V. Metabolomics: Perspectives on potential biomarkers in organ transplantation and immunosuppressant toxicity. Clin. Biochem. 2016, 49, 377–384. [Google Scholar] [CrossRef]
- Muhrez, K.; Nadal-Desbarats, L.; Halimi, J.-M.; Dieme, B.; Büchler, M.; Emond, P.; Blasco, H.; Le Guellec, C. Elucidating time-dependent changes in the urinary metabolome of renal transplant patients by a combined 1H NMR and GC-MS approach. Mol. Biosyst. 2015, 11, 2493–2510. [Google Scholar] [CrossRef]
- Banas, M.C.; Böhmig, G.A.; Viklicky, O.; Rostaing, L.P.; Jouve, T.; Guirado, L.; Facundo, C.; Bestard, O.; Gröne, H.J.; Kobayashi, K.; et al. A Prospective Multicenter Trial to Evaluate Urinary Metabolomics for Non-invasive Detection of Renal Allograft Rejection (PARASOL): Study Protocol and Patient Recruitment. Front. Med. 2021, 8, 780585. [Google Scholar] [CrossRef]
- Li, L.; Sui, W.; Che, W.; Li, W.; Chen, J.; Li, H.; Tang, D.; Li, M.; Dai, Y. 1H NMR-based metabolic profiling of human serum before and after renal transplantation. ASAIO J. 2013, 59, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Schroeder, A.W.; Yang, J.Y.C.; Sarwal, R.D.; Liberto, J.M.; Sarwal, M.M. Targeted Urine Metabolomics for Monitoring Renal Allograft Injury and Immunosuppression in Pediatric Patients. J. Clin. Med. 2020, 9, 2341. [Google Scholar] [CrossRef] [PubMed]
- Blydt-Hansen, T.D.; Sharma, A.; Gibson, I.W.; Mandal, R.; Wishart, D.S. Urinary metabolomics for noninvasive detection of borderline and acute T cell-mediated rejection in children after kidney transplantation. Am. J. Transplant. 2014, 14, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, J.; Ye, L.; Xu, G. Serum metabolomics study of the acute graft rejection in human renal transplantation based on liquid chromatography-mass spectrometry. J. Proteome Res. 2014, 13, 2659–2667. [Google Scholar] [CrossRef]
- Chen, J.; Wen, H.; Liu, J.; Yu, C.; Zhao, X.; Shi, X.; Xu, G. Metabonomics study of the acute graft rejection in rat renal transplantation using reversed-phase liquid chromatography and hydrophilic interaction chromatography coupled with mass spectrometry. Mol. Biosyst. 2012, 8, 871–878. [Google Scholar] [CrossRef]
- Missailidis, C.; Hallqvist, J.; Qureshi, A.R.; Barany, P.; Heimburger, O.; Lindholm, B.; Stenvinkel, P.; Bergman, P. Serum Trimethylamine-N-Oxide Is Strongly Related to Renal Function and Predicts Outcome in Chronic Kidney Disease. PLoS ONE 2016, 11, e0141738. [Google Scholar] [CrossRef]
- Nkuipou-Kenfack, E.; Duranton, F.; Gayrard, N.; Argilés, À.; Lundin, U.; Weinberger, K.M.; Dakna, M.; Delles, C.; Mullen, W.; Husi, H.; et al. Assessment of metabolomic and proteomic biomarkers in detection and prognosis of progression of renal function in chronic kidney disease. PLoS ONE 2014, 9, e96955. [Google Scholar] [CrossRef]
- Corrêa, R.R.; Machado, J.R.; da Silva, M.V.; Helmo, F.R.; Guimarães, C.S.; Rocha, L.P.; Faleiros, A.C.; dos Reis, M.A. The importance of C4d in biopsies of kidney transplant recipients. Clin. Dev. Immunol. 2013, 2013, 678180. [Google Scholar] [CrossRef][Green Version]
- Sis, B.; Dadras, F.; Khoshjou, F.; Cockfield, S.; Mihatsch, M.J.; Solez, K. Reproducibility studies on arteriolar hyaline thickening scoring in calcineurin inhibitor-treated renal allograft recipients. Am. J. Transplant. 2006, 6, 1444–1450. [Google Scholar] [CrossRef]
- Yagisawa, T.; Mieno, M.; Ichimaru, N.; Morita, K.; Nakamura, M.; Hotta, K.; Kenmochi, T.; Yuzawa, K. Trends of kidney transplantation in Japan in 2018: Data from the kidney transplant registry. Ren. Replace. Ther. 2019, 5, 1–14. [Google Scholar] [CrossRef]
- Pallardó Mateu, L.M.; Sancho Calabuig, A.; Capdevila Plaza, L.; Franco Esteve, A. Acute rejection and late renal transplant failure: Risk factors and prognosis. Nephrol. Dial. Transplant. 2004, 19 (Suppl. 3), iii38–iii42. [Google Scholar] [CrossRef] [PubMed]
- Cole, E.H.; Johnston, O.; Rose, C.L.; Gill, J.S. Impact of acute rejection and new-onset diabetes on long-term transplant graft and patient survival. Clin. J. Am. Soc. Nephrol. CJASN 2008, 3, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Bentata, Y. Tacrolimus: 20 years of use in adult kidney transplantation. What we should know about its nephrotoxicity. Artif. Organs 2020, 44, 140–152. [Google Scholar] [CrossRef] [PubMed]
- Baranovicova, E.; Vnucak, M.; Granak, K.; Lehotsky, J.; Kadasova, N.; Miklusica, J.; Dedinska, I. Circulating Metabolites in Relation to the Kidney Allograft Function in Posttransplant Patients. Metabolites 2022, 12, 661. [Google Scholar] [CrossRef]
- van de Poll, M.C.; Soeters, P.B.; Deutz, N.E.; Fearon, K.C.; Dejong, C.H. Renal metabolism of amino acids: Its role in interorgan amino acid exchange. Am. J. Clin. Nutr. 2004, 79, 185–197. [Google Scholar] [CrossRef]
- Møller, N.; Meek, S.; Bigelow, M.; Andrews, J.; Nair, K. The kidney is an important site for in vivo phenylalanine-to-tyrosine conversion in adult humans: A metabolic role of the kidney. Proc. Natl. Acad. Sci. USA 2000, 97, 1242–1246. [Google Scholar] [CrossRef]
- Busch, M.; Fleck, C.; Wolf, G.; Stein, G. Asymmetrical (ADMA) and symmetrical dimethylarginine (SDMA) as potential risk factors for cardiovascular and renal outcome in chronic kidney disease—Possible candidates for paradoxical epidemiology? Amino Acids 2006, 30, 225–232. [Google Scholar] [CrossRef]
- Frenay, A.R.; van den Berg, E.; de Borst, M.H.; Beckmann, B.; Tsikas, D.; Feelisch, M.; Navis, G.; Bakker, S.J.; van Goor, H. Plasma ADMA associates with all-cause mortality in renal transplant recipients. Amino Acids 2015, 47, 1941–1949. [Google Scholar] [CrossRef][Green Version]
- Said, M.Y.; Douwes, R.M.; van Londen, M.; Minović, I.; Frenay, A.R.; de Borst, M.H.; van den Berg, E.; Heiner-Fokkema, M.R.; Kayacelebi, A.A.; Bollenbach, A.; et al. Effect of renal function on homeostasis of asymmetric dimethylarginine (ADMA): Studies in donors and recipients of renal transplants. Amino Acids 2019, 51, 565–575. [Google Scholar] [CrossRef]
- Cheng, T.H.; Ma, M.C.; Liao, M.T.; Zheng, C.M.; Lu, K.C.; Liao, C.H.; Hou, Y.C.; Liu, W.C.; Lu, C.L. Indoxyl Sulfate, a Tubular Toxin, Contributes to the Development of Chronic Kidney Disease. Toxins 2020, 12, 684. [Google Scholar] [CrossRef]
- Niwa, T.; Ise, M. Indoxyl sulfate, a circulating uremic toxin, stimulates the progression of glomerular sclerosis. J. Lab. Clin. Med. 1994, 124, 96–104. [Google Scholar] [PubMed]
- Gatarek, P.; Kaluzna-Czaplinska, J. Trimethylamine N-oxide (TMAO) in human health. EXCLI J. 2021, 20, 301–319. [Google Scholar] [CrossRef]
- Sis, B.; Mengel, M.; Haas, M.; Colvin, R.B.; Halloran, P.F.; Racusen, L.C.; Solez, K.; Baldwin, W.M., 3rd; Bracamonte, E.R.; Broecker, V.; et al. Banff ’09 meeting report: Antibody mediated graft deterioration and implementation of Banff working groups. Am. J. Transplant. 2010, 10, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, S.; Imai, E.; Horio, M.; Yasuda, Y.; Tomita, K.; Nitta, K.; Yamagata, K.; Tomino, Y.; Yokoyama, H.; Hishida, A. Revised equations for estimated GFR from serum creatinine in Japan. Am. J. Kidney Dis. 2009, 53, 982–992. [Google Scholar] [CrossRef]
- Yamano, E.; Sugimoto, M.; Hirayama, A.; Kume, S.; Yamato, M.; Jin, G.; Tajima, S.; Goda, N.; Iwai, K.; Fukuda, S.; et al. Index markers of chronic fatigue syndrome with dysfunction of TCA and urea cycles. Sci. Rep. 2016, 6, 34990. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, H.; Katsumata, K.; Iwabuchi, A.; Udo, R.; Tago, T.; Kasahara, K.; Mazaki, J.; Enomoto, M.; Ishizaki, T.; Soya, R.; et al. Salivary metabolomics with machine learning for colorectal cancer detection. Cancer Sci. 2022, 113, 3234–3243. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, T.; Katsumata, K.; Udo, R.; Tago, T.; Kasahara, K.; Mazaki, J.; Kuwabara, H.; Kawakita, H.; Enomoto, M.; Ishizaki, T.; et al. Validation of Urinary Charged Metabolite Profiles in Colorectal Cancer Using Capillary Electrophoresis-Mass Spectrometry. Metabolites 2022, 12, 59. [Google Scholar] [CrossRef] [PubMed]
- Udo, R.; Katsumata, K.; Kuwabara, H.; Enomoto, M.; Ishizaki, T.; Sunamura, M.; Nagakawa, Y.; Soya, R.; Sugimoto, M.; Tsuchida, A. Urinary charged metabolite profiling of colorectal cancer using capillary electrophoresis-mass spectrometry. Sci. Rep. 2020, 10, 21057. [Google Scholar] [CrossRef]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
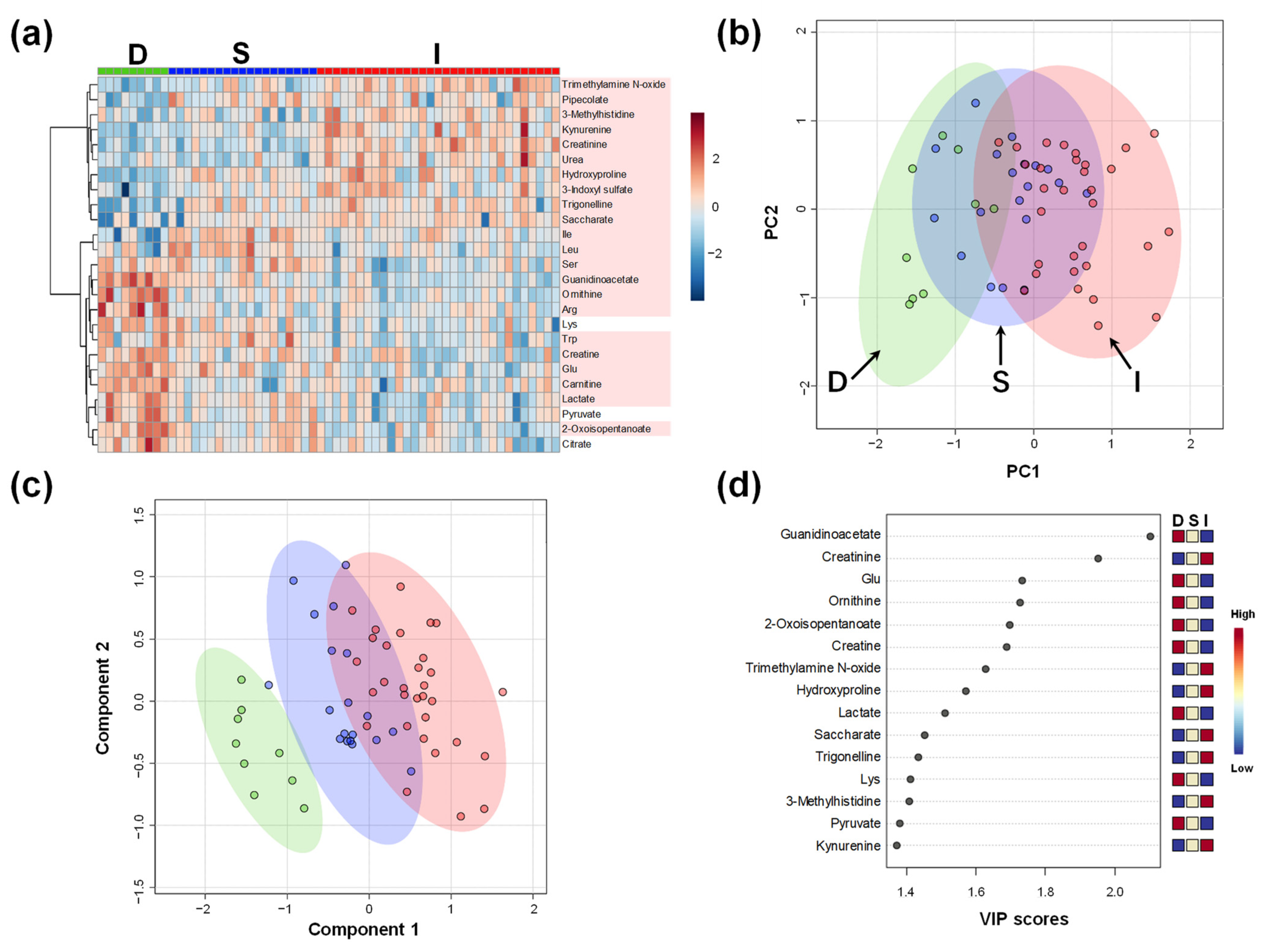
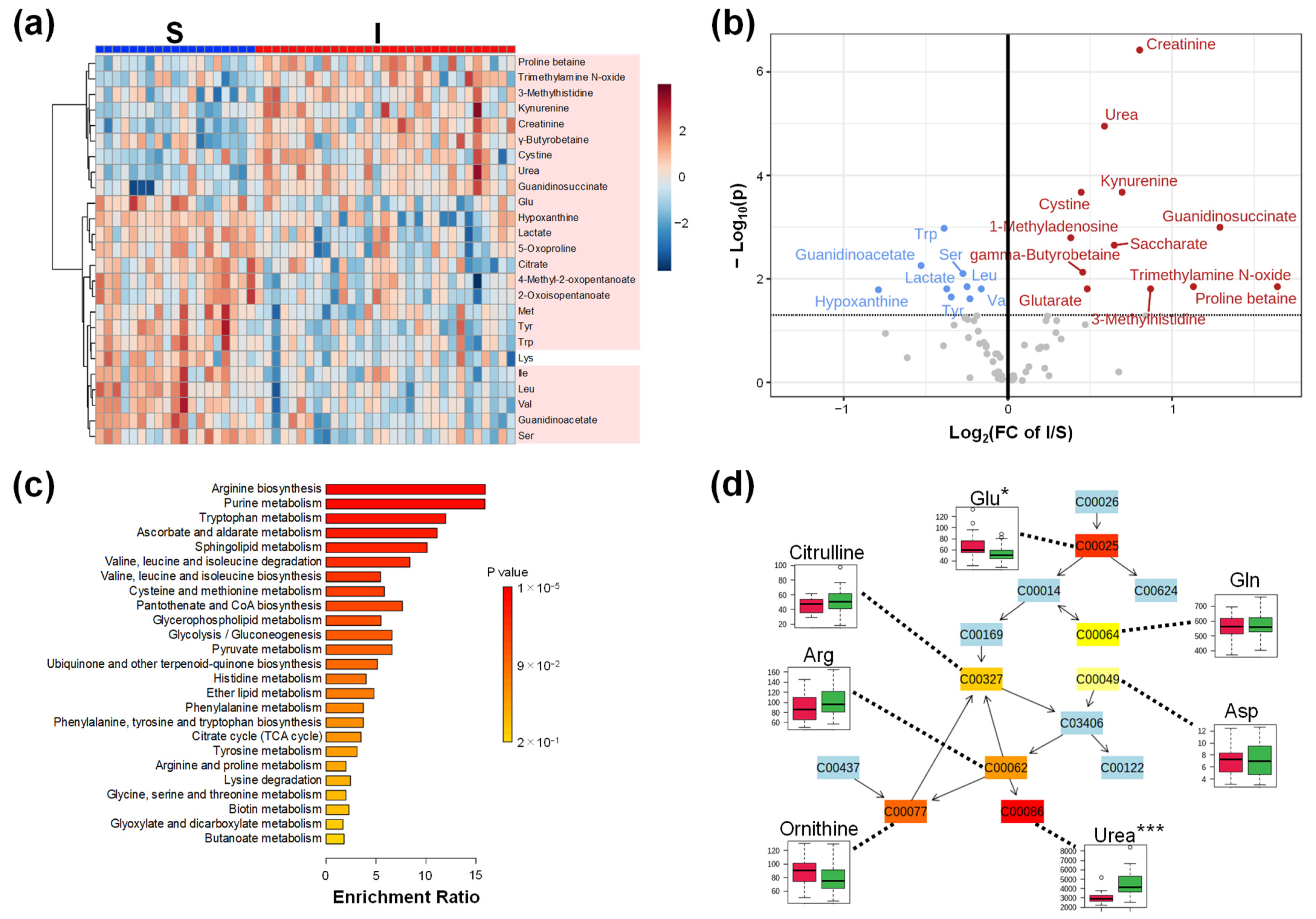
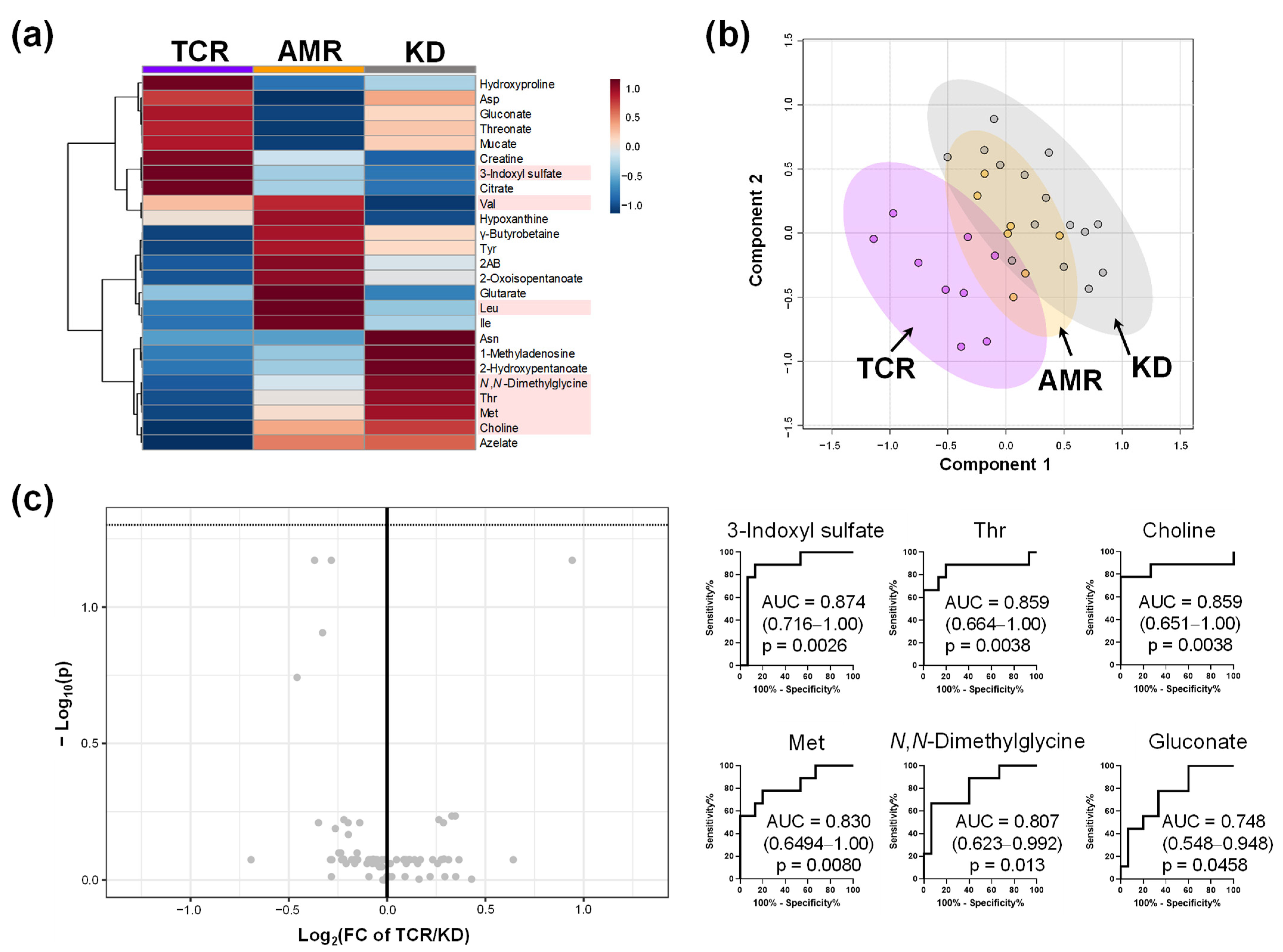

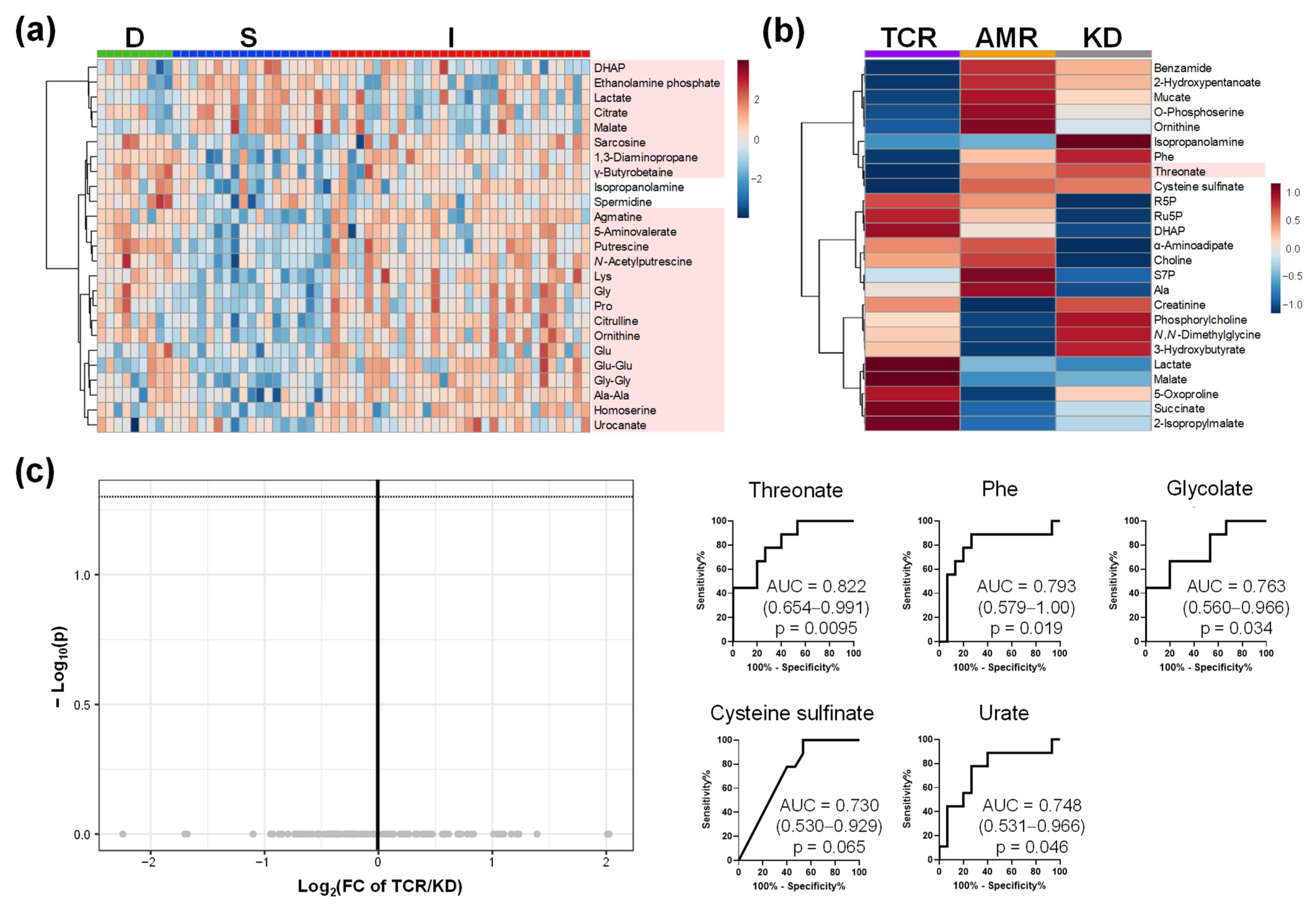
| Item | D | S | I | p-Value |
|---|---|---|---|---|
| n | 9 | 19 | 31 | |
| Sex (Male/Female) | 3/6 | 11/8 | 22/9 | 0.12 a |
| Age | 64 (43–67) | 49 (23–74) | 47 (30–68) | 0.024 *b |
| BMI | 23.9 (±3.73) | 22.2 (±3.49) | 23.2 (±3.18) | 0.40 b |
| Creatinine, mg/dL | 0.68 (±0.15) | 1.1 (±0.35) | 2.1 (±0.71) | <0.0001 ***b |
| eGFR, mL/min/1.73 m2 | 82.2 (±18.7) | 57.2 (±12.2) | 28.8 (±9.01) | |
| Donor’s age | 47 (20–68) | 59 (5–72) | 0.026 *c | |
| Living-donor (%) | 18 (94.7) | 30 (96.8) | 0.72 a | |
| DM (%) | 6 (31.6) | 12 (38.7) | 0.46 a | |
| ABO-I (%) | 4 (21.1) | 12 (38.7) | 0.13 a | |
| HD duration, months | 16 (1–262) | 5.5 (1–250) | 0.17 c | |
| HLA A, B, DR mismatch | 2 (±1.6) | 3.2 (±1.4) | 0.0092 **c | |
| HD (%) | 10 (52.6) | 20 (64.5) | 0.55 a | |
| PEKT (%) | 8 (42.1) | 9 (29.0) | 0.30 a | |
| Tac (%) | 11 (57.9) | 15 (48.4) | 0.51 a | |
| MMF (%) | 19 (100) | 30 (96.8) | 0.43 a |
| Item | TCR | AMR | KD | p-Value |
|---|---|---|---|---|
| n | 9 | 7 | 15 | |
| Sex (Male/Female) | 6/3 | 4/3 | 12/3 | 0.52 a |
| Age | 47 (34–65) | 60 (42–66) | 47 (30–68) | 0.48 b |
| Donor’s age | 54 (5–72) | 55 (36–66) | 61 (38–71) | 0.32 b |
| BMI | 23.4 (±3.71) | 24.0 (±4.16) | 23.7 (±2.41) | 0.69 b |
| Banff score | ||||
| i + t | 4.3 (±0.5) | 1.7 (±0.8) | 1.5 (±1.2) | |
| c4d | 0.7 (±1.0) | 1.3 (±1.1) | 0.9 (±1.2) | |
| aah | 0 (±0) | 0.3 (±0.8) | 0.5 (±0.8) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwamoto, H.; Okihara, M.; Akashi, I.; Kihara, Y.; Konno, O.; Kawachi, S.; Sunamura, M.; Sugimoto, M. Metabolomic Profiling of Plasma, Urine, and Saliva of Kidney Transplantation Recipients. Int. J. Mol. Sci. 2022, 23, 13938. https://doi.org/10.3390/ijms232213938
Iwamoto H, Okihara M, Akashi I, Kihara Y, Konno O, Kawachi S, Sunamura M, Sugimoto M. Metabolomic Profiling of Plasma, Urine, and Saliva of Kidney Transplantation Recipients. International Journal of Molecular Sciences. 2022; 23(22):13938. https://doi.org/10.3390/ijms232213938
Chicago/Turabian StyleIwamoto, Hitoshi, Masaaki Okihara, Isao Akashi, Yu Kihara, Osamu Konno, Shigeyuki Kawachi, Makoto Sunamura, and Masahiro Sugimoto. 2022. "Metabolomic Profiling of Plasma, Urine, and Saliva of Kidney Transplantation Recipients" International Journal of Molecular Sciences 23, no. 22: 13938. https://doi.org/10.3390/ijms232213938
APA StyleIwamoto, H., Okihara, M., Akashi, I., Kihara, Y., Konno, O., Kawachi, S., Sunamura, M., & Sugimoto, M. (2022). Metabolomic Profiling of Plasma, Urine, and Saliva of Kidney Transplantation Recipients. International Journal of Molecular Sciences, 23(22), 13938. https://doi.org/10.3390/ijms232213938






