2’-Fucosyllactose Inhibits Coxsackievirus Class A Type 9 Infection by Blocking Virus Attachment and Internalisation
Abstract
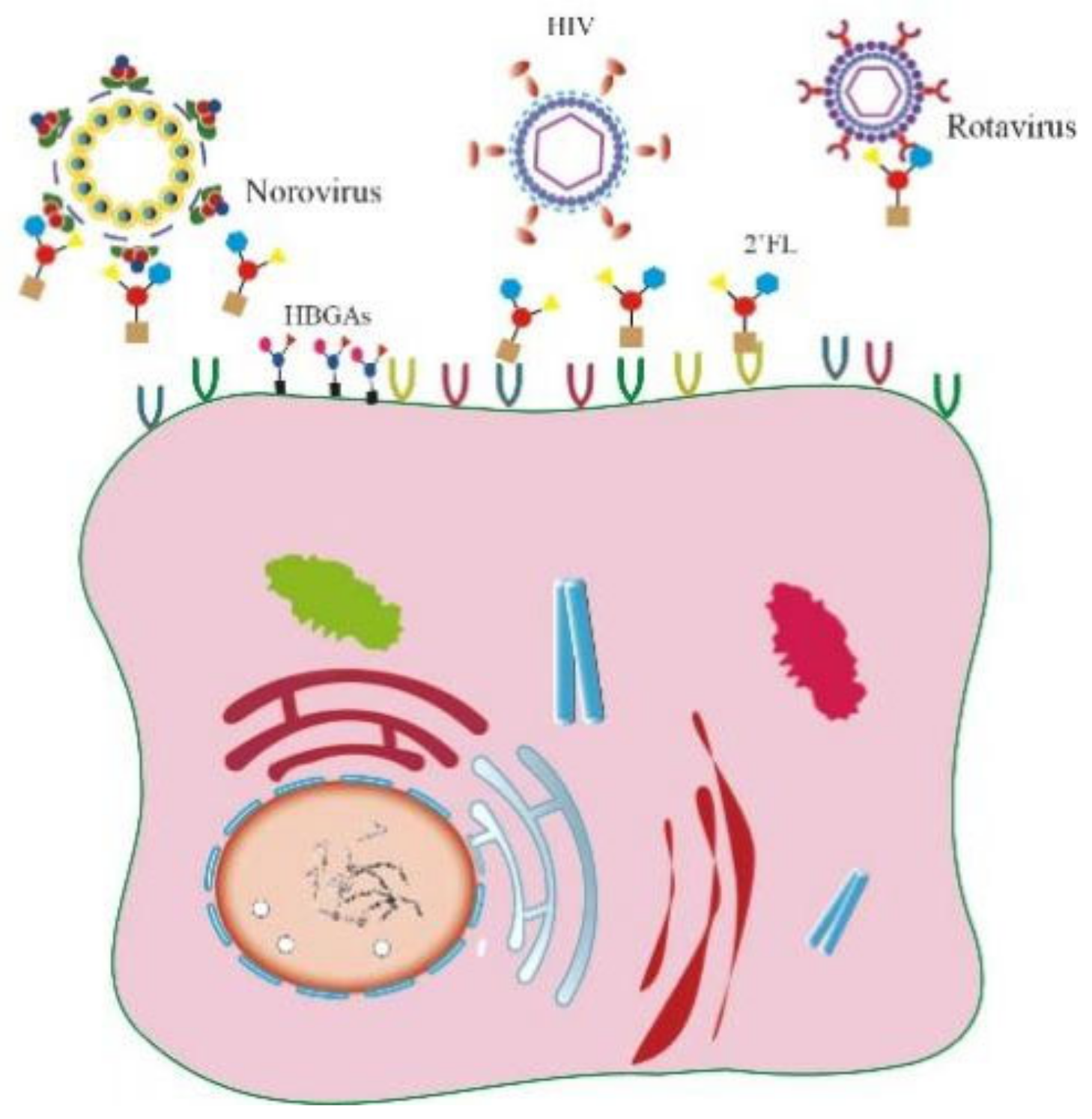
1. Introduction
2. Results
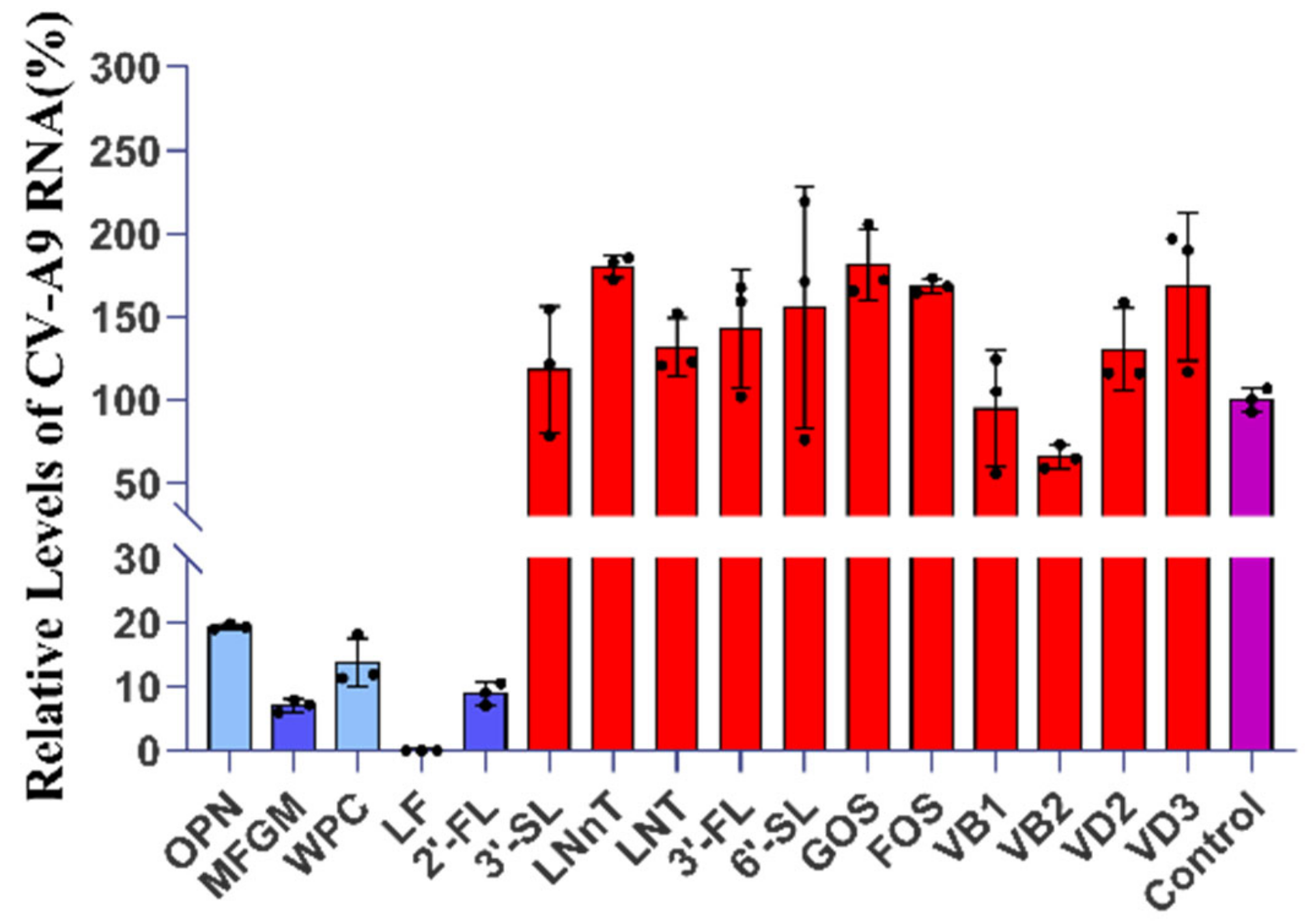
2.1. Five Components with Significant Anti-CV-A9 Effect Were Screened from Breast Milk
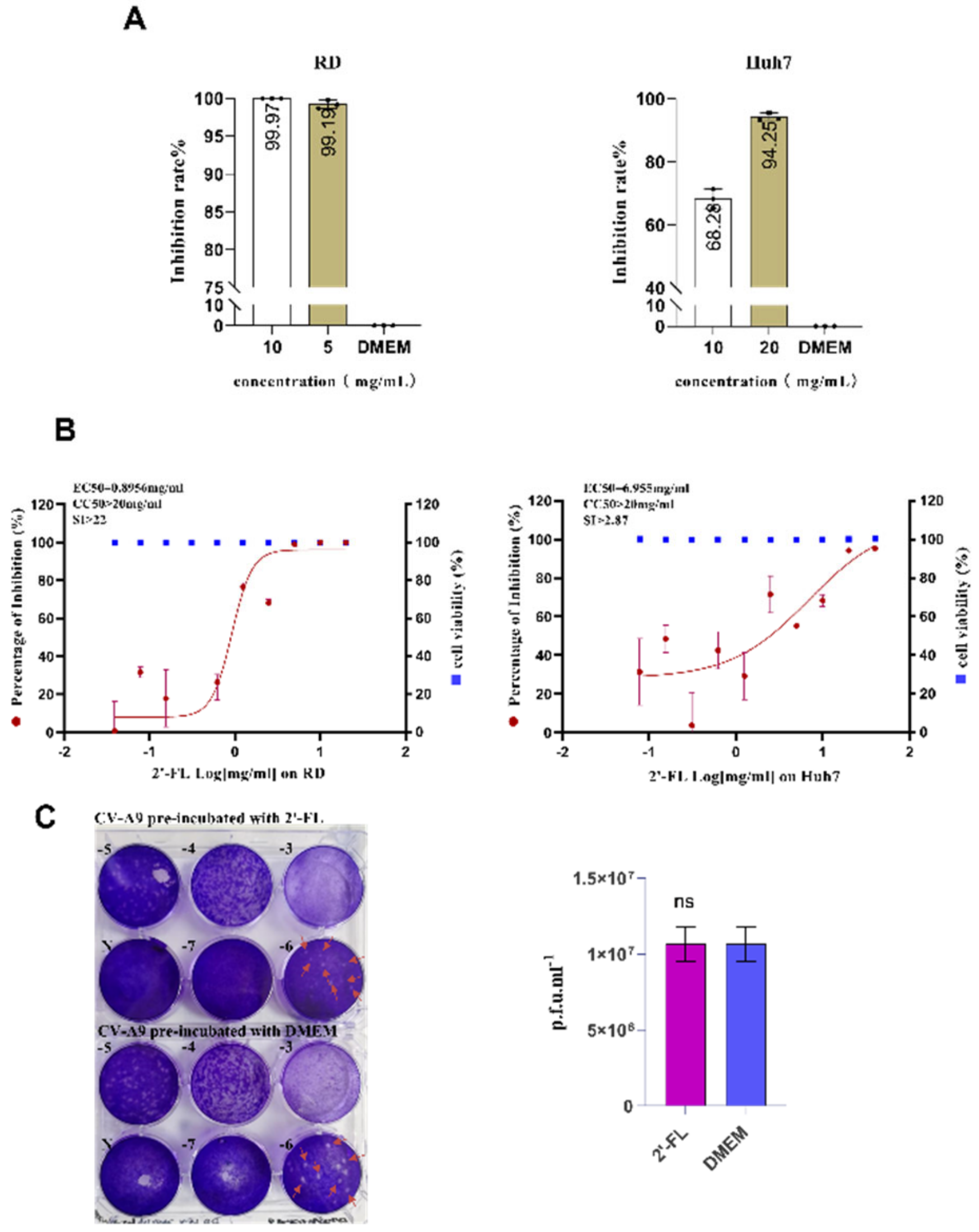
2.2. Oligosaccharide 2’-FL Targets the Host Cell Instead of the Virus Particle to Block CV-A9 Infection
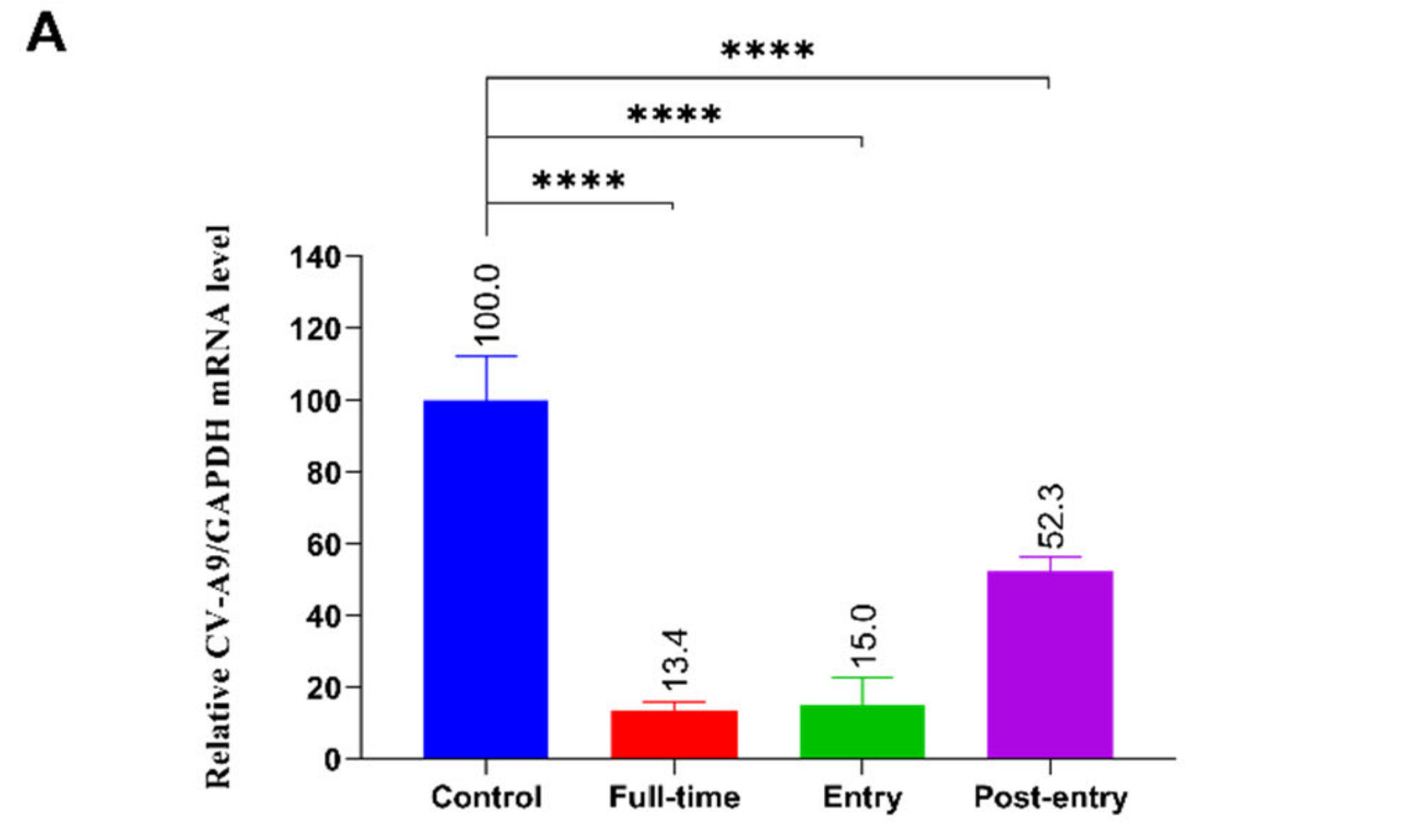
2.3. The Effect of 2’-FL on CV-A9 Life Cycle
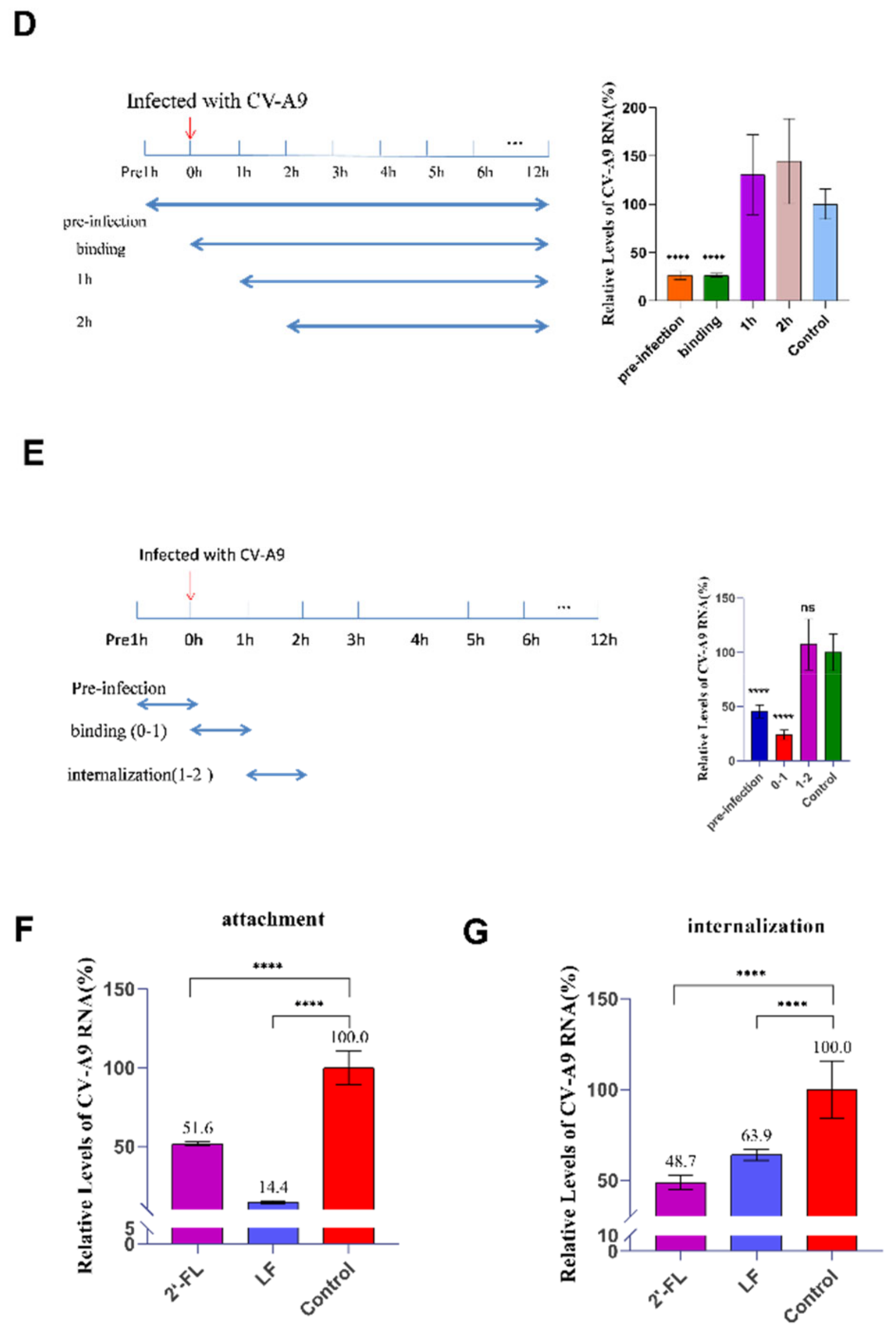
2.4. Oligosaccharide 2’-FL Inhibits CV-A9 Binding and Internalisation
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Virus Infection
4.2. Preliminary Drug Screening In Vitro
4.3. Antiviral and Cytotoxicity Assays
4.4. Time-of-Addition Assay
4.5. Viral RNA Extraction and qRT-PCR
4.6. Virus Attachment and Internalisation Assay
4.7. Plaque Assay
4.8. Time Course and Single-Step Growth Curve Experiment
4.9. Molecular Docking Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Duijts, L.; Jaddoe, V.W.; Hofman, A.; Moll, H.A. Prolonged and exclusive breastfeeding reduces the risk of infectious diseases in infancy. Pediatrics 2010, 126, e18–e25. [Google Scholar] [CrossRef]
- Conzelmann, C.; Groß, R.; Meister, T.L.; Todt, D.; Krawczyk, A.; Dittmer, U.; Stenger, S.; Münch, J.; Steinmann, E.; Müller, J.A.; et al. Pasteurization Inactivates SARS-CoV-2-Spiked Breast Milk. Pediatrics 2021, 147, e2020031690. [Google Scholar] [CrossRef]
- Pfaender, S.; Heyden, J.; Friesland, M.; Ciesek, S.; Ejaz, A.; Steinmann, J.; Steinmann, J.; Malarski, A.; Stoiber, H.; Tsiavaliaris, G.; et al. Inactivation of hepatitis C virus infectivity by human breast milk. J. Infect. Dis. 2013, 208, 1943–1952. [Google Scholar] [CrossRef]
- Kell, D.B.; Heyden, E.L.; Pretorius, E. The Biology of Lactoferrin, an Iron-Binding Protein That Can Help Defend Against Viruses and Bacteria. Front. Immunol. 2020, 11, 1221. [Google Scholar] [CrossRef] [PubMed]
- Prentice, A.; Jarjou, L.M.; Drury, P.J.; Dewit, O.; Crawford, M.A. Breast-milk fatty acids of rural Gambian mothers: Effects of diet and maternal parity. J. Pediatr. Gastroenterol. Nutr. 1989, 8, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.; Marino, J.; Amanat, F.; Krammer, F.; Hahn-Holbrook, J.; Zolla-Pazner, S.; Powell, R.L. Robust and Specific Secretory IgA Against SARS-CoV-2 Detected in Human Milk. iScience 2020, 23, 101735. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Hong, B.; Luo, Y.; Peng, Q.; Wang, L.; Jin, X.; Chen, Y.; Hu, Y.; Shi, Y.; Li, T.; et al. The effect of whey protein on viral infection and replication of SARS-CoV-2 and pangolin coronavirus in vitro. Signal Transduct. Target. Ther. 2020, 5, 275. [Google Scholar] [CrossRef] [PubMed]
- Laucirica, D.R.; Triantis, V.; Schoemaker, R.; Estes, M.K.; Ramani, S. Milk Oligosaccharides Inhibit Human Rotavirus Infectivity in MA104 Cells. J. Nutr. 2017, 147, 1709–1714. [Google Scholar] [CrossRef]
- Drobni, P.; Näslund, J.; Evander, M. Lactoferrin inhibits human papillomavirus binding and uptake in vitro. Antivir. Res. 2004, 64, 63–68. [Google Scholar] [CrossRef]
- Puddu, P.; Borghi, P.; Gessani, S.; Valenti, P.; Belardelli, F.; Seganti, L. Antiviral effect of bovine lactoferrin saturated with metal ions on early steps of human immunodeficiency virus type 1 infection. Int. J. Biochem. Cell Biol. 1998, 30, 1055–1062. [Google Scholar] [CrossRef]
- Superti, F.; Siciliano, R.; Rega, B.; Giansanti, F.; Valenti, P.; Antonini, G. Involvement of bovine lactoferrin metal saturation, sialic acid and protein fragments in the inhibition of rotavirus infection. Biochim. Biophys. Acta 2001, 1528, 107–115. [Google Scholar] [CrossRef]
- Carvalho, C.A.M.; Casseb, S.M.M.; Gonçalves, R.B.; Silva, E.V.P.; Gomes, A.M.O.; Vasconcelos, P.F.C. Bovine lactoferrin activity against Chikungunya and Zika viruses. J. Gen. Virol. 2017, 98, 1749–1754. [Google Scholar] [CrossRef] [PubMed]
- Weng, T.Y.; Chen, L.C.; Shyu, H.W.; Chen, S.H.; Wang, J.R.; Yu, C.K.; Lei, H.Y.; Yeh, T.M. Lactoferrin inhibits enterovirus 71 infection by binding to VP1 protein and host cells. Antivir. Res. 2005, 67, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Shoemark, D.K.; Colenso, C.K.; Toelzer, C.; Gupta, K.; Sessions, R.B.; Davidson, A.D.; Berger, I.; Schaffitzel, C.; Spencer, J.; Mulholland, A.J. Molecular Simulations suggest Vitamins, Retinoids and Steroids as Ligands of the Free Fatty Acid Pocket of the SARS-CoV-2 Spike Protein. Angew. Chem. Int. Ed. Engl. 2021, 60, 7098–7110. [Google Scholar] [CrossRef] [PubMed]
- Schroten, H.; Hanisch, F.G.; Hansman, G.S. Human Norovirus Interactions with Histo-Blood Group Antigens and Human Milk Oligosaccharides. J. Virol. 2016, 90, 5855–5859. [Google Scholar] [CrossRef]
- Hong, P.; Ninonuevo, M.R.; Lee, B.; Lebrilla, C.; Bode, L. Human milk oligosaccharides reduce HIV-1-gp120 binding to dendritic cell-specific ICAM3-grabbing non-integrin (DC-SIGN). Br. J. Nutr. 2009, 101, 482–486. [Google Scholar] [CrossRef]
- Zevgiti, S.; Zabala, J.G.; Darji, A.; Dietrich, U.; Panou-Pomonis, E.; Sakarellos-Daitsiotis, M. Sialic acid and sialyl-lactose glyco-conjugates: Design, synthesis and binding assays to lectins and swine influenza H1N1 virus. J. Pept. Sci. 2012, 18, 52–58. [Google Scholar] [CrossRef]
- Marjomäki, V.; Turkki, P.; Huttunen, M. Infectious Entry Pathway of Enterovirus B Species. Viruses 2015, 7, 6387–6399. [Google Scholar] [CrossRef]
- Nikonov, O.S.; Chernykh, E.S.; Garber, M.B.; Nikonova, E.Y. Enteroviruses: Classification, Diseases They Cause, and Approaches to Development of Antiviral Drugs. Biochemistry 2017, 82, 1615–1631. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Wakabayashi, H.; Oda, H.; Yamauchi, K.; Abe, F. Lactoferrin for prevention of common viral infections. J. Infect. Chemother. 2014, 20, 666–671. [Google Scholar] [CrossRef]
- Berlutti, F.; Pantanella, F.; Natalizi, T.; Frioni, A.; Paesano, R.; Polimeni, A.; Valenti, P. Antiviral properties of lactoferrin--a natural immunity molecule. Molecules 2011, 16, 6992–7018. [Google Scholar] [CrossRef] [PubMed]
- Campione, E.; Cosio, T.; Rosa, L.; Lanna, C.; Di Girolamo, S.; Gaziano, R.; Valenti, P.; Bianchi, L. Lactoferrin as Protective Natural Barrier of Respiratory and Intestinal Mucosa against Coronavirus Infection and Inflammation. Int. J. Mol. Sci. 2020, 21, E4903. [Google Scholar] [CrossRef] [PubMed]
- Angeloni, S.; Ridet, J.L.; Kusy, N.; Gao, H.; Crevoisier, F.; Guinchard, S.; Kochhar, S.; Sigrist, H.; Sprenger, N. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology 2005, 15, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Huttunen, M.; Waris, M.; Kajander, R.; Hyypiä, T.; Marjomäki, V. Coxsackievirus A9 infects cells via nonacidic multivesicular bodies. J. Virol. 2014, 88, 5138–5151. [Google Scholar] [CrossRef]
- Shakeel, S.; Seitsonen, J.J.; Kajander, T.; Laurinmäki, P.; Hyypiä, T.; Susi, P.; Butcher, S.J. Structural and functional analysis of coxsackievirus A9 integrin αvβ6 binding and uncoating. J. Virol. 2013, 87, 3943–3951. [Google Scholar] [CrossRef]
- He, S.T.; Qin, H.; Guan, L.; Liu, K.; Hong, B.; Zhang, X.; Lou, F.; Li, M.; Lin, W.; Chen, Y.; et al. Bovine lactoferrin inhibits SARS-CoV-2 and SARS-CoV-1 by targeting the RdRp complex and alleviates viral infection in the hamster model. J. Med. Virol. 2022. [CrossRef]
- Salaris, C.; Scarpa, M.; Elli, M.; Bertolini, A.; Guglielmetti, S.; Pregliasco, F.; Blandizzi, C.; Brun, P.; Castagliuolo, I. Protective Effects of Lactoferrin against SARS-CoV-2 Infection In Vitro. Nutrients 2021, 13, 328. [Google Scholar] [CrossRef]
- Ikeda, M.; Sugiyama, K.; Tanaka, T.; Tanaka, K.; Sekihara, H.; Shimotohno, K.; Kato, N. Lactoferrin markedly inhibits hepatitis C virus infection in cultured human hepatocytes. Biochem. Biophys. Res. Commun. 1998, 245, 549–553. [Google Scholar] [CrossRef]
- Hara, K.; Ikeda, M.; Saito, S.; Matsumoto, S.; Numata, K.; Kato, N.; Tanaka, K.; Sekihara, H. Lactoferrin inhibits hepatitis B virus infection in cultured human hepatocytes. Hepatol. Res. 2002, 24, 228. [Google Scholar] [CrossRef]
- Swart, P.J.; Harmsen, M.C.; Kuipers, M.E.; Van Dijk, A.A.; Van Der Strate, B.W.; Van Berkel, P.H.; Nuijens, J.H.; Smit, C.; Witvrouw, M.; De Clercq, E.; et al. Charge modification of plasma and milk proteins results in antiviral active compounds. J. Pept. Sci. 1999, 5, 563–576. [Google Scholar] [CrossRef]
- Hasegawa, K.; Motsuchi, W.; Tanaka, S.; Dosako, S. Inhibition with lactoferrin of in vitro infection with human herpes virus. Jpn. J. Med. Sci. Biol. 1994, 47, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.E.; Kariwa, H.; Mizutani, T.; Yoshimatsu, K.; Arikawa, J.; Takashima, I. In vitro antiviral activity of lactoferrin and ribavirin upon hantavirus. Arch. Virol. 2000, 145, 1571–1582. [Google Scholar] [CrossRef] [PubMed]
- Tinari, A.; Pietrantoni, A.; Ammendolia, M.G.; Valenti, P.; Superti, F. Inhibitory activity of bovine lactoferrin against echovirus induced programmed cell death in vitro. Int. J. Antimicrob. Agents 2005, 25, 433–438. [Google Scholar] [CrossRef]
- Pietrantoni, A.; Di Biase, A.M.; Tinari, A.; Marchetti, M.; Valenti, P.; Seganti, L.; Superti, F. Bovine lactoferrin inhibits adenovirus infection by interacting with viral structural polypeptides. Antimicrob. Agents Chemother. 2003, 47, 2688–2691. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Sato, R.; Aoki, T.; Omoe, K.; Inanami, O.; Hankanga, C.; Yamada, Y.; Tomizawa, N.; Yasuda, J.; Sasaki, J. Effect of bovine lactoferrin on functions of activated feline peripheral blood mononuclear cells during chronic feline immunodeficiency virus infection. J. Vet. Med. Sci. 2008, 70, 429–435. [Google Scholar] [CrossRef]
- Taha, S.H.; Mehrez, M.A.; Sitohy, M.Z.; Abou Dawood, A.G.; Abd-El Hamid, M.M.; Kilany, W.H. Effectiveness of esterified whey proteins fractions against Egyptian Lethal Avian Influenza A (H5N1). Virol. J. 2010, 7, 330. [Google Scholar] [CrossRef]
- Inagaki, M.; Muranishi, H.; Yamada, K.; Kakehi, K.; Uchida, K.; Suzuki, T.; Yabe, T.; Nakagomi, T.; Nakagomi, O.; Kanamaru, Y. Bovine κ-casein inhibits human rotavirus (HRV) infection via direct binding of glycans to HRV. J. Dairy Sci. 2014, 97, 2653–2661. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.B.; Cheung, R.C.; Wong, J.H.; Wang, Y.; Ip, D.T.; Wan, D.C.; Xia, J. Antiviral activities of whey proteins. Appl. Microbiol. Biotechnol. 2015, 99, 6997–7008. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, D.L.; Doster, R.S.; Weitkamp, J.H.; Aronoff, D.M.; Gaddy, J.A.; Townsend, S.D. Human Milk Oligosaccharides Exhibit Antimicrobial and Antibiofilm Properties against Group B Streptococcus. ACS Infect. Dis. 2017, 3, 595–605. [Google Scholar] [CrossRef]
- Ackerman, D.L.; Craft, K.M.; Doster, R.S.; Weitkamp, J.H.; Aronoff, D.M.; Gaddy, J.A.; Townsend, S.D. Antimicrobial and Antibiofilm Activity of Human Milk Oligosaccharides against Streptococcus agalactiae, Staphylococcus aureus, and Acinetobacter baumannii. ACS Infect. Dis. 2018, 4, 315–324. [Google Scholar] [CrossRef]
- Li, M.; Monaco, M.H.; Wang, M.; Comstock, S.S.; Kuhlenschmidt, T.B.; Fahey, G.C.; Miller, M.J.; Kuhlenschmidt, M.S.; Donovan, S.M. Human milk oligosaccharides shorten rotavirus-induced diarrhea and modulate piglet mucosal immunity and colonic microbiota. ISME J. 2014, 8, 1609–1620. [Google Scholar] [CrossRef]
- Weichert, S.; Koromyslova, A.; Singh, B.K.; Hansman, S.; Jennewein, S.; Schroten, H.; Hansman, G.S. Structural Basis for Norovirus Inhibition by Human Milk Oligosaccharides. J. Virol. 2016, 90, 4843–4848. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.E.; Xu, L.L.; Townsend, S.D. Prospecting Human Milk Oligosaccharides as a Defense Against Viral Infections. ACS Infect. Dis. 2021, 7, 254–263. [Google Scholar] [CrossRef]
- Bode, L.; Kuhn, L.; Kim, H.Y.; Hsiao, L.; Nissan, C.; Sinkala, M.; Kankasa, C.; Mwiya, M.; Thea, D.M.; Aldrovandi, G.M. Human milk oligosaccharide concentration and risk of postnatal transmission of HIV through breastfeeding. Am. J. Clin. Nutr. 2012, 96, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Van Liempt, E.; Bank, C.M.; Mehta, P.; Garciá-Vallejo, J.J.; Kawar, Z.S.; Geyer, R.; Alvarez, R.A.; Cummings, R.D.; van Kooyk, Y.; van Die, I. Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett. 2006, 580, 6123–6131. [Google Scholar] [CrossRef] [PubMed]
- Ramani, S.; Stewart, C.J.; Laucirica, D.R.; Ajami, N.J.; Robertson, B.; Autran, C.A.; Shinge, D.; Rani, S.; Anandan, S.; Hu, L.; et al. Human milk oligosaccharides, milk microbiome and infant gut microbiome modulate neonatal rotavirus infection. Nat. Commun. 2018, 9, 5010. [Google Scholar] [CrossRef]
- Heikkilä, O.; Merilahti, P.; Hakanen, M.; Karelehto, E.; Alanko, J.; Sukki, M.; Kiljunen, S.; Susi, P. Integrins are not essential for entry of coxsackievirus A9 into SW480 human colon adenocarcinoma cells. Virol. J. 2016, 13, 171. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, G.; Liu, S.; Chen, X.; Peng, R.; Dai, L.; Qu, X.; Li, S.; Song, H.; Gao, Z.; et al. Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B. Cell 2019, 177, 1553–1565.e16. [Google Scholar] [CrossRef]


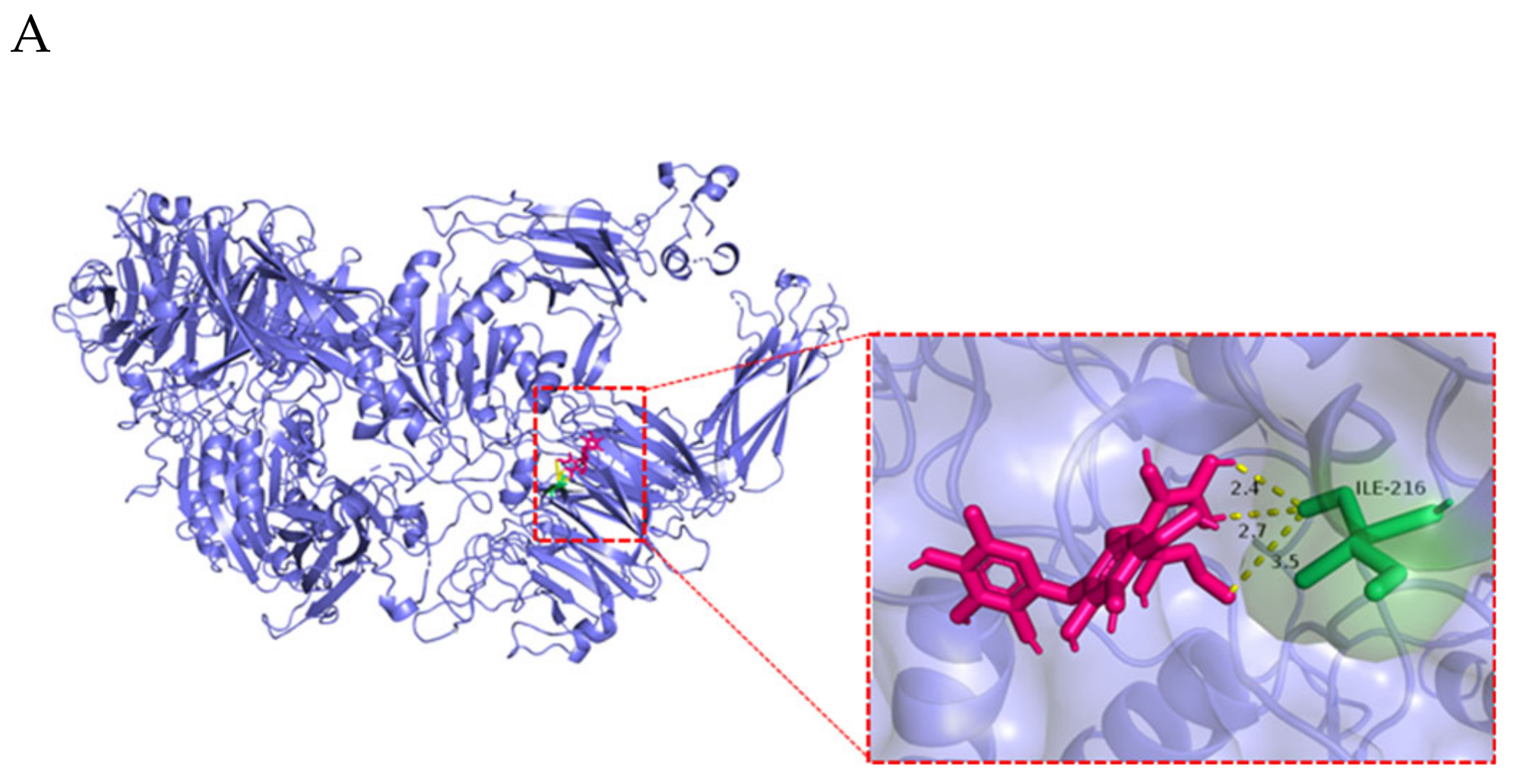
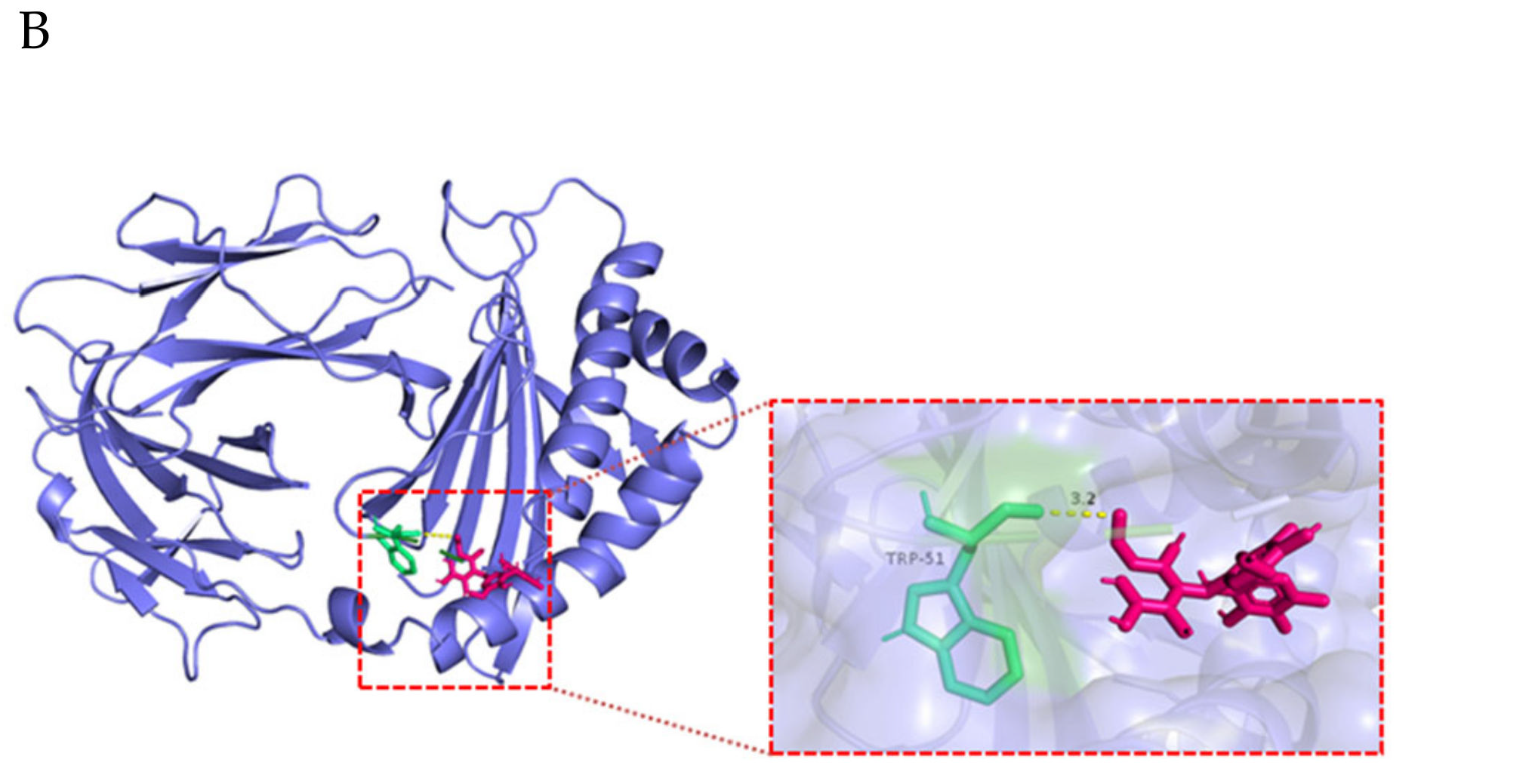
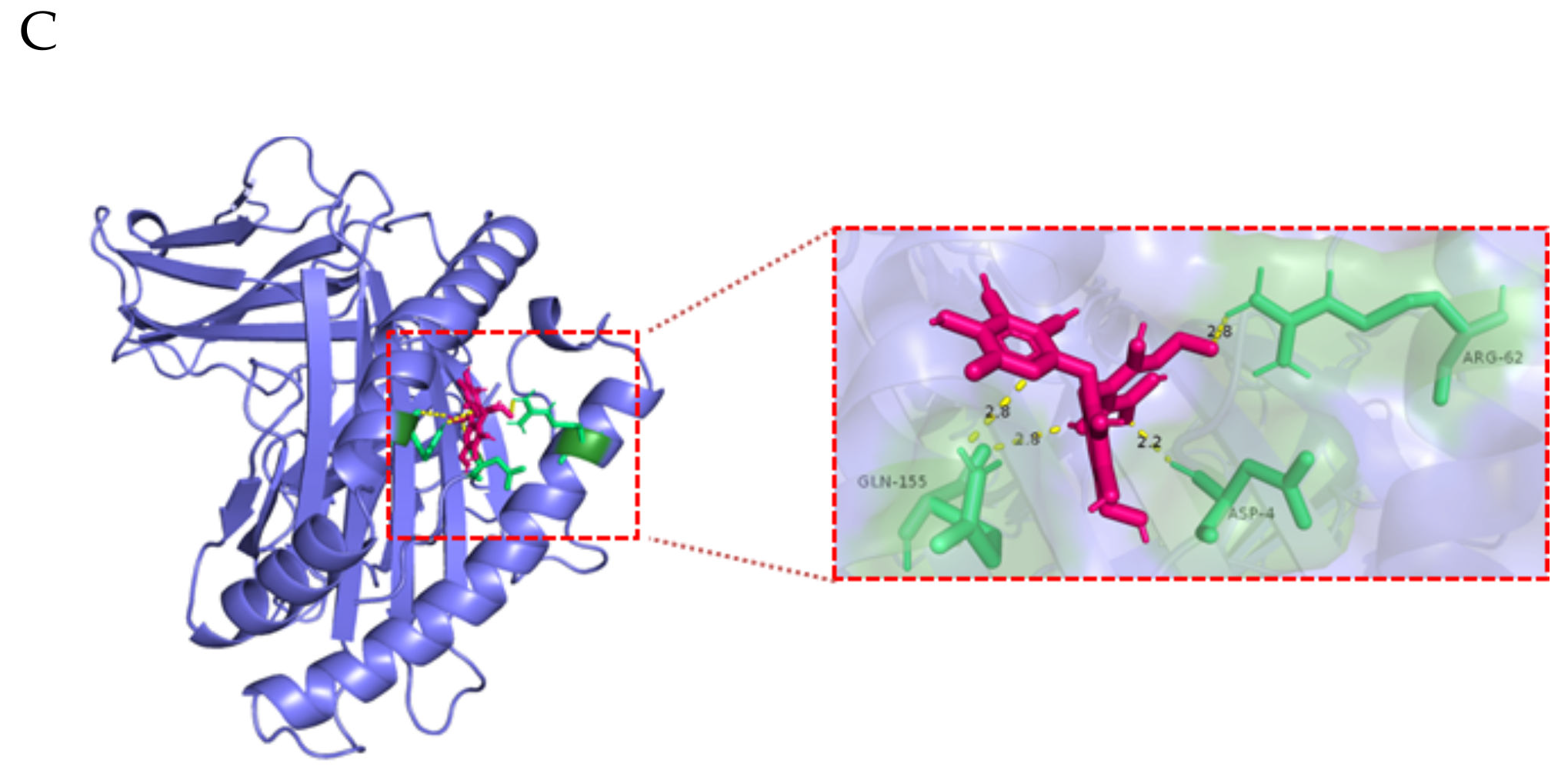
| Ligand | Protein | Binding Energy (ΔG) kcal/mol | Ligand Efficiency |
|---|---|---|---|
| 2’-FL (C18H32O15) | FCGRT | −2.34 | −0.07 |
| β2M | −3.19 | −0.1 | |
| ITGB6 (αvβ6) | −2.14 | −0.06 |
| Oligonucleotide Name | Sequence |
|---|---|
| CV-A9 -F | TCATGACACCAGCTGATAAG |
| CV-A9-R | TGCTCATCTGCTCTGAAGTATC |
| GAPDH-F | AGCCTCAAGATCATCAGCAATG |
| GAPDH-R | ATGGACTGTGGTCATGAGTCCTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lou, F.; Hu, R.; Chen, Y.; Li, M.; An, X.; Song, L.; Tong, Y.; Fan, H. 2’-Fucosyllactose Inhibits Coxsackievirus Class A Type 9 Infection by Blocking Virus Attachment and Internalisation. Int. J. Mol. Sci. 2022, 23, 13727. https://doi.org/10.3390/ijms232213727
Lou F, Hu R, Chen Y, Li M, An X, Song L, Tong Y, Fan H. 2’-Fucosyllactose Inhibits Coxsackievirus Class A Type 9 Infection by Blocking Virus Attachment and Internalisation. International Journal of Molecular Sciences. 2022; 23(22):13727. https://doi.org/10.3390/ijms232213727
Chicago/Turabian StyleLou, Fuxing, Ruolan Hu, Yangzhen Chen, Mengzhe Li, Xiaoping An, Lihua Song, Yigang Tong, and Huahao Fan. 2022. "2’-Fucosyllactose Inhibits Coxsackievirus Class A Type 9 Infection by Blocking Virus Attachment and Internalisation" International Journal of Molecular Sciences 23, no. 22: 13727. https://doi.org/10.3390/ijms232213727
APA StyleLou, F., Hu, R., Chen, Y., Li, M., An, X., Song, L., Tong, Y., & Fan, H. (2022). 2’-Fucosyllactose Inhibits Coxsackievirus Class A Type 9 Infection by Blocking Virus Attachment and Internalisation. International Journal of Molecular Sciences, 23(22), 13727. https://doi.org/10.3390/ijms232213727






