Aberrant Cortical Layer Development of Brain Organoids Derived from Noonan Syndrome-iPSCs
Abstract
1. Introduction
2. Results
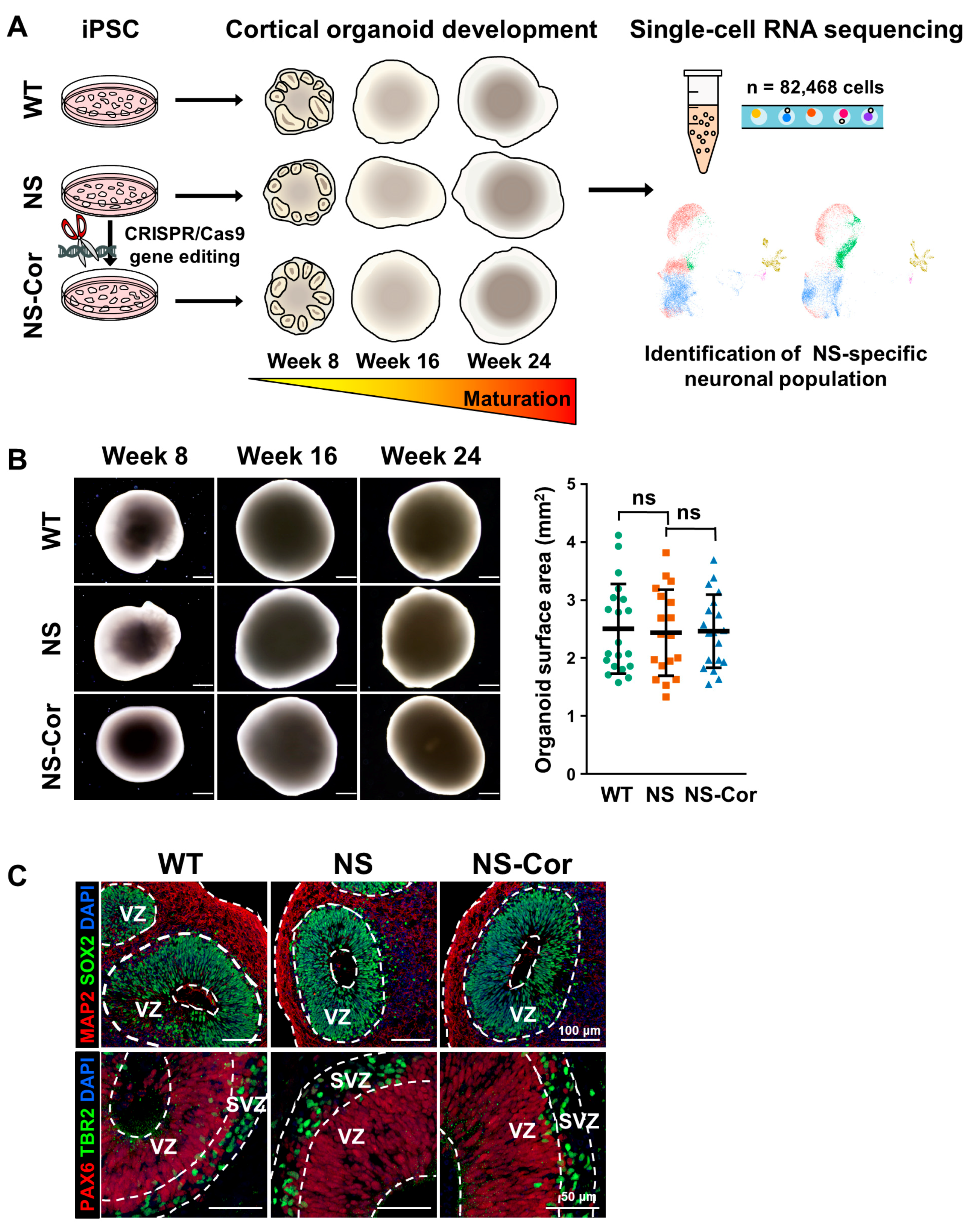
2.1. Normal Development of NS-iPSCs into Cortical Organoids
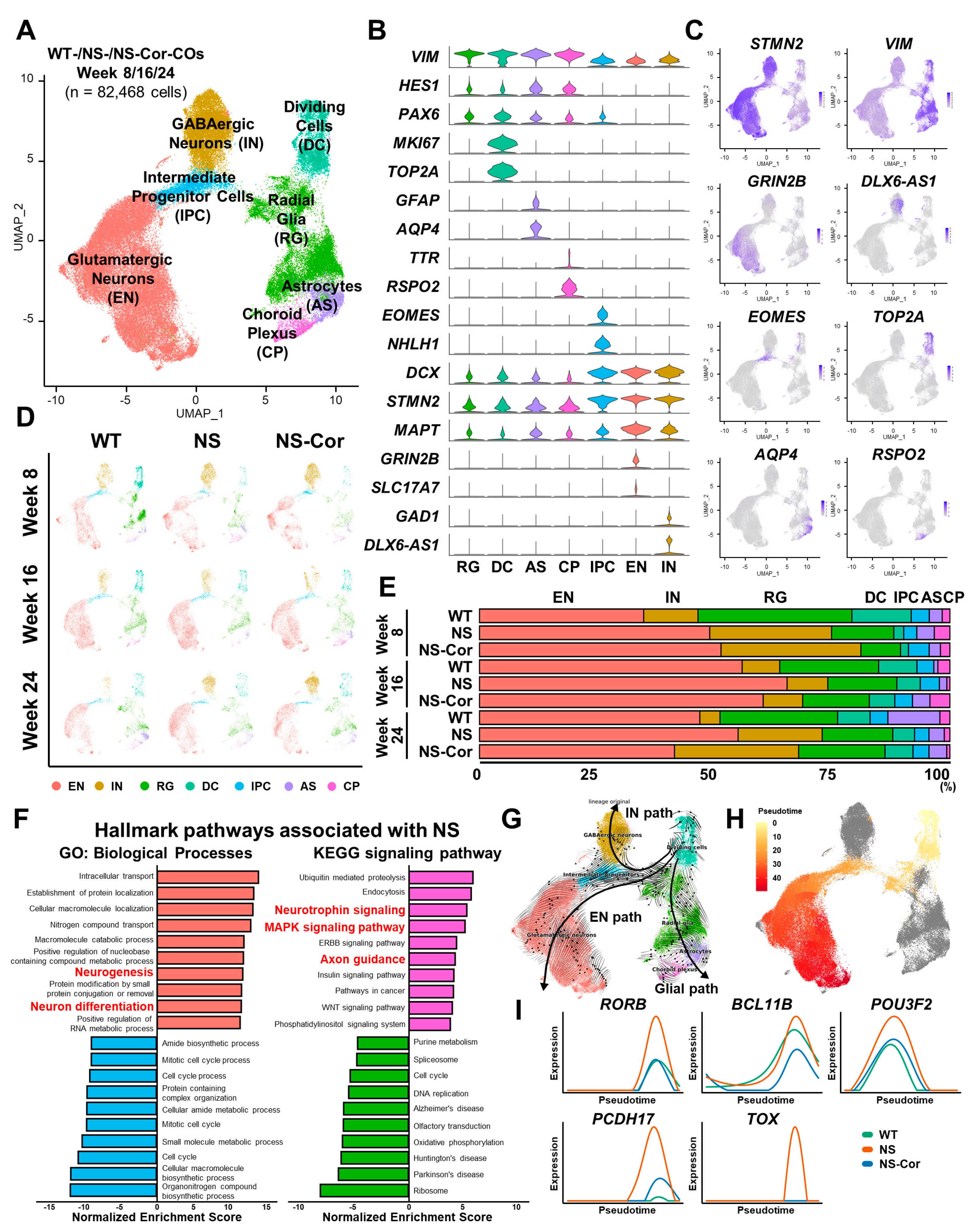
2.2. Aberrant Glutamatergic Neurogenesis in NS-COs
2.3. Defective Cortical Layer Formation in NS-COs
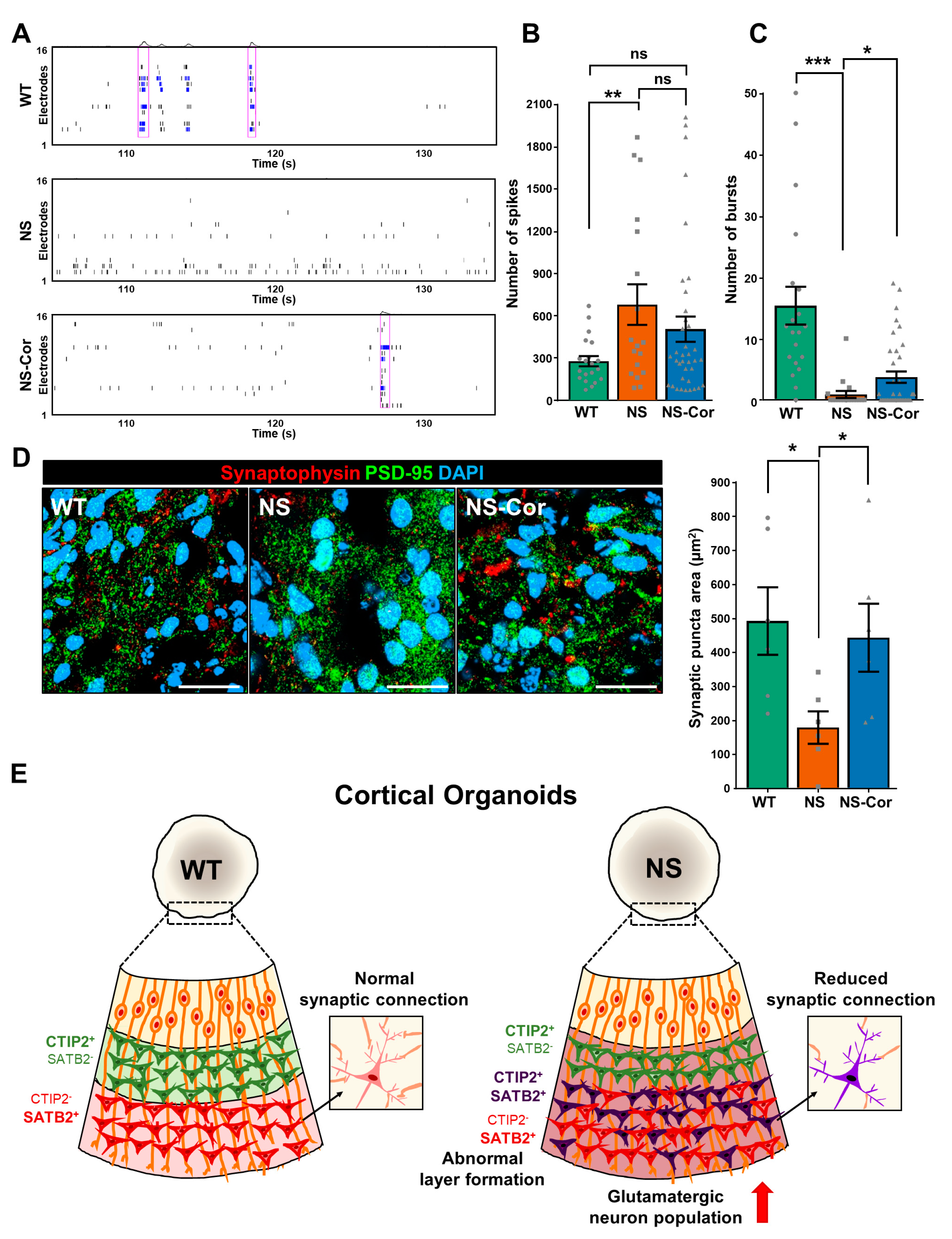
2.4. Impaired Neuronal Connectivity in NS-COs
3. Discussion
4. Materials and Methods
4.1. Maintenance of Human Induced Pluripotent Stem Cells (hiPSCs)
4.2. Genomic Correction of NS-iPSCs
4.3. Cortical Organoid Differentiation
4.4. Real-Time Quantitative PCR
4.5. Western Blotting
4.6. Single-Cell RNA Sequencing (scRNA-seq) Library Preparation
4.7. Analysis of scRNA-seq Data
4.8. Immunostaining
4.9. Multi-Electrode Array (MEA)
4.10. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roberts, A.E.; Allanson, J.E.; Tartaglia, M.; Gelb, B.D. Noonan syndrome. Lancet 2013, 381, 333–342. [Google Scholar] [CrossRef]
- Bhambhani, V.; Muenke, M. Noonan syndrome. Am. Fam. Physician 2014, 89, 37–43. [Google Scholar] [PubMed]
- Lee, D.; Portnoy, S.; Hill, P.; Gillberg, C.; Patton, M. Psychological profile of children with Noonan syndrome. Dev. Med. Child Neurol. 2005, 47, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, W.; Wingbermühle, E.; Egger, J.; Van der Burgt, I.; Tuinier, S. Noonan syndrome: Psychological and psychiatric aspects. Am. J. Med. Genet. Part A 2008, 146, 191–196. [Google Scholar] [CrossRef]
- Pierpont, E.I.; Pierpont, M.E.; Mendelsohn, N.J.; Roberts, A.E.; Tworog-Dube, E.; Seidenberg, M.S. Genotype differences in cognitive functioning in Noonan syndrome. Genes Brain Behav. 2009, 8, 275–282. [Google Scholar] [CrossRef]
- Cesarini, L.; Alfieri, P.; Pantaleoni, F.; Vasta, I.; Cerutti, M.; Petrangeli, V.; Mariotti, P.; Leoni, C.; Ricci, D.; Vicari, S. Cognitive profile of disorders associated with dysregulation of the RAS/MAPK signaling cascade. Am. J. Med. Genet. Part A 2009, 149, 140–146. [Google Scholar] [CrossRef]
- Roberts, A.E. Noonan Syndrome. In GeneReviews(®); Copyright © 1993–2022, University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved.: Seattle (WA); Adam, M.P., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA.
- Gauthier, A.S.; Furstoss, O.; Araki, T.; Chan, R.; Neel, B.G.; Kaplan, D.R.; Miller, F.D. Control of CNS cell-fate decisions by SHP-2 and its dysregulation in Noonan syndrome. Neuron 2007, 54, 245–262. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Ehninger, D.; Zhou, M.; Oh, J.-Y.; Kang, M.; Kwak, C.; Ryu, H.-H.; Butz, D.; Araki, T.; Cai, Y. Mechanism and treatment for learning and memory deficits in mouse models of Noonan syndrome. Nat. Neurosci. 2014, 17, 1736–1743. [Google Scholar] [CrossRef]
- Altmüller, F.; Pothula, S.; Annamneedi, A.; Nakhaei-Rad, S.; Montenegro-Venegas, C.; Pina-Fernández, E.; Marini, C.; Santos, M.; Schanze, D.; Montag, D. Aberrant neuronal activity-induced signaling and gene expression in a mouse model of RASopathy. PLoS Genet. 2017, 13, e1006684. [Google Scholar] [CrossRef]
- Levy, A.D.; Xiao, X.; Shaw, J.E.; Devi, S.P.S.; Katrancha, S.M.; Bennett, A.M.; Greer, C.A.; Howe, J.R.; Machida, K.; Koleske, A.J. Noonan syndrome-associated SHP2 dephosphorylates GluN2B to regulate NMDA receptor function. Cell Rep. 2018, 24, 1523–1535. [Google Scholar] [CrossRef]
- Ryu, H.-H.; Kim, T.; Kim, J.-W.; Kang, M.; Park, P.; Kim, Y.G.; Kim, H.; Ha, J.; Choi, J.E.; Lee, J. Excitatory neuron–specific SHP2-ERK signaling network regulates synaptic plasticity and memory. Sci. Signal. 2019, 12, eaau5755. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Ai, H.; Xue, F.; Luan, Y.; Zhang, B. The Noonan syndrome-associated D61G variant of the protein tyrosine phosphatase SHP2 prevents synaptic down-scaling. J. Biol. Chem. 2020, 295, 10023–10031. [Google Scholar] [CrossRef]
- Mason, J.O.; Price, D.J. Building brains in a dish: Prospects for growing cerebral organoids from stem cells. Neuroscience 2016, 334, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Mak, I.W.; Evaniew, N.; Ghert, M. Lost in translation: Animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014, 6, 114. [Google Scholar] [PubMed]
- Chan, W.K.; Griffiths, R.; Price, D.J.; Mason, J.O. Cerebral organoids as tools to identify the developmental roots of autism. Mol. Autism 2020, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Yang, J.; Xiang, Y. Modeling human neurodevelopmental diseases with brain organoids. Cell Regen. 2022, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, S.-L.; Yang, M.; Herrlinger, S.; Shao, Q.; Collar, J.L.; Fierro, E.; Shi, Y.; Liu, A.; Lu, H. Modeling microcephaly with cerebral organoids reveals a WDR62–CEP170–KIF2A pathway promoting cilium disassembly in neural progenitors. Nat. Commun. 2019, 10, 2612. [Google Scholar] [CrossRef]
- Bershteyn, M.; Nowakowski, T.J.; Pollen, A.A.; Di Lullo, E.; Nene, A.; Wynshaw-Boris, A.; Kriegstein, A.R. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell 2017, 20, 435–449.e4. [Google Scholar] [CrossRef]
- Mariani, J.; Coppola, G.; Zhang, P.; Abyzov, A.; Provini, L.; Tomasini, L.; Amenduni, M.; Szekely, A.; Palejev, D.; Wilson, M. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 2015, 162, 375–390. [Google Scholar] [CrossRef]
- Qian, X.; Su, Y.; Adam, C.D.; Deutschmann, A.U.; Pather, S.R.; Goldberg, E.M.; Su, K.; Li, S.; Lu, L.; Jacob, F. Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell 2020, 26, 766–781.e9. [Google Scholar] [CrossRef]
- Trujillo, C.A.; Adams, J.W.; Negraes, P.D.; Carromeu, C.; Tejwani, L.; Acab, A.; Tsuda, B.; Thomas, C.A.; Sodhi, N.; Fichter, K.M. Pharmacological reversal of synaptic and network pathology in human MECP2-KO neurons and cortical organoids. EMBO Mol. Med. 2021, 13, e12523. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-C.; Jang, S.-Y.; Lee, D.; Lee, J.; Kang, U.; Chang, H.; Kim, H.J.; Han, S.-H.; Seo, J.; Choi, M. A logical network-based drug-screening platform for Alzheimer’s disease representing pathological features of human brain organoids. Nat. Commun. 2021, 12, 280. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Park, J.S.; Kim, D.; Kim, B.; Lee, J.H.; Nam, Y.; Yoo, H.-W.; Lee, B.H.; Han, Y.-M. SHP2 mutations induce precocious gliogenesis of Noonan syndrome-derived iPSCs during neural development in vitro. Stem Cell Res. Ther. 2020, 11, 209. [Google Scholar] [CrossRef]
- Steinberg, D.J.; Repudi, S.; Saleem, A.; Kustanovich, I.; Viukov, S.; Abudiab, B.; Banne, E.; Mahajnah, M.; Hanna, J.H.; Stern, S. Modeling genetic epileptic encephalopathies using brain organoids. EMBO Mol. Med. 2021, 13, e13610. [Google Scholar] [CrossRef] [PubMed]
- Noctor, S.C.; Flint, A.C.; Weissman, T.A.; Dammerman, R.S.; Kriegstein, A.R. Neurons derived from radial glial cells establish radial units in neocortex. Nature 2001, 409, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.-P.; Miyawaki, A.; Gage, F.H.; Jan, Y.N.; Jan, L.Y. Local generation of glia is a major astrocyte source in postnatal cortex. Nature 2012, 484, 376–380. [Google Scholar] [CrossRef]
- Zeng, H.; Shen, E.H.; Hohmann, J.G.; Oh, S.W.; Bernard, A.; Royall, J.J.; Glattfelder, K.J.; Sunkin, S.M.; Morris, J.A.; Guillozet-Bongaarts, A.L. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell 2012, 149, 483–496. [Google Scholar] [CrossRef]
- Harb, K.; Magrinelli, E.; Nicolas, C.S.; Lukianets, N.; Frangeul, L.; Pietri, M.; Sun, T.; Sandoz, G.; Grammont, F.; Jabaudon, D. Area-specific development of distinct projection neuron subclasses is regulated by postnatal epigenetic modifications. Elife 2016, 5, e09531. [Google Scholar] [CrossRef]
- Nowakowski, T.J.; Bhaduri, A.; Pollen, A.A.; Alvarado, B.; Mostajo-Radji, M.A.; Di Lullo, E.; Haeussler, M.; Sandoval-Espinosa, C.; Liu, S.J.; Velmeshev, D. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 2017, 358, 1318–1323. [Google Scholar] [CrossRef]
- Cadwell, C.R.; Bhaduri, A.; Mostajo-Radji, M.A.; Keefe, M.G.; Nowakowski, T.J. Development and arealization of the cerebral cortex. Neuron 2019, 103, 980–1004. [Google Scholar] [CrossRef]
- Trujillo, C.A.; Gao, R.; Negraes, P.D.; Gu, J.; Buchanan, J.; Preissl, S.; Wang, A.; Wu, W.; Haddad, G.G.; Chaim, I.A. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell 2019, 25, 558–569.e7. [Google Scholar] [CrossRef] [PubMed]
- Zafeiriou, M.-P.; Bao, G.; Hudson, J.; Halder, R.; Blenkle, A.; Schreiber, M.-K.; Fischer, A.; Schild, D.; Zimmermann, W.-H. Developmental GABA polarity switch and neuronal plasticity in Bioengineered Neuronal Organoids. Nat. Commun. 2020, 11, 3791. [Google Scholar] [CrossRef] [PubMed]
- Pierpont, E.I. Neuropsychological functioning in individuals with Noonan syndrome: A systematic literature review with educational and treatment recommendations. J. Pediatric Neuropsychol. 2016, 2, 14–33. [Google Scholar] [CrossRef]
- Francis, F.; Meyer, G.; Fallet-Bianco, C.; Moreno, S.; Kappeler, C.; Socorro, A.C.; Tuy, F.P.D.; Beldjord, C.; Chelly, J. Human disorders of cortical development: From past to present. Eur. J. Neurosci. 2006, 23, 877–893. [Google Scholar] [CrossRef] [PubMed]
- Barkovich, A.J.; Guerrini, R.; Kuzniecky, R.I.; Jackson, G.D.; Dobyns, W.B. A developmental and genetic classification for malformations of cortical development: Update 2012. Brain 2012, 135, 1348–1369. [Google Scholar] [CrossRef]
- Brancati, G.; Treutlein, B.; Camp, J.G. Resolving neurodevelopmental and vision disorders using organoid single-cell multi-omics. Neuron 2020, 107, 1000–1013. [Google Scholar] [CrossRef]
- Klaus, J.; Kanton, S.; Kyrousi, C.; Ayo-Martin, A.C.; Di Giaimo, R.; Riesenberg, S.; O’Neill, A.C.; Camp, J.G.; Tocco, C.; Santel, M. Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat. Med. 2019, 25, 561–568. [Google Scholar] [CrossRef]
- Tang, X.-Y.; Xu, L.; Wang, J.; Hong, Y.; Wang, Y.; Zhu, Q.; Wang, D.; Zhang, X.-Y.; Liu, C.-Y.; Fang, K.-H. DSCAM/PAK1 pathway suppression reverses neurogenesis deficits in iPSC-derived cerebral organoids from patients with Down syndrome. J. Clin. Investig. 2021, 131, e135763. [Google Scholar] [CrossRef]
- Villa, C.E.; Cheroni, C.; Dotter, C.P.; López-Tóbon, A.; Oliveira, B.; Sacco, R.; Yahya, A.Ç.; Morandell, J.; Gabriele, M.; Tavakoli, M.R. CHD8 haploinsufficiency links autism to transient alterations in excitatory and inhibitory trajectories. Cell Rep. 2022, 39, 110615. [Google Scholar] [CrossRef]
- Xing, L.; Larsen, R.S.; Bjorklund, G.R.; Li, X.; Wu, Y.; Philpot, B.D.; Snider, W.D.; Newbern, J.M. Layer specific and general requirements for ERK/MAPK signaling in the developing neocortex. Elife 2016, 5, e11123. [Google Scholar] [CrossRef]
- Greig, L.C.; Woodworth, M.B.; Galazo, M.J.; Padmanabhan, H.; Macklis, J.D. Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 2013, 14, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Kwan, K.Y.; Šestan, N.; Anton, E. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development 2012, 139, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, K.; Leone, D.P.; Bateson, R.K.; Dobreva, G.; Kohwi, Y.; Kohwi-Shigematsu, T.; Grosschedl, R.; McConnell, S.K. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc. Natl. Acad. Sci. USA 2012, 109, 19071–19078. [Google Scholar] [CrossRef] [PubMed]
- Alcamo, E.A.; Chirivella, L.; Dautzenberg, M.; Dobreva, G.; Fariñas, I.; Grosschedl, R.; McConnell, S.K. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 2008, 57, 364–377. [Google Scholar] [CrossRef] [PubMed]
- Britanova, O.; de Juan Romero, C.; Cheung, A.; Kwan, K.Y.; Schwark, M.; Gyorgy, A.; Vogel, T.; Akopov, S.; Mitkovski, M.; Agoston, D. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 2008, 57, 378–392. [Google Scholar] [CrossRef]
- Baranek, C.; Dittrich, M.; Parthasarathy, S.; Bonnon, C.G.; Britanova, O.; Lanshakov, D.; Boukhtouche, F.; Sommer, J.E.; Colmenares, C.; Tarabykin, V. Protooncogene Ski cooperates with the chromatin-remodeling factor Satb2 in specifying callosal neurons. Proc. Natl. Acad. Sci. USA 2012, 109, 3546–3551. [Google Scholar] [CrossRef]
- Leone, D.P.; Heavner, W.E.; Ferenczi, E.A.; Dobreva, G.; Huguenard, J.R.; Grosschedl, R.; McConnell, S.K. Satb2 regulates the differentiation of both callosal and subcerebral projection neurons in the developing cerebral cortex. Cereb. Cortex 2015, 25, 3406–3419. [Google Scholar] [CrossRef]
- Choi, G.B.; Yim, Y.S.; Wong, H.; Kim, S.; Kim, H.; Kim, S.V.; Hoeffer, C.A.; Littman, D.R.; Huh, J.R. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 2016, 351, 933–939. [Google Scholar] [CrossRef]
- Stoner, R.; Chow, M.L.; Boyle, M.P.; Sunkin, S.M.; Mouton, P.R.; Roy, S.; Wynshaw-Boris, A.; Colamarino, S.A.; Lein, E.S.; Courchesne, E. Patches of disorganization in the neocortex of children with autism. N. Engl. J. Med. 2014, 370, 1209–1219. [Google Scholar] [CrossRef]
- Paolino, A.; Fenlon, L.R.; Suárez, R.; Richards, L.J. Transcriptional control of long-range cortical projections. Curr. Opin. Neurobiol. 2018, 53, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Arlotta, P.; Molyneaux, B.J.; Chen, J.; Inoue, J.; Kominami, R.; Macklis, J.D. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 2005, 45, 207–221. [Google Scholar] [CrossRef] [PubMed]
- McFadden, K.; Minshew, N.J. Evidence for dysregulation of axonal growth and guidance in the etiology of ASD. Front. Hum. Neurosci. 2013, 7, 671. [Google Scholar] [CrossRef] [PubMed]
- Minshew, N.J.; Williams, D.L. The new neurobiology of autism: Cortex, connectivity, and neuronal organization. Arch. Neurol. 2007, 64, 945–950. [Google Scholar] [CrossRef]
- Kana, R.K.; Libero, L.E.; Moore, M.S. Disrupted cortical connectivity theory as an explanatory model for autism spectrum disorders. Phys. Life Rev. 2011, 8, 410–437. [Google Scholar] [CrossRef]
- Rane, P.; Cochran, D.; Hodge, S.M.; Haselgrove, C.; Kennedy, D.; Frazier, J.A. Connectivity in autism: A review of MRI connectivity studies. Harv. Rev. Psychiatry 2015, 23, 223. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.L.; Shrestha, S.B.; Reiss, A.L.; Saggar, M.; Green, T. Altered canonical and striatal-frontal resting state functional connectivity in children with pathogenic variants in the Ras/mitogen-activated protein kinase pathway. Mol. Psychiatry 2022, 27, 1542–1551. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Bae, S.; Kim, J.-S. Cas-Designer: A web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 2015, 31, 4014–4016. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.B.; Lee, J.; Kang, M.; Kim, B.; Ju, Y.; Do, H.-S.; Yoo, H.-W.; Lee, B.H.; Han, Y.-M. Dysregulated ECM remodeling proteins lead to aberrant osteogenesis of Costello syndrome iPSCs. Stem Cell Rep. 2021, 16, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Paşca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.-Y.; O’rourke, N.A.; Nguyen, K.D. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., III; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.-R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast gene set enrichment analysis. BioRxiv 2021, 060012. [Google Scholar]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lönnerberg, P.; Furlan, A. RNA velocity of single cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef]
- Cao, J.; Packer, J.S.; Ramani, V.; Cusanovich, D.A.; Huynh, C.; Daza, R.; Qiu, X.; Lee, C.; Furlan, S.N.; Steemers, F.J. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 2017, 357, 661–667. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, B.; Koh, Y.; Do, H.; Ju, Y.; Choi, J.B.; Cho, G.; Yoo, H.-W.; Lee, B.H.; Han, J.; Park, J.-E.; et al. Aberrant Cortical Layer Development of Brain Organoids Derived from Noonan Syndrome-iPSCs. Int. J. Mol. Sci. 2022, 23, 13861. https://doi.org/10.3390/ijms232213861
Kim B, Koh Y, Do H, Ju Y, Choi JB, Cho G, Yoo H-W, Lee BH, Han J, Park J-E, et al. Aberrant Cortical Layer Development of Brain Organoids Derived from Noonan Syndrome-iPSCs. International Journal of Molecular Sciences. 2022; 23(22):13861. https://doi.org/10.3390/ijms232213861
Chicago/Turabian StyleKim, Bumsoo, Yongjun Koh, Hyunsu Do, Younghee Ju, Jong Bin Choi, Gahyang Cho, Han-Wook Yoo, Beom Hee Lee, Jinju Han, Jong-Eun Park, and et al. 2022. "Aberrant Cortical Layer Development of Brain Organoids Derived from Noonan Syndrome-iPSCs" International Journal of Molecular Sciences 23, no. 22: 13861. https://doi.org/10.3390/ijms232213861
APA StyleKim, B., Koh, Y., Do, H., Ju, Y., Choi, J. B., Cho, G., Yoo, H.-W., Lee, B. H., Han, J., Park, J.-E., & Han, Y.-M. (2022). Aberrant Cortical Layer Development of Brain Organoids Derived from Noonan Syndrome-iPSCs. International Journal of Molecular Sciences, 23(22), 13861. https://doi.org/10.3390/ijms232213861





