Chalcogen Bond as a Factor Stabilizing Ligand Conformation in the Binding Pocket of Carbonic Anhydrase IX Receptor Mimic
Abstract
1. Introduction
2. Results and Discussion
2.1. Metric and Spectroscopic Signatures of Non-Covalent Interactions
2.1.1. Car–Parrinello Molecular Dynamics Description of the AZM ligand and Its Complexes with Amino Acids
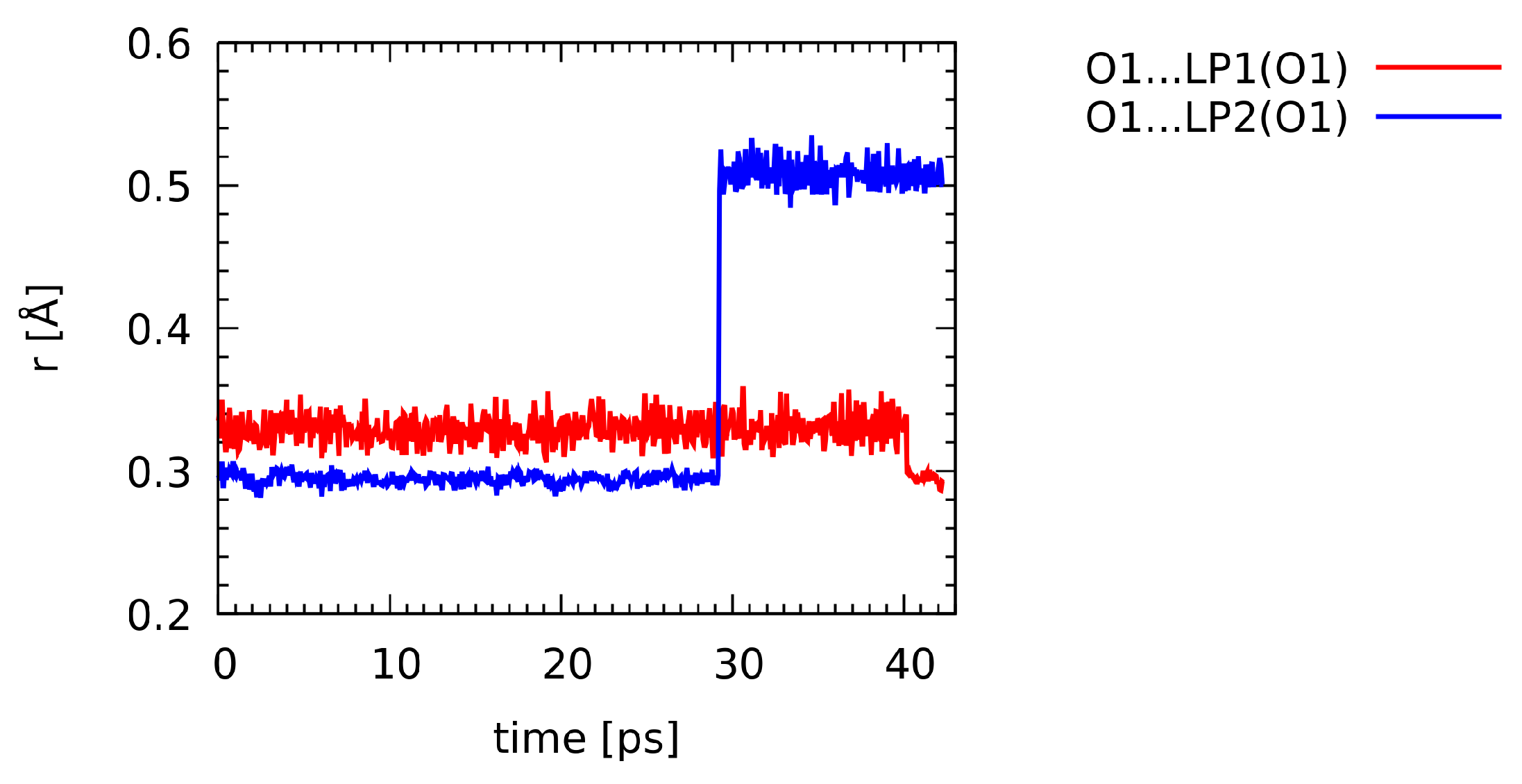
2.1.2. Nuclear Quantum Effects: Car-Parrinello vs. Path Integral Molecular Dynamics
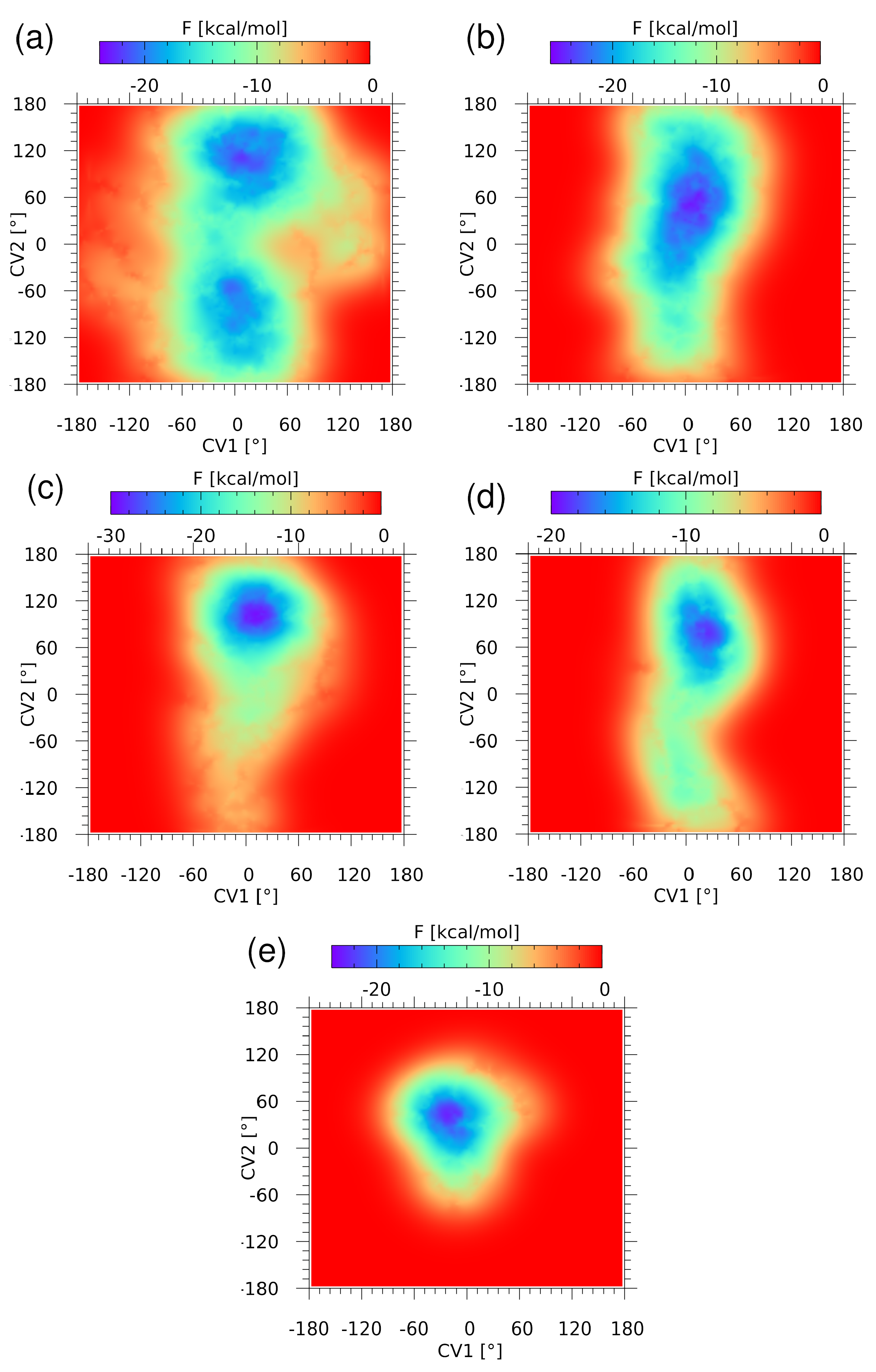
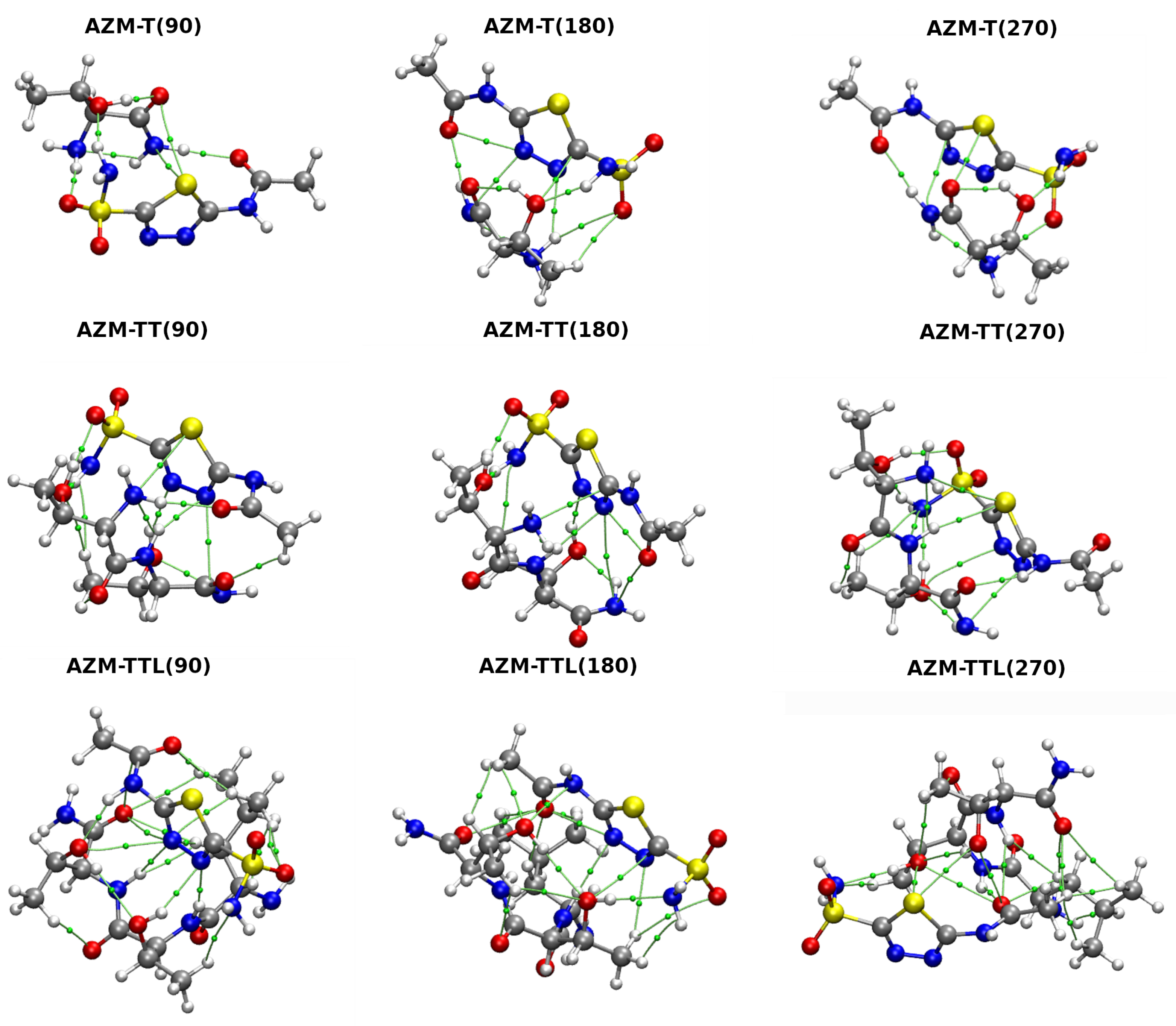
2.1.3. Free Energy Landscapes Derived from CPMD-Based Metadynamics
2.2. Electronic Structure Evidences of the Non-Covalent Network of Intra- and Intermolecular Interactions
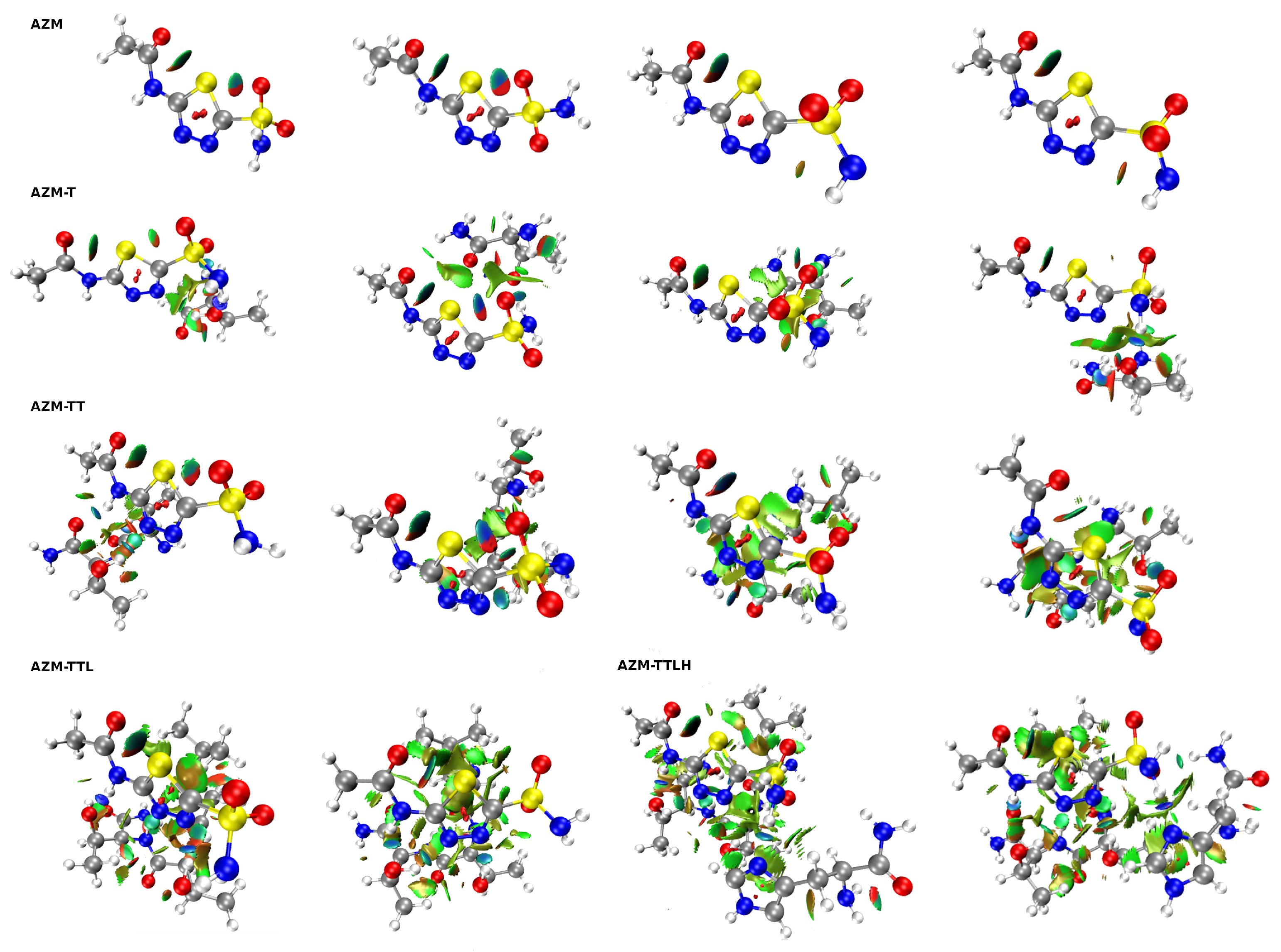
2.3. Non-Covalent Interactions Analysis Based on the NCI Index
2.4. Atoms in Molecules Analysis of Intra- and Intermolecular Non-Covalent Bonds
2.5. Partitioning of the Interaction Energy
3. Materials and Methods
3.1. First-Principle Molecular Dynamics and Metadynamics Methods
3.1.1. Car–Parrinello Molecular Dynamics (CPMD)
3.1.2. Path Integral Molecular Dynamics (PIMD)
3.1.3. Metadynamics (METD)
3.1.4. Post-Processing of the Results Based on Time-Evolution Methods
3.2. Density Functional Theory (DFT) and Post-Hartree–Fock Methods
3.3. Symmetry-Adapted Perturbation Theory (SAPT)
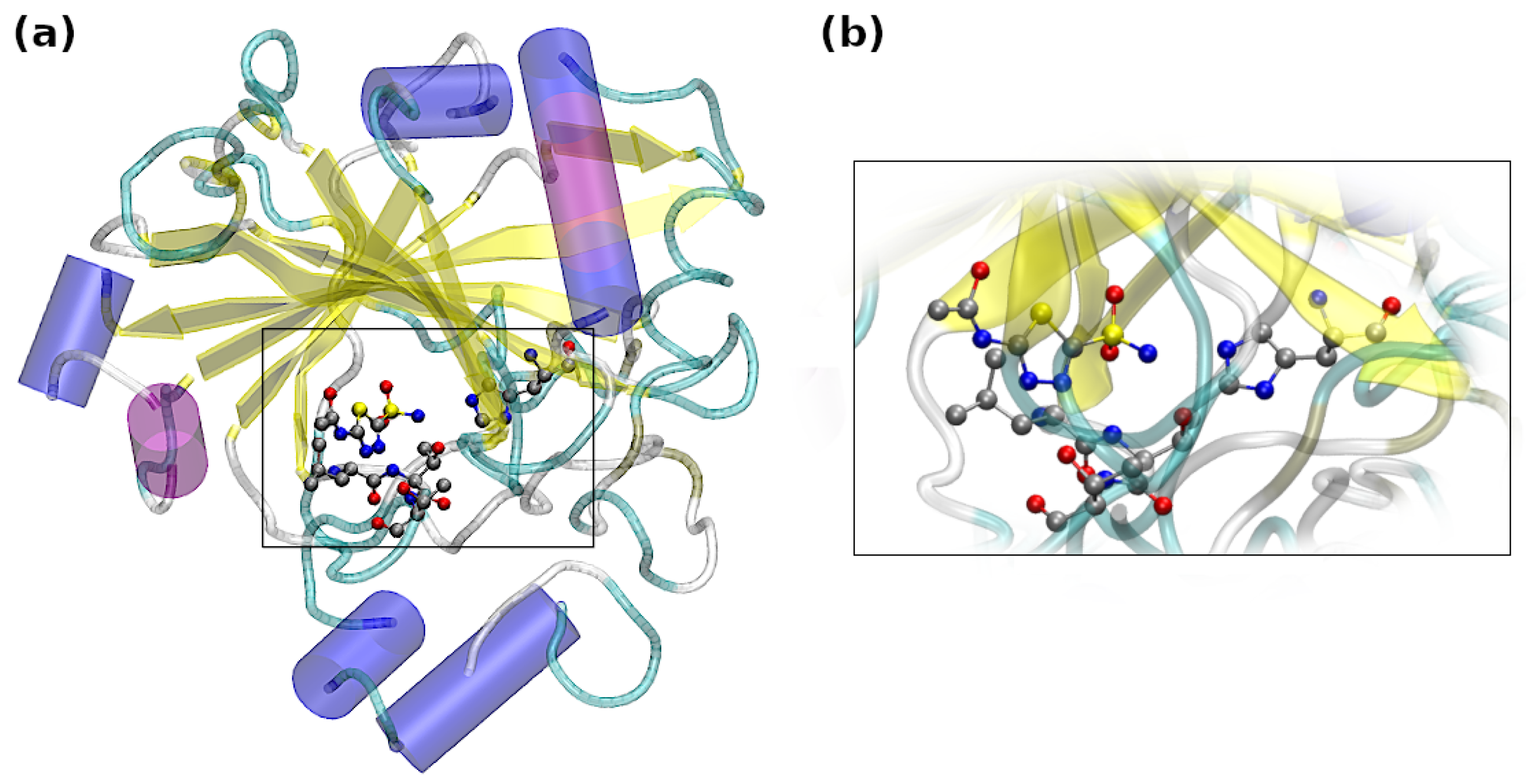
- Extracted from the Carbonic Anhydrase IX mimic–AZM complex (PDB deposit 3DC3) and
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AZM | Acetazolamide |
| FPMD | First-Principle Molecular Dynamics |
| METD | Metadynamics |
| DFT | Density Functional Theory |
| NCI | Non-Covalent Interaction |
| NBO | Natural Bond Orbital |
| AIM | Atoms In Molecules |
| SAPT | Symmetry-Adapted Perturbation Theory |
| CPMD | Car–Parrinello Molecular Dynamics |
| PIMD | Path Integral Molecular Dynamics |
| HB | Hydrogen bond |
| EDA | Energy Decomposition Analysis |
| PDB | Protein Data Bank |
| CV | Collective Variable |
| PES | Potential Energy Surface |
| BCP | Bond Critical Point |
| RCP | Ring Critical Point |
| MEP | Molecular Electrostatic Potential |
| BSSE | Basis Set Superposition Error |
| NQE | Nuclear Quantum Effects |
References
- Müller-Dethlefs, K.; Hobza, P. Noncovalent Interactions: A Challenge for Experiment and Theory. Chem. Rev. 1999, 100, 143–168. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, O. Noncovalent interactions in biocomplexes. Phys. Sci. Rev. 2016, 1, 20160001. [Google Scholar] [CrossRef]
- McNaught, A.D.; Wilkinson, A. Lewis acid. In The IUPAC Compendium of Chemical Terminology; International Union of Pure and Applied Chemistry (IUPAC), Blackwell Science: Cambridge, UK, 2014. [Google Scholar] [CrossRef]
- Lewis, G.N. Valence and the Structure of Atoms and Molecules; American Chemical Society. Monograph Series; Chemical Catalog Company: New York, NY, USA, 1923; ISBN 9780598985408. [Google Scholar]
- Alkorta, I.; Elguero, J.; Frontera, A. Not Only Hydrogen Bonds: Other Noncovalent Interactions. Crystals 2020, 10, 180. [Google Scholar] [CrossRef]
- Politzer, P.; Riley, K.E.; Bulat, F.A.; Murray, J.S. Perspectives on halogen bonding and other σ-hole interactions: Lex parsimoniae (Occam’s Razor). Comput. Theor. Chem. 2012, 998, 2–8. [Google Scholar] [CrossRef]
- Price, A.J.A.; Bryenton, K.R.; Johnson, E.R. Requirements for an accurate dispersion-corrected density functional. J. Chem. Phys. 2021, 154, 230902. [Google Scholar] [CrossRef] [PubMed]
- Pauling, L.; Corey, R.B.; Branson, H.R. The structure of proteins: Two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. USA 1951, 37, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Pauling, L.; Corey, R.B. A Proposed Structure For The Nucleic Acids. Proc. Natl. Acad. Sci. USA 1953, 39, 84–97. [Google Scholar] [CrossRef]
- Fersht, A.R. The hydrogen bond in molecular recognition. Trends Biochem. Sci. 1987, 12, 301–304. [Google Scholar] [CrossRef]
- Dong, J.; Davis, A.P. Molecular Recognition Mediated by Hydrogen Bonding in Aqueous Media. Angew. Chem. Int. Ed. 2020, 60, 8035–8048. [Google Scholar] [CrossRef]
- Ishida, T. Low-Barrier Hydrogen Bond Hypothesis in the Catalytic Triad Residue of Serine Proteases: Correlation between Structural Rearrangement and Chemical Shifts in the Acylation Process. Biochemistry 2006, 45, 5413–5420. [Google Scholar] [CrossRef]
- Franconetti, A.; de Gonzalo, G. Recent Developments on Supported Hydrogen-bond Organocatalysts. ChemCatChem 2018, 10, 5554–5572. [Google Scholar] [CrossRef]
- Herschlag, D.; Pinney, M.M. Hydrogen Bonds: Simple after All? Biochemistry 2018, 57, 3338–3352. [Google Scholar] [CrossRef] [PubMed]
- Morozov, D.; Groenhof, G. Hydrogen Bond Fluctuations Control Photochromism in a Reversibly Photo-Switchable Fluorescent Protein. Angew. Chem. Int. Ed. 2015, 55, 576–578. [Google Scholar] [CrossRef] [PubMed]
- Clark, T.; Hennemann, M.; Murray, J.S.; Politzer, P. Halogen bonding: The σ-hole. J. Mol. Model. 2006, 13, 291–296. [Google Scholar] [CrossRef]
- Murray, J.S.; Politzer, P. Molecular electrostatic potentials and noncovalent interactions. Wiley Interdiscip. Rev. Comput. Mol. Sci. WIRES Comput. Mol. Sci. 2017, 7, e1326. [Google Scholar] [CrossRef]
- Murray, J.S.; Lane, P.; Politzer, P. A predicted new type of directional noncovalent interaction. Int. J. Quantum Chem. 2007, 107, 2286–2292. [Google Scholar] [CrossRef]
- Murray, J.S.; Resnati, G.; Politzer, P. Close contacts and noncovalent interactions in crystals. Faraday Discuss. 2017, 203, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.S.; Lane, P.; Clark, T.; Politzer, P. σ-hole bonding: Molecules containing group VI atoms. J. Mol. Model. 2007, 13, 1033–1038. [Google Scholar] [CrossRef]
- Murray, J.S.; Lane, P.; Clark, T.; Riley, K.E.; Politzer, P. σ-Holes, π-holes and electrostatically-driven interactions. J. Mol. Model. 2011, 18, 541–548. [Google Scholar] [CrossRef]
- Politzer, P.; Murray, J.S. σ-holes and π-holes: Similarities and differences. J. Comput. Chem. 2017, 39, 464–471. [Google Scholar] [CrossRef]
- Clark, T.; Murray, J.S.; Politzer, P. A perspective on quantum mechanics and chemical concepts in describing noncovalent interactions. Phys. Chem. Chem. Phys. 2018, 20, 30076–30082. [Google Scholar] [CrossRef]
- Politzer, P.; Murray, J.S.; Clark, T.; Resnati, G. The σ-hole revisited. Phys. Chem. Chem. Phys. 2017, 19, 32166–32178. [Google Scholar] [CrossRef]
- Politzer, P.; Murray, J.S.; Clark, T. Interpretation. In Topics in Current Chemistry; Springer International Publishing: Berlin/Heidelberg, Germany, 2014; pp. 19–42. [Google Scholar] [CrossRef]
- Scheiner, S. Principles Guiding the Square Bonding Motif Containing a Pair of Chalcogen Bonds between Chalcogenadiazoles. J. Phys. Chem. A 2022, 126, 1194–1203. [Google Scholar] [CrossRef]
- Frontera, A.; Bauza, A. On the Importance of Pnictogen and Chalcogen Bonding Interactions in Supramolecular Catalysis. Int. J. Mol. Sci. 2021, 22, 12550. [Google Scholar] [CrossRef]
- Grabowski, S. Triel Bonds, π-Hole-π-Electrons Interactions in Complexes of Boron and Aluminium Trihalides and Trihydrides with Acetylene and Ethylene. Molecules 2015, 20, 11297–11316. [Google Scholar] [CrossRef]
- Grabowski, S. New Type of Halogen Bond: Multivalent Halogen Interacting with π- and σ-Electrons. Molecules 2017, 22, 2150. [Google Scholar] [CrossRef]
- Grabowski, S. Tetrel Bonds with π-Electrons Acting as Lewis Bases—Theoretical Results and Experimental Evidences. Molecules 2018, 23, 1183. [Google Scholar] [CrossRef] [PubMed]
- Zierkiewicz, W.; Michalczyk, M.; Scheiner, S. Noncovalent Bonds through Sigma and Pi-Hole Located on the Same Molecule. Guiding Principles and Comparisons. Molecules 2021, 26, 1740. [Google Scholar] [CrossRef] [PubMed]
- Bauzá, A.; Frontera, A. Aerogen Bonding Interaction: A New Supramolecular Force? Angew. Chem. Int. Ed. 2015, 54, 7340–7343. [Google Scholar] [CrossRef] [PubMed]
- Cavallo, G.; Metrangolo, P.; Milani, R.; Pilati, T.; Priimagi, A.; Resnati, G.; Terraneo, G. The Halogen Bond. Chem. Rev. 2016, 116, 2478–2601. [Google Scholar] [CrossRef]
- Scheiner, S. Participation of S and Se in hydrogen and chalcogen bonds. CrystEngComm 2021, 23, 6821–6837. [Google Scholar] [CrossRef]
- Lu, J.; Scheiner, S. Effects of Halogen, Chalcogen, Pnicogen, and Tetrel Bonds on IR and NMR Spectra. Molecules 2019, 24, 2822. [Google Scholar] [CrossRef]
- Vogel, L.; Wonner, P.; Huber, S.M. Chalcogen Bonding: An Overview. Angew. Chem. Int. Ed. 2018, 58, 1880–1891. [Google Scholar] [CrossRef]
- Meyer, E.A.; Castellano, R.K.; Diederich, F. Interactions with Aromatic Rings in Chemical and Biological Recognition. Angew. Chem. Int. Ed. 2003, 42, 1210–1250. [Google Scholar] [CrossRef]
- Bauzá, A.; Frontera, A. Halogen and Chalcogen Bond Energies Evaluated Using Electron Density Properties. ChemPhysChem 2019, 21, 26–31. [Google Scholar] [CrossRef]
- Brammer, L. Halogen bonding, chalcogen bonding, pnictogen bonding, tetrel bonding: Origins, current status and discussion. Faraday Discuss. 2017, 203, 485–507. [Google Scholar] [CrossRef]
- Wang, W.; Ji, B.; Zhang, Y. Chalcogen Bond: A Sister Noncovalent Bond to Halogen Bond. J. Phys. Chem. A 2009, 113, 8132–8135. [Google Scholar] [CrossRef]
- Genis, C.; Sippel, K.H.; Case, N.; Cao, W.; Avvaru, B.S.; Tartaglia, L.J.; Govindasamy, L.; Tu, C.; Agbandje-McKenna, M.; Silverman, D.N.; et al. Design of a Carbonic Anhydrase IX Active-Site Mimic To Screen Inhibitors for Possible Anticancer Properties. Biochemistry 2009, 48, 1322–1331. [Google Scholar] [CrossRef] [PubMed]
- Potter, C.P.S.; Harris, A.L. Diagnostic, prognostic and therapeutic implications of carbonic anhydrases in cancer. Br. J. Cancer 2003, 89, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Thiry, A.; Dogné, J.M.; Masereel, B.; Supuran, C.T. Targeting tumor-associated carbonic anhydrase IX in cancer therapy. Trends Pharmacol. Sci. 2006, 27, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T.; Briganti, F.; Tilli, S.; Chegwidden, W.; Scozzafava, A. Carbonic anhydrase inhibitors: Sulfonamides as antitumor agents? Bioorg. Med. Chem. 2001, 9, 703–714. [Google Scholar] [CrossRef]
- Parkkila, S.; Rajaniemi, H.; Parkkila, A.K.; Kivelä, J.; Waheed, A.; Pastoreková, S.; Pastorek, J.; Sly, W.S. Carbonic anhydrase inhibitor suppresses invasion of renal cancer cells in-vitro. Proc. Natl. Acad. Sci. USA 2000, 97, 2220–2224. [Google Scholar] [CrossRef] [PubMed]
- Vivo, M.D.; Masetti, M.; Bottegoni, G.; Cavalli, A. Role of Molecular Dynamics and Related Methods in Drug Discovery. J. Med. Chem. 2016, 59, 4035–4061. [Google Scholar] [CrossRef]
- Car, R.; Parrinello, M. Unified Approach for Molecular Dynamics and Density-Functional Theory. Phys. Rev. Lett. 1985, 55, 2471–2474. [Google Scholar] [CrossRef] [PubMed]
- Tuckerman, M.E.; Marx, D.; Klein, M.L.; Parrinello, M. Efficient and general algorithms for path integral Car-Parrinello molecular dynamics. J. Chem. Phys. 1996, 104, 5579–5588. [Google Scholar] [CrossRef]
- Feynman, R.P. Space-Time Approach to Non-Relativistic Quantum Mechanics. Rev. Mod. Phys. 1948, 20, 367–387. [Google Scholar] [CrossRef]
- Laio, A.; Parrinello, M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA 2002, 99, 12562–12566. [Google Scholar] [CrossRef]
- Bader, R.F.W.; Nguyen-Dang, T.T.; Tal, Y. Quantum topology of molecular charge distributions. II. Molecular structure and its change. J. Chem. Phys. 1979, 70, 4316–4329. [Google Scholar] [CrossRef]
- Bader, R.F.W. Quantum topology of molecular charge distributions. III. The mechanics of an atom in a molecule. J. Chem. Phys. 1980, 73, 2871–2883. [Google Scholar] [CrossRef]
- Bader, R. Atoms in Molecules: A Quantum Theory; International Series of Monographs on Chemistry; Clarendon Press: Oxford, UK, 1994. [Google Scholar]
- Weinhold, F.; Landis, C.R. Natural Bond Orbitals and extensions of localized bonding concepts. Chem. Educ. Res. Pract. 2001, 2, 91–104. [Google Scholar] [CrossRef]
- Weinhold, F.; Landis, C.R. Discovering Chemistry With Natural Bond Orbitals; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Johnson, E.R.; Keinan, S.; Mori-Sánchez, P.; Contreras-García, J.; Cohen, A.J.; Yang, W. Revealing Noncovalent Interactions. J. Am. Chem. Soc. 2010, 132, 6498–6506. [Google Scholar] [CrossRef]
- Jeziorski, B.; Moszynski, R.; Szalewicz, K. Perturbation Theory Approach to Intermolecular Potential Energy Surfaces of van der Waals Complexes. Chem. Rev. 1994, 94, 1887–1930. [Google Scholar] [CrossRef]
- Szalewicz, K. Symmetry-adapted perturbation theory of intermolecular forces. Wiley Interdiscip. Rev. Comput. Mol. Sci. WIRES Comput. Mol. Sci. 2011, 2, 254–272. [Google Scholar] [CrossRef]
- Zhao, L.; von Hopffgarten, M.; Andrada, D.M.; Frenking, G. Energy decomposition analysis. Wiley Interdiscip. Rev. Comput. Mol. Sci. WIRES Comput. Mol. Sci. 2017, 8, e1345. [Google Scholar] [CrossRef]
- Marx, D.; Parrinello, M. The Effect of Quantum and Thermal Fluctuations on the Structure of the Floppy Molecule C2H3+. Science 1996, 271, 179–181. [Google Scholar] [CrossRef]
- Tuckerman, M.E.; Marx, D. Heavy-Atom Skeleton Quantization and Proton Tunneling in “Intermediate-Barrier” Hydrogen Bonds. Phys. Rev. Lett. 2001, 86, 4946–4949. [Google Scholar] [CrossRef]
- Espinosa, E.; Molins, E.; Lecomte, C. Hydrogen bond strengths revealed by topological analyses of experimentally observed electron densities. Chem. Phys. Lett. 1998, 285, 170–173. [Google Scholar] [CrossRef]
- Vener, M.V.; Egorova, A.N.; Churakov, A.V.; Tsirelson, V.G. Intermolecular hydrogen bond energies in crystals evaluated using electron density properties: DFT computations with periodic boundary conditions. J. Comput. Chem. 2012, 33, 2303–2309. [Google Scholar] [CrossRef]
- Consortium. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar] [CrossRef]
- Dunning, T.H. Gaussian basis sets for use in correlated molecular calculations. I. The atoms boron through neon and hydrogen. J. Chem. Phys. 1989, 90, 1007–1023. [Google Scholar] [CrossRef]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1996, 77, 3865–3868. [Google Scholar] [CrossRef]
- Grimme, S. Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 2006, 27, 1787–1799. [Google Scholar] [CrossRef]
- Troullier, N.; Martins, J.L. Efficient pseudopotentials for plane-wave calculations. Phys. Rev. B 1991, 43, 1993–2006. [Google Scholar] [CrossRef]
- Schlegel, H.B. Estimating the hessian for gradient-type geometry optimizations. Theor. Chem. Acc. 1984, 66, 333–340. [Google Scholar] [CrossRef]
- Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef]
- Marzari, N.; Vanderbilt, D. Maximally localized generalized Wannier functions for composite energy bands. Phys. Rev. B 1997, 56, 12847–12865. [Google Scholar] [CrossRef]
- CPMD Version 4.3-4610, Copyright IBM Corp. (1990–2004) Copyright MPI für Festkoerperforschung Stuttgart (1997–2001). Available online: http://www.cpmd.org (accessed on 28 December 2021).
- Tuckerman, M.E.; Berne, B.J.; Martyna, G.J.; Klein, M.L. Efficient molecular dynamics and hybrid Monte Carlo algorithms for path integrals. J. Chem. Phys. 1993, 99, 2796–2808. [Google Scholar] [CrossRef]
- Iannuzzi, M.; Laio, A.; Parrinello, M. Efficient Exploration of Reactive Potential Energy Surfaces Using Car-Parrinello Molecular Dynamics. Phys. Rev. Lett. 2003, 90. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.; Kelley, C.; Lang, R.; Kotz, D.; Campbell, J.; Elber, G.; Woo, A. Gnuplot 4.4: An Interactive Plotting Program. 2010. Available online: http://gnuplot.sourceforge.net/ (accessed on 28 December 2021).
- Humphrey, W.; Dalke, A.; Schulten, K. VMD—Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Backus, J.W.; Stern, H.; Ziller, I.; Hughes, R.A.; Nutt, R.; Beeber, R.J.; Best, S.; Goldberg, R.; Haibt, L.M.; Herrick, H.L.; et al. The FORTRAN automatic coding system. In IRE-AIEE-ACM ’57 (Western): Papers Presented at the February 26-28, 1957, Western Joint Computer Conference: Techniques for Reliability; ACM Press: New York, NY, USA, 1957. [Google Scholar] [CrossRef]
- Hohenberg, P.; Kohn, W. Inhomogeneous Electron Gas. Phys. Rev. 1964, 136, B864–B871. [Google Scholar] [CrossRef]
- Kohn, W.; Sham, L.J. Self-Consistent Equations Including Exchange and Correlation Effects. Phys. Rev. 1965, 140, A1133–A1138. [Google Scholar] [CrossRef]
- Scheiner, S. Dissection of the Origin of π-Holes and the Noncovalent Bonds in Which They Engage. J. Phys. Chem. A 2021, 125, 6514–6528. [Google Scholar] [CrossRef]
- Pacios, L.F. Change with the intermolecular distance of electron properties of hydrogen bond dimers at equilibrium and non-equilibrium geometries. Struct. Chem. 2005, 16, 223–241. [Google Scholar] [CrossRef]
- Nanayakkara, S.; Kraka, E. A new way of studying chemical reactions: A hand-in-hand URVA and QTAIM approach. Phys. Chem. Chem. Phys. 2019, 21, 15007–15018. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian~16 Revision A.03; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Pastorczak, E.; Corminboeuf, C. Perspective: Found in translation: Quantum chemical tools for grasping non-covalent interactions. J. Chem. Phys. 2017, 146, 120901. [Google Scholar] [CrossRef]
- Foster, J.P.; Weinhold, F. Natural hybrid orbitals. J. Am. Chem. Soc. 1980, 102, 7211–7218. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Quantitative analysis of molecular surface based on improved Marching Tetrahedra algorithm. J. Molec. Graph. Model. 2012, 38, 314–323. [Google Scholar] [CrossRef]
- Glendening, E.D.; Reed, A.E.; Carpenter, J.E.; Weinhold, F. NBO, version 3.1; as contained within the Gaussian 16 program, Gaussian Inc.: Pittsburgh, PA, USA, 2016. [Google Scholar]
- Adamo, C.; Barone, V. Toward reliable density functional methods without adjustable parameters: The PBE0 model. J. Chem. Phys. 1999, 110, 6158–6170. [Google Scholar] [CrossRef]
- Van Lenthe, E.; Baerends, E.J. Optimized Slater-type basis sets for the elements 1–118. J. Comput. Chem. 2003, 24, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- Kitaura, K.; Morokuma, K. A new energy decomposition scheme for molecular interactions within the Hartree-Fock approximation. Int. J. Quantum Chem. 1976, 10, 325–340. [Google Scholar] [CrossRef]
- te Velde, G.; Bickelhaupt, F.M.; Baerends, E.J.; Guerra, C.F.; van Gisbergen, S.J.A.; Snijders, J.G.; Ziegler, T. Chemistry with ADF. J. Comput. Chem. 2001, 22, 931–967. [Google Scholar] [CrossRef]
- ADF2014, SCM, Theoretical Chemistry; Vrije Universiteit: Amsterdam, The Netherlands, 2014; Available online: http://www.scm.com (accessed on 28 December 2021).
- Koch, U.; Popelier, P.L.A. Characterization of C-H-O Hydrogen Bonds on the Basis of the Charge Density. J. Phys. Chem. 1995, 99, 9747–9754. [Google Scholar] [CrossRef]
- Grabowski, S.J.; Sokalski, W.A.; Dyguda, E.; Leszczyński, J. Quantitative Classification of Covalent and Noncovalent H-Bonds. J. Phys. Chem. B 2006, 110, 6444–6446. [Google Scholar] [CrossRef]
- Boys, S.; Bernardi, F. The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol. Phys. 1970, 19, 553–566. [Google Scholar] [CrossRef]
- Smith, D.G.A.; Burns, L.A.; Simmonett, A.C.; Parrish, R.M.; Schieber, M.C.; Galvelis, R.; Kraus, P.; Kruse, H.; Di Remigio, R.; Alenaizan, A.; et al. PSI4 1.4: Open-source software for high-throughput quantum chemistry. J. Chem. Phys. 2020, 152, 184108. [Google Scholar] [CrossRef]
- Michalczyk, M.; Zierkiewicz, W.; Wysokiński, R.; Scheiner, S. Theoretical Studies of IR and NMR Spectral Changes Induced by Sigma-Hole Hydrogen, Halogen, Chalcogen, Pnicogen, and Tetrel Bonds in a Model Protein Environment. Molecules 2019, 24, 3329. [Google Scholar] [CrossRef]


| Complex | Origin | VCP | GCP | E1 | E2 | ||
|---|---|---|---|---|---|---|---|
| AZM | DFT | 0.020 | 0.066 | −0.014 | 0.015 | 4.478 | 4.132 |
| CPMD | 0.045 | 0.152 | −0.040 | 0.039 | 12.490 | 10.486 | |
| AZM-T | DFT | 0.020 | 0.065 | −0.014 | 0.015 | 4.426 | 4.086 |
| CPMD | 0.049 | 0.163 | −0.045 | 0.043 | 14.018 | 11.490 | |
| AZM-TT | DFT | 0.021 | 0.067 | −0.015 | 0.016 | 4.568 | 4.210 |
| CPMD | 0.053 | 0.173 | −0.049 | 0.046 | 15.249 | 12.370 | |
| AZM-TTL | DFT | 0.022 | 0.071 | −0.016 | 0.017 | 4.940 | 4.520 |
| CPMD | 0.052 | 0.164 | −0.047 | 0.044 | 14.605 | 11.773 | |
| AZM-TTLH | DFT | 0.022 | 0.072 | −0.016 | 0.017 | 4.953 | 4.533 |
| CPMD | 0.054 | 0.170 | −0.049 | 0.046 | 15.445 | 12.341 |
| Complex | Electrostatics | Exchange | Induction | Dispersion | SAPT0 | SAPT2 |
|---|---|---|---|---|---|---|
| Experimental structure | ||||||
| AZM-T | −5.846 | 13.985 | −2.603 | −7.157 | −2.979 | −1.621 |
| AZM-TT | −9.853 | 19.973 | −4.291 | −11.171 | −6.599 | −5.342 |
| AZM-TTL | −18.854 | 30.558 | −8.371 | −20.057 | −19.657 | −16.723 |
| AZM-TTLH, SAPT0 | −12.516 | 29.249 | −8.447 | −22.552 | −14.266 | – |
| Optimized structure | ||||||
| AZM-T | −22.738 | 26.825 | −8.096 | −13.031 | −20.543 | −17.040 |
| AZM-TT | −40.280 | 51.954 | −15.134 | −26.006 | −36.737 | −29.465 |
| AZM-TTL | −40.443 | 51.785 | −15.710 | −30.315 | −41.111 | −34.684 |
| AZM-TTLH, SAPT0 | −68.386 | 69.536 | −26.657 | −40.273 | −65.780 | – |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wojtkowiak, K.; Michalczyk, M.; Zierkiewicz, W.; Jezierska, A.; Panek, J.J. Chalcogen Bond as a Factor Stabilizing Ligand Conformation in the Binding Pocket of Carbonic Anhydrase IX Receptor Mimic. Int. J. Mol. Sci. 2022, 23, 13701. https://doi.org/10.3390/ijms232213701
Wojtkowiak K, Michalczyk M, Zierkiewicz W, Jezierska A, Panek JJ. Chalcogen Bond as a Factor Stabilizing Ligand Conformation in the Binding Pocket of Carbonic Anhydrase IX Receptor Mimic. International Journal of Molecular Sciences. 2022; 23(22):13701. https://doi.org/10.3390/ijms232213701
Chicago/Turabian StyleWojtkowiak, Kamil, Mariusz Michalczyk, Wiktor Zierkiewicz, Aneta Jezierska, and Jarosław J. Panek. 2022. "Chalcogen Bond as a Factor Stabilizing Ligand Conformation in the Binding Pocket of Carbonic Anhydrase IX Receptor Mimic" International Journal of Molecular Sciences 23, no. 22: 13701. https://doi.org/10.3390/ijms232213701
APA StyleWojtkowiak, K., Michalczyk, M., Zierkiewicz, W., Jezierska, A., & Panek, J. J. (2022). Chalcogen Bond as a Factor Stabilizing Ligand Conformation in the Binding Pocket of Carbonic Anhydrase IX Receptor Mimic. International Journal of Molecular Sciences, 23(22), 13701. https://doi.org/10.3390/ijms232213701







