The HIV-1 Integrase C-Terminal Domain Induces TAR RNA Structural Changes Promoting Tat Binding
Abstract
1. Introduction
2. Results
2.1. IN Binds TAR RNA with No Apparent Structural Specificity
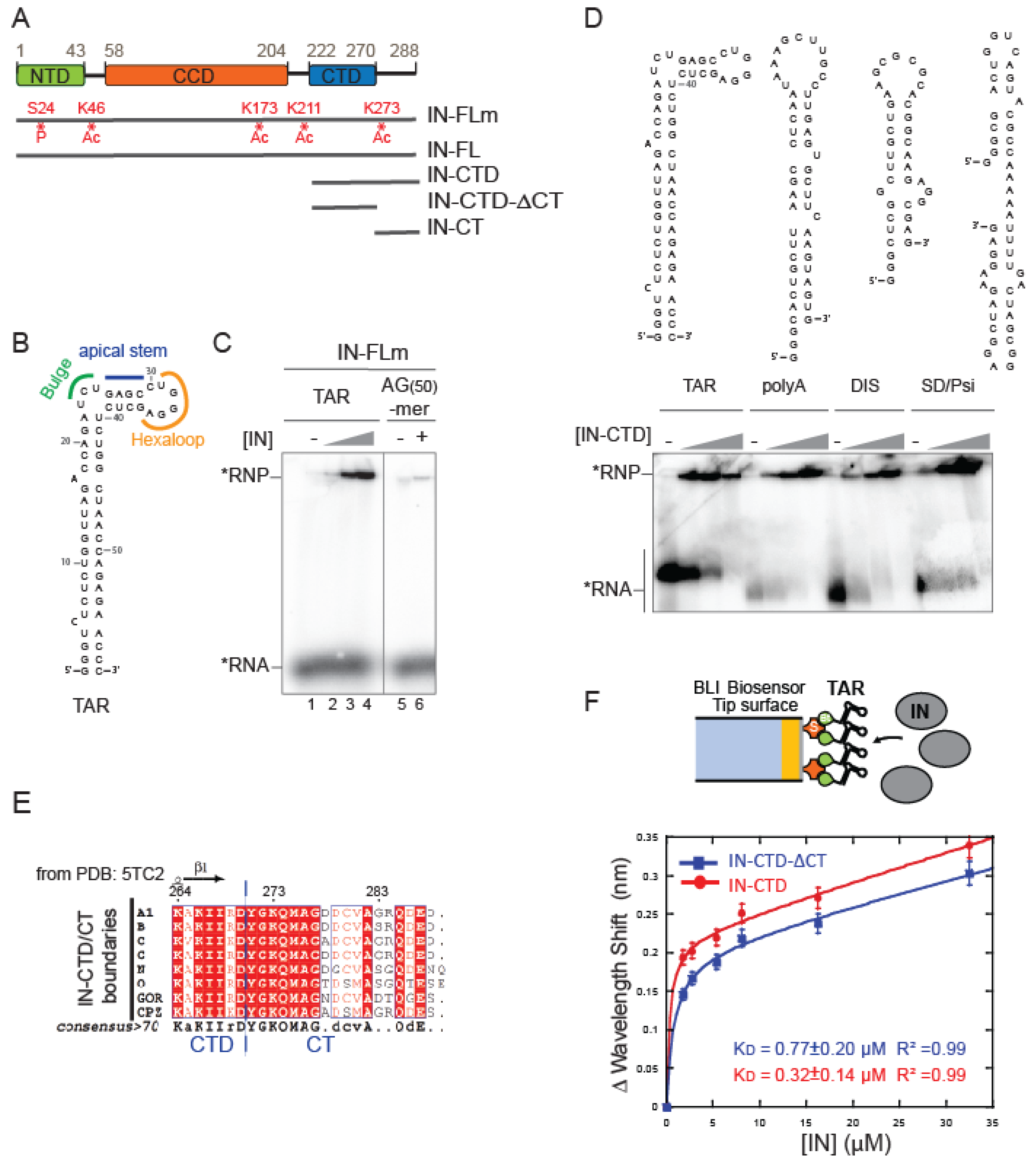
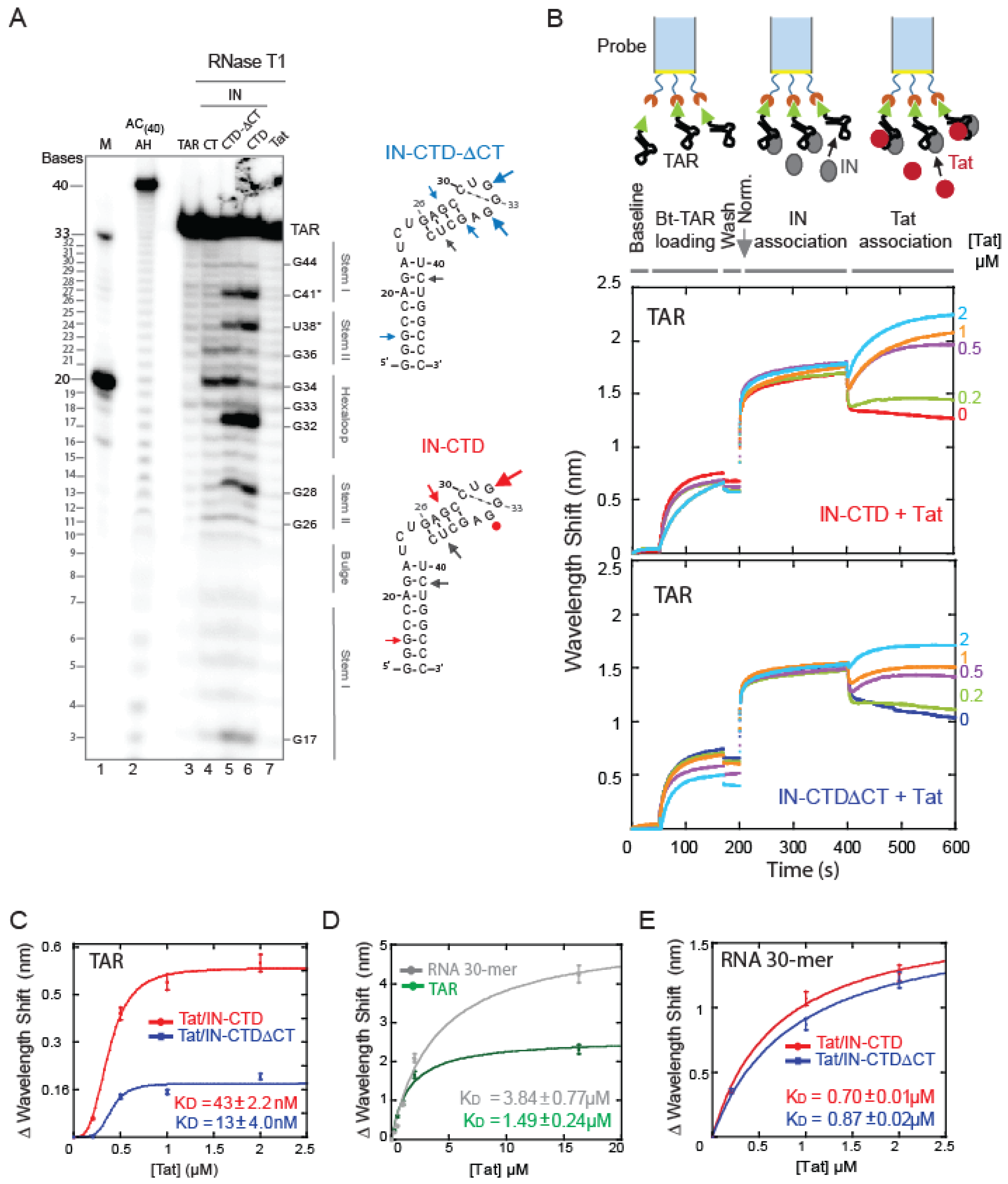
2.2. The C-Terminal Tail Senses the TAR RNA Shape
2.3. IN-CTD Deeply Affects the Structure of TAR Favoring Tat Interaction
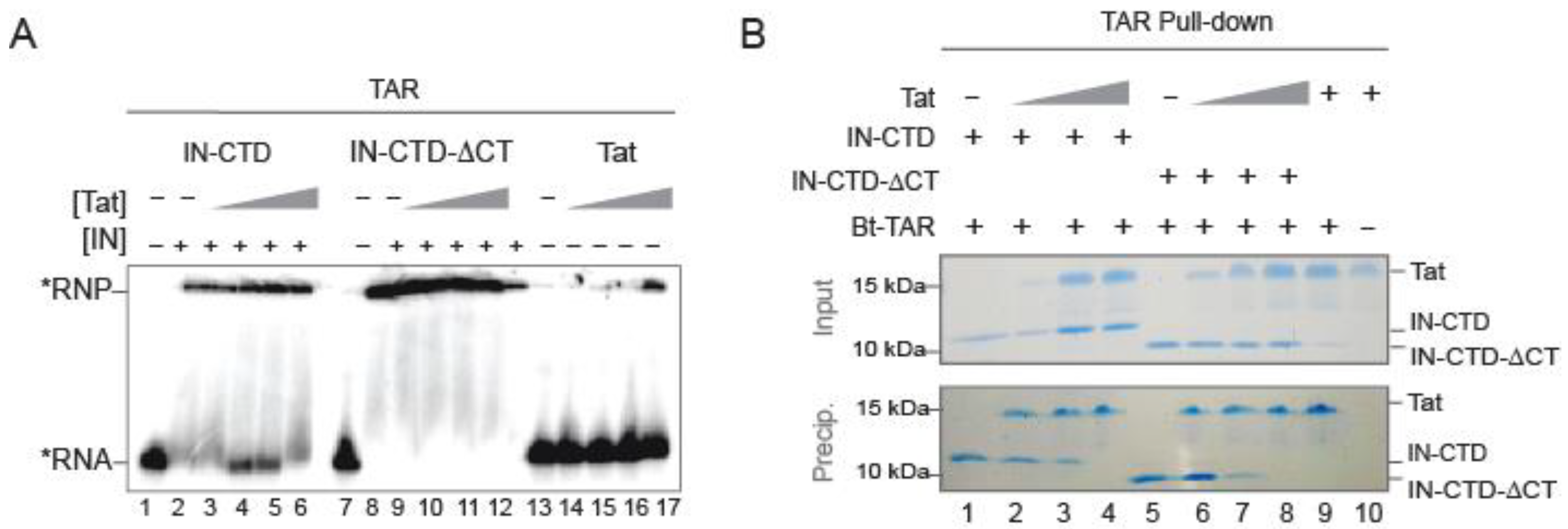
2.4. Tat Competes with IN-CTD and Displaces It from TAR
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. In Vitro RNA Synthesis, Purification, and Radiolabeling
4.3. Electrophoretic Mobility Shift Assays (EMSA)
4.4. RNA Structural Probing
4.5. Pulldown Assay
4.6. Bio-Layer Interferometry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beckmann, B.M.; Castello, A.; Medenbach, J. The expanding universe of ribonucleoproteins: Of novel RNA-binding proteins and unconventional interactions. Pflugers Arch.-Eur. J. Physiol. 2016, 468, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, S.; Jana, T.; Mittelman, K.; Chapal, M.; Kumar, D.K.; Carmi, M.; Barkai, N. Intrinsically Disordered Regions Direct Transcription Factor In Vivo Binding Specificity. Mol. Cell 2020, 79, 459–471.e4. [Google Scholar] [CrossRef]
- Cassiday, L.A.; Maher, L.J., 3rd. Having it both ways: Transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002, 30, 4118–4126. [Google Scholar] [CrossRef]
- Basu, S.; Bahadur, R.P. A structural perspective of RNA recognition by intrinsically disordered proteins. Cell. Mol. Life Sci. 2016, 73, 4075–4084. [Google Scholar] [CrossRef]
- Garcia-Moreno, M.; Jarvelin, A.I.; Castello, A. Unconventional RNA-binding proteins step into the virus-host battlefront. Wiley Interdiscip. Rev. RNA 2018, 9, e1498. [Google Scholar] [CrossRef]
- Kessl, J.J.; Kutluay, S.B.; Townsend, D.; Rebensburg, S.; Slaughter, A.; Larue, R.C.; Shkriabai, N.; Bakouche, N.; Fuchs, J.R.; Bieniasz, P.D.; et al. HIV-1 Integrase Binds the Viral RNA Genome and Is Essential during Virion Morphogenesis. Cell 2016, 166, 1257–1268.e12. [Google Scholar] [CrossRef]
- Maertens, G.N.; Engelman, A.N.; Cherepanov, P. Structure and function of retroviral integrase. Nat. Rev. Microbiol. 2022, 20, 22–34. [Google Scholar] [CrossRef]
- Sakai, H.; Kawamura, M.; Sakuragi, J.; Sakuragi, S.; Shibata, R.; Ishimoto, A.; Ono, N.; Ueda, S.; Adachi, A. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J. Virol. 1993, 67, 1169–1174. [Google Scholar] [CrossRef]
- Rocchi, C.; Gouet, P.; Parissi, V.; Fiorini, F. The C-Terminal Domain of HIV-1 Integrase: A Swiss Army Knife for the Virus? Viruses 2022, 14, 1397. [Google Scholar] [CrossRef] [PubMed]
- Esposito, D.; Craigie, R. HIV integrase structure and function. Adv. Virus Res. 1999, 52, 319–333. [Google Scholar] [PubMed]
- Dyda, F.; Hickman, A.B.; Jenkins, T.M.; Engelman, A.; Craigie, R.; Davies, D.R. Crystal structure of the catalytic domain of HIV-1 integrase: Similarity to other polynucleotidyl transferases. Science 1994, 266, 1981–1986. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Zheng, R.; Caffrey, M.; Craigie, R.; Clore, G.M.; Gronenborn, A.M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. Struct. Biol. 1997, 4, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.P.; Xiao, J.; Knutson, J.R.; Lewis, M.S.; Han, M.K. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry 1997, 36, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Jenkins, T.M.; Craigie, R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. USA 1996, 93, 13659–13664. [Google Scholar] [CrossRef]
- Eijkelenboom, A.P.; Lutzke, R.A.; Boelens, R.; Plasterk, R.H.; Kaptein, R.; Hard, K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat. Struct. Biol. 1995, 2, 807–810. [Google Scholar] [CrossRef]
- Engelman, A.; Hickman, A.B.; Craigie, R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J. Virol. 1994, 68, 5911–5917. [Google Scholar] [CrossRef]
- Jenkins, T.M.; Engelman, A.; Ghirlando, R.; Craigie, R. A soluble active mutant of HIV-1 integrase: Involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 1996, 271, 7712–7718. [Google Scholar] [CrossRef]
- Woerner, A.M.; Marcus-Sekura, C.J. Characterization of a DNA binding domain in the C-terminus of HIV-1 integrase by deletion mutagenesis. Nucleic Acids Res. 1993, 21, 3507–3511. [Google Scholar] [CrossRef]
- Lutzke, R.A.; Vink, C.; Plasterk, R.H. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 1994, 22, 4125–4131. [Google Scholar] [CrossRef] [PubMed]
- Vink, C.; Oude Groeneger, A.M.; Plasterk, R.H. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res. 1993, 21, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Dar, M.J.; Monel, B.; Krishnan, L.; Shun, M.C.; Di Nunzio, F.; Helland, D.E.; Engelman, A. Biochemical and virological analysis of the 18-residue C-terminal tail of HIV-1 integrase. Retrovirology 2009, 6, 94. [Google Scholar] [CrossRef] [PubMed]
- De Houwer, S.; Demeulemeester, J.; Thys, W.; Taltynov, O.; Zmajkovicova, K.; Christ, F.; Debyser, Z. Identification of residues in the C-terminal domain of HIV-1 integrase that mediate binding to the transportin-SR2 protein. J. Biol. Chem. 2012, 287, 34059–34068. [Google Scholar] [CrossRef]
- Mauro, E.; Lesbats, P.; Lapaillerie, D.; Chaignepain, S.; Maillot, B.; Oladosu, O.; Robert, X.; Fiorini, F.; Kieffer, B.; Bouaziz, S.; et al. Human H4 tail stimulates HIV-1 integration through binding to the carboxy-terminal domain of integrase. Nucleic Acids Res. 2019, 47, 3607–3618. [Google Scholar] [CrossRef]
- Mohammed, K.D.; Topper, M.B.; Muesing, M.A. Sequential deletion of the integrase (Gag-Pol) carboxyl terminus reveals distinct phenotypic classes of defective HIV-1. J. Virol. 2011, 85, 4654–4666. [Google Scholar] [CrossRef]
- Elliott, J.L.; Kutluay, S.B. Going beyond Integration: The Emerging Role of HIV-1 Integrase in Virion Morphogenesis. Viruses 2020, 12, 1005. [Google Scholar] [CrossRef]
- Madison, M.K.; Lawson, D.Q.; Elliott, J.; Ozanturk, A.N.; Koneru, P.C.; Townsend, D.; Errando, M.; Kvaratskhelia, M.; Kutluay, S.B. Allosteric HIV-1 Integrase Inhibitors Lead to Premature Degradation of the Viral RNA Genome and Integrase in Target Cells. J. Virol. 2017, 91, e00821-17. [Google Scholar] [CrossRef]
- Fontana, J.; Jurado, K.A.; Cheng, N.; Ly, N.L.; Fuchs, J.R.; Gorelick, R.J.; Engelman, A.N.; Steven, A.C. Distribution and Redistribution of HIV-1 Nucleocapsid Protein in Immature, Mature, and Integrase-Inhibited Virions: A Role for Integrase in Maturation. J. Virol. 2015, 89, 9765–9780. [Google Scholar] [CrossRef]
- Cereseto, A.; Manganaro, L.; Gutierrez, M.I.; Terreni, M.; Fittipaldi, A.; Lusic, M.; Marcello, A.; Giacca, M. Acetylation of HIV-1 integrase by p300 regulates viral integration. EMBO J. 2005, 24, 3070–3081. [Google Scholar] [CrossRef]
- Manganaro, L.; Lusic, M.; Gutierrez, M.I.; Cereseto, A.; Del Sal, G.; Giacca, M. Concerted action of cellular JNK and Pin1 restricts HIV-1 genome integration to activated CD4+ T lymphocytes. Nat. Med. 2010, 16, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Zamborlini, A.; Coiffic, A.; Beauclair, G.; Delelis, O.; Paris, J.; Koh, Y.; Magne, F.; Giron, M.L.; Tobaly-Tapiero, J.; Deprez, E.; et al. Impairment of human immunodeficiency virus type-1 integrase SUMOylation correlates with an early replication defect. J. Biol. Chem. 2011, 286, 21013–21022. [Google Scholar] [CrossRef] [PubMed]
- Kanja, M.; Cappy, P.; Levy, N.; Oladosu, O.; Schmidt, S.; Rossolillo, P.; Winter, F.; Gasser, R.; Moog, C.; Ruff, M.; et al. NKNK: A New Essential Motif in the C-Terminal Domain of HIV-1 Group M Integrases. J. Virol. 2020, 94, e01035-20. [Google Scholar] [CrossRef] [PubMed]
- Shema Mugisha, C.; Dinh, T.; Kumar, A.; Tenneti, K.; Eschbach, J.E.; Davis, K.; Gifford, R.; Kvaratskhelia, M.; Kutluay, S.B. Emergence of Compensatory Mutations Reveals the Importance of Electrostatic Interactions between HIV-1 Integrase and Genomic RNA. mBio 2022, 13, e00431-22. [Google Scholar] [CrossRef]
- Winans, S.; Goff, S.P. Mutations altering acetylated residues in the CTD of HIV-1 integrase cause defects in proviral transcription at early times after integration of viral DNA. PLoS Pathog. 2020, 16, e1009147. [Google Scholar] [CrossRef]
- Jonkers, I.; Lis, J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015, 16, 167–177. [Google Scholar] [CrossRef]
- Mousseau, G.; Valente, S.T. Role of Host Factors on the Regulation of Tat-Mediated HIV-1 Transcription. Curr. Pharm. Des. 2017, 23, 4079–4090. [Google Scholar] [CrossRef]
- He, N.; Liu, M.; Hsu, J.; Xue, Y.; Chou, S.; Burlingame, A.; Krogan, N.J.; Alber, T.; Zhou, Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell 2010, 38, 428–438. [Google Scholar] [CrossRef]
- Pham, V.V.; Salguero, C.; Khan, S.N.; Meagher, J.L.; Brown, W.C.; Humbert, N.; de Rocquigny, H.; Smith, J.L.; D’Souza, V.M. HIV-1 Tat interactions with cellular 7SK and viral TAR RNAs identifies dual structural mimicry. Nat. Commun. 2018, 9, 4266. [Google Scholar] [CrossRef]
- Schulze-Gahmen, U.; Hurley, J.H. Structural mechanism for HIV-1 TAR loop recognition by Tat and the super elongation complex. Proc. Natl. Acad. Sci. USA 2018, 115, 12973–12978. [Google Scholar] [CrossRef]
- Sobhian, B.; Laguette, N.; Yatim, A.; Nakamura, M.; Levy, Y.; Kiernan, R.; Benkirane, M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell 2010, 38, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Garber, M.E.; Fang, S.M.; Fischer, W.H.; Jones, K.A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 1998, 92, 451–462. [Google Scholar] [CrossRef]
- Ne, E.; Palstra, R.J.; Mahmoudi, T. Transcription: Insights From the HIV-1 Promoter. Int. Rev. Cell Mol. Biol. 2018, 335, 191–243. [Google Scholar] [PubMed]
- Craigie, R. The molecular biology of HIV integrase. Future Virol. 2012, 7, 679–686. [Google Scholar] [CrossRef]
- Levy, N.; Eiler, S.; Pradeau-Aubreton, K.; Maillot, B.; Stricher, F.; Ruff, M. Production of unstable proteins through the formation of stable core complexes. Nat. Commun. 2016, 7, 10932. [Google Scholar] [CrossRef]
- Eijkelenboom, A.P.; Sprangers, R.; Hard, K.; Puras Lutzke, R.A.; Plasterk, R.H.; Boelens, R.; Kaptein, R. Refined solution structure of the C-terminal DNA-binding domain of human immunovirus-1 integrase. Proteins 1999, 36, 556–564. [Google Scholar] [CrossRef]
- Lodi, P.J.; Ernst, J.A.; Kuszewski, J.; Hickman, A.B.; Engelman, A.; Craigie, R.; Clore, G.M.; Gronenborn, A.M. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry 1995, 34, 9826–9833. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neubock, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Honer Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Muller-Esparza, H.; Osorio-Valeriano, M.; Steube, N.; Thanbichler, M.; Randau, L. Bio-Layer Interferometry Analysis of the Target Binding Activity of CRISPR-Cas Effector Complexes. Front. Mol. Biosci. 2020, 7, 98. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B.; Jeang, K.T. Detailed mutational analysis of TAR RNA: Critical spacing between the bulge and loop recognition domains. Nucleic Acids Res. 1991, 19, 6169–6176. [Google Scholar] [CrossRef] [PubMed]
- Churcher, M.J.; Lamont, C.; Hamy, F.; Dingwall, C.; Green, S.M.; Lowe, A.D.; Butler, J.G.; Gait, M.J.; Karn, J. High affinity binding of TAR RNA by the human immunodeficiency virus type-1 tat protein requires base-pairs in the RNA stem and amino acid residues flanking the basic region. J. Mol. Biol. 1993, 230, 90–110. [Google Scholar] [CrossRef] [PubMed]
- Delling, U.; Reid, L.S.; Barnett, R.W.; Ma, M.Y.; Climie, S.; Sumner-Smith, M.; Sonenberg, N. Conserved nucleotides in the TAR RNA stem of human immunodeficiency virus type 1 are critical for Tat binding and trans activation: Model for TAR RNA tertiary structure. J. Virol. 1992, 66, 3018–3025. [Google Scholar] [CrossRef]
- Roy, S.; Delling, U.; Chen, C.H.; Rosen, C.A.; Sonenberg, N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev. 1990, 4, 1365–1373. [Google Scholar] [CrossRef]
- Dixit, U.; Bhutoria, S.; Wu, X.; Qiu, L.; Spira, M.; Mathew, S.; Harris, R.; Adams, L.J.; Cahill, S.; Pathak, R.; et al. INI1/SMARCB1 Rpt1 domain mimics TAR RNA in binding to integrase to facilitate HIV-1 replication. Nat. Commun. 2021, 12, 2743. [Google Scholar] [CrossRef]
- Liu, S.; Koneru, P.C.; Li, W.; Pathirage, C.; Engelman, A.N.; Kvaratskhelia, M.; Musier-Forsyth, K. HIV-1 integrase binding to genomic RNA 5′-UTR induces local structural changes in vitro and in virio. Retrovirology 2021, 18, 37. [Google Scholar] [CrossRef]
- Dingwall, C.; Ernberg, I.; Gait, M.J.; Green, S.M.; Heaphy, S.; Karn, J.; Lowe, A.D.; Singh, M.; Skinner, M.A.; Valerio, R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl. Acad. Sci. USA 1989, 86, 6925–6929. [Google Scholar] [CrossRef]
- Lu, R.; Ghory, H.Z.; Engelman, A. Genetic analyses of conserved residues in the carboxyl-terminal domain of human immunodeficiency virus type 1 integrase. J. Virol. 2005, 79, 10356–10368. [Google Scholar] [CrossRef]
- Dubois, N.; Khoo, K.K.; Ghossein, S.; Seissler, T.; Wolff, P.; McKinstry, W.J.; Mak, J.; Paillart, J.C.; Marquet, R.; Bernacchi, S. The C-terminal p6 domain of the HIV-1 Pr55(Gag) precursor is required for specific binding to the genomic RNA. RNA Biol. 2018, 15, 923–936. [Google Scholar] [CrossRef]
- Cordingley, M.G.; LaFemina, R.L.; Callahan, P.L.; Condra, J.H.; Sardana, V.V.; Graham, D.J.; Nguyen, T.M.; LeGrow, K.; Gotlib, L.; Schlabach, A.J.; et al. Sequence-specific interaction of Tat protein and Tat peptides with the transactivation-responsive sequence element of human immunodeficiency virus type 1 in vitro. Proc. Natl. Acad. Sci. USA 1990, 87, 8985–8989. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, J.D.; Tan, R.; Calnan, B.J.; Frankel, A.D.; Williamson, J.R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science 1992, 257, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Weeks, K.M.; Ampe, C.; Schultz, S.C.; Steitz, T.A.; Crothers, D.M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science 1990, 249, 1281–1285. [Google Scholar] [CrossRef] [PubMed]
- Comas-Garcia, M.; Davis, S.R.; Rein, A. On the Selective Packaging of Genomic RNA by HIV-1. Viruses 2016, 8, 246. [Google Scholar] [CrossRef]
- Mayol, K.; Munier, S.; Beck, A.; Verrier, B.; Guillon, C. Design and characterization of an HIV-1 Tat mutant: Inactivation of viral and cellular functions but not antigenicity. Vaccine 2007, 25, 6047–6060. [Google Scholar] [CrossRef]
- Guillon, C.; Stankovic, K.; Ataman-Onal, Y.; Biron, F.; Verrier, B. Evidence for CTL-mediated selection of Tat and Rev mutants after the onset of the asymptomatic period during HIV type 1 infection. AIDS Res. Hum. Retrovir. 2006, 22, 1283–1292. [Google Scholar] [CrossRef]
- Foucault, M.; Mayol, K.; Receveur-Brechot, V.; Bussat, M.C.; Klinguer-Hamour, C.; Verrier, B.; Beck, A.; Haser, R.; Gouet, P.; Guillon, C. UV and X-ray structural studies of a 101-residue long Tat protein from a HIV-1 primary isolate and of its mutated, detoxified, vaccine candidate. Proteins 2010, 78, 1441–1456. [Google Scholar] [CrossRef]
- Fiorini, F.; Bonneau, F.; Le Hir, H. Biochemical characterization of the RNA helicase UPF1 involved in nonsense-mediated mRNA decay. Methods Enzymol. 2012, 511, 255–274. [Google Scholar]
- Fiorini, F.; Boudvillain, M.; Le Hir, H. Tight intramolecular regulation of the human Upf1 helicase by its N- and C-terminal domains. Nucleic Acids Res. 2013, 41, 2404–2415. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocchi, C.; Louvat, C.; Miele, A.E.; Batisse, J.; Guillon, C.; Ballut, L.; Lener, D.; Negroni, M.; Ruff, M.; Gouet, P.; et al. The HIV-1 Integrase C-Terminal Domain Induces TAR RNA Structural Changes Promoting Tat Binding. Int. J. Mol. Sci. 2022, 23, 13742. https://doi.org/10.3390/ijms232213742
Rocchi C, Louvat C, Miele AE, Batisse J, Guillon C, Ballut L, Lener D, Negroni M, Ruff M, Gouet P, et al. The HIV-1 Integrase C-Terminal Domain Induces TAR RNA Structural Changes Promoting Tat Binding. International Journal of Molecular Sciences. 2022; 23(22):13742. https://doi.org/10.3390/ijms232213742
Chicago/Turabian StyleRocchi, Cecilia, Camille Louvat, Adriana Erica Miele, Julien Batisse, Christophe Guillon, Lionel Ballut, Daniela Lener, Matteo Negroni, Marc Ruff, Patrice Gouet, and et al. 2022. "The HIV-1 Integrase C-Terminal Domain Induces TAR RNA Structural Changes Promoting Tat Binding" International Journal of Molecular Sciences 23, no. 22: 13742. https://doi.org/10.3390/ijms232213742
APA StyleRocchi, C., Louvat, C., Miele, A. E., Batisse, J., Guillon, C., Ballut, L., Lener, D., Negroni, M., Ruff, M., Gouet, P., & Fiorini, F. (2022). The HIV-1 Integrase C-Terminal Domain Induces TAR RNA Structural Changes Promoting Tat Binding. International Journal of Molecular Sciences, 23(22), 13742. https://doi.org/10.3390/ijms232213742







