Maximizing the Production of Recombinant Proteins in Plants: From Transcription to Protein Stability
Abstract
1. Introduction
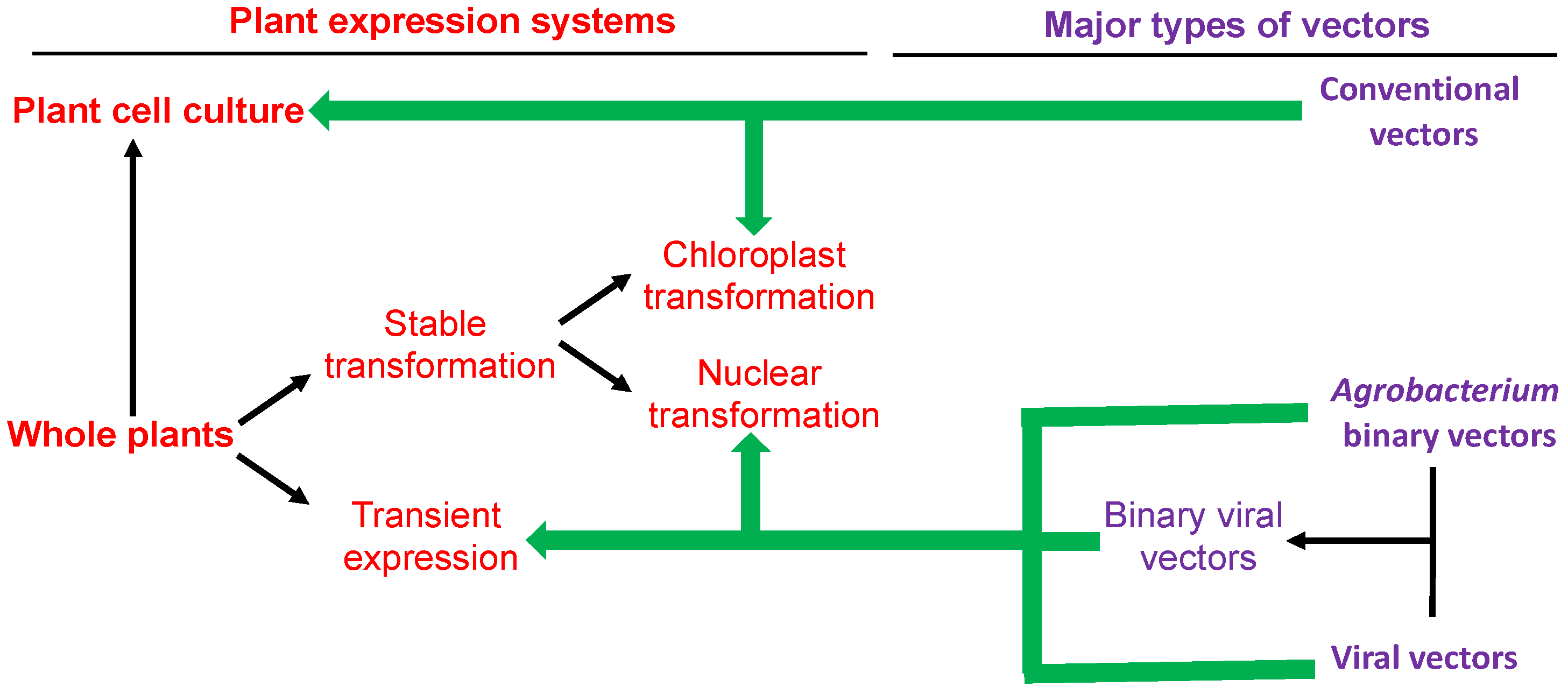
2. Major Types of Plant Systems and Vectors for Recombinant Protein Production
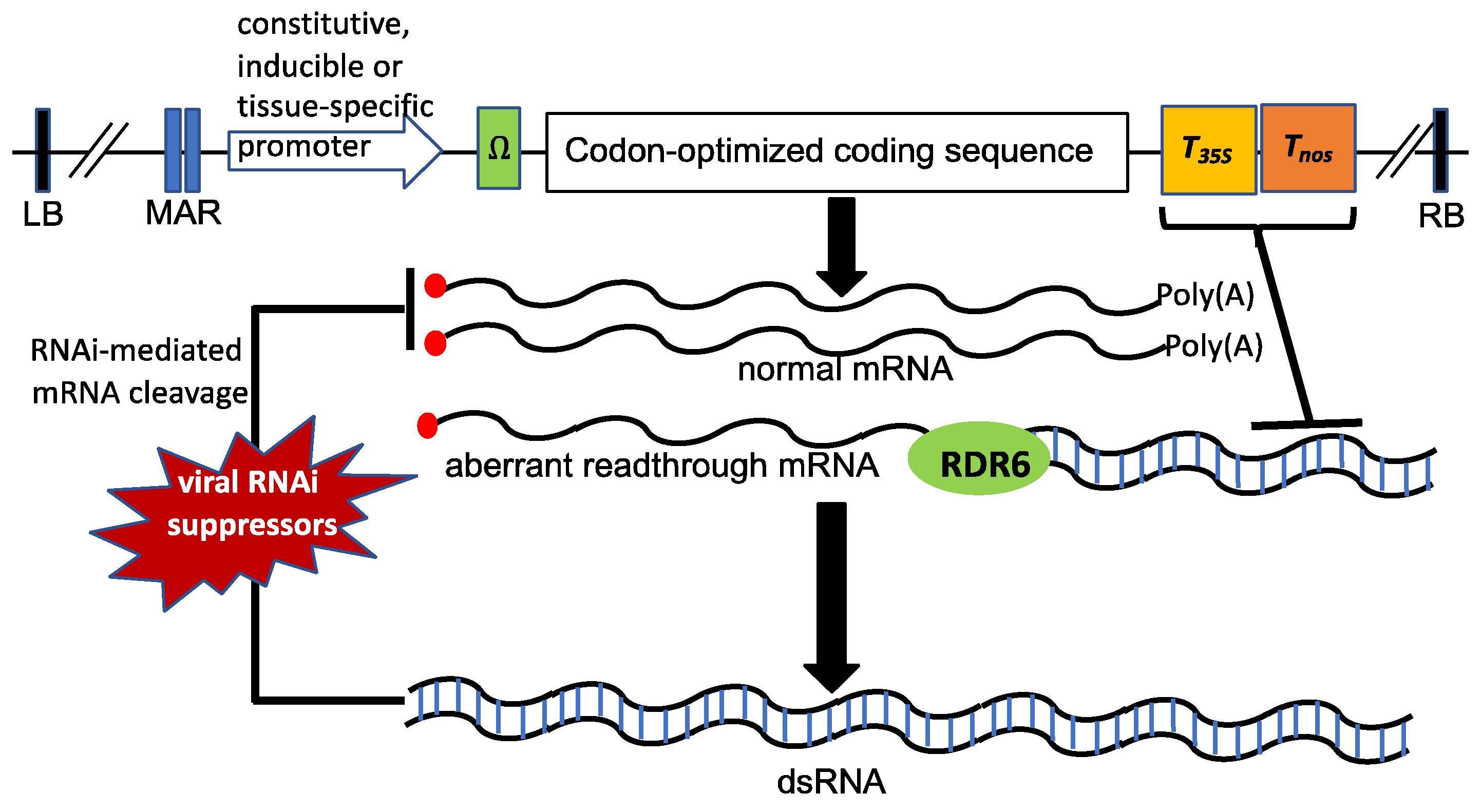
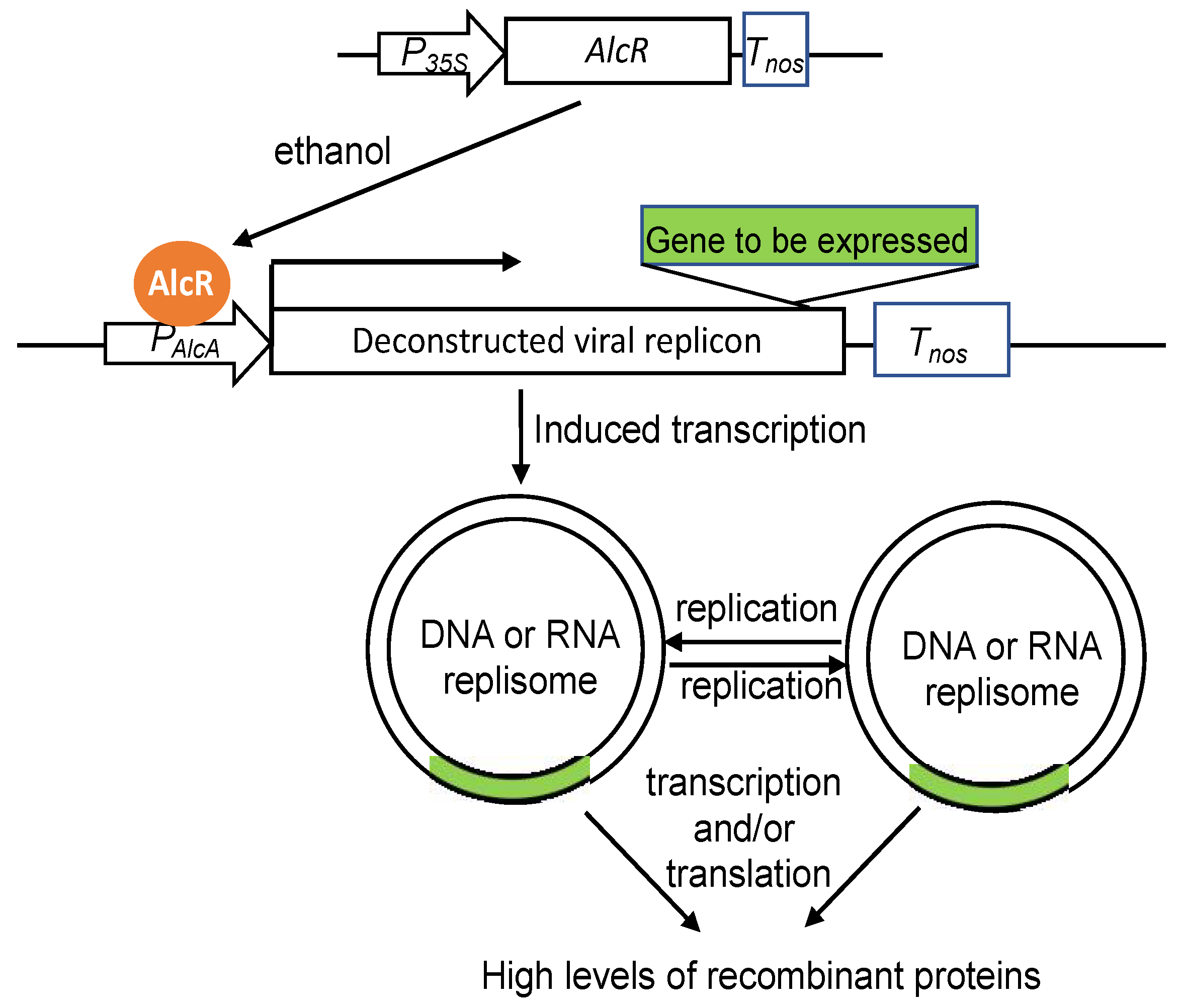
3. Elements of Genetic Constructs for Transcriptional Enhancement and Control
4. Minimizing Post-Transcriptional Gene Silencing (PTGS)
5. Enhancing the Translation of Recombinant Proteins
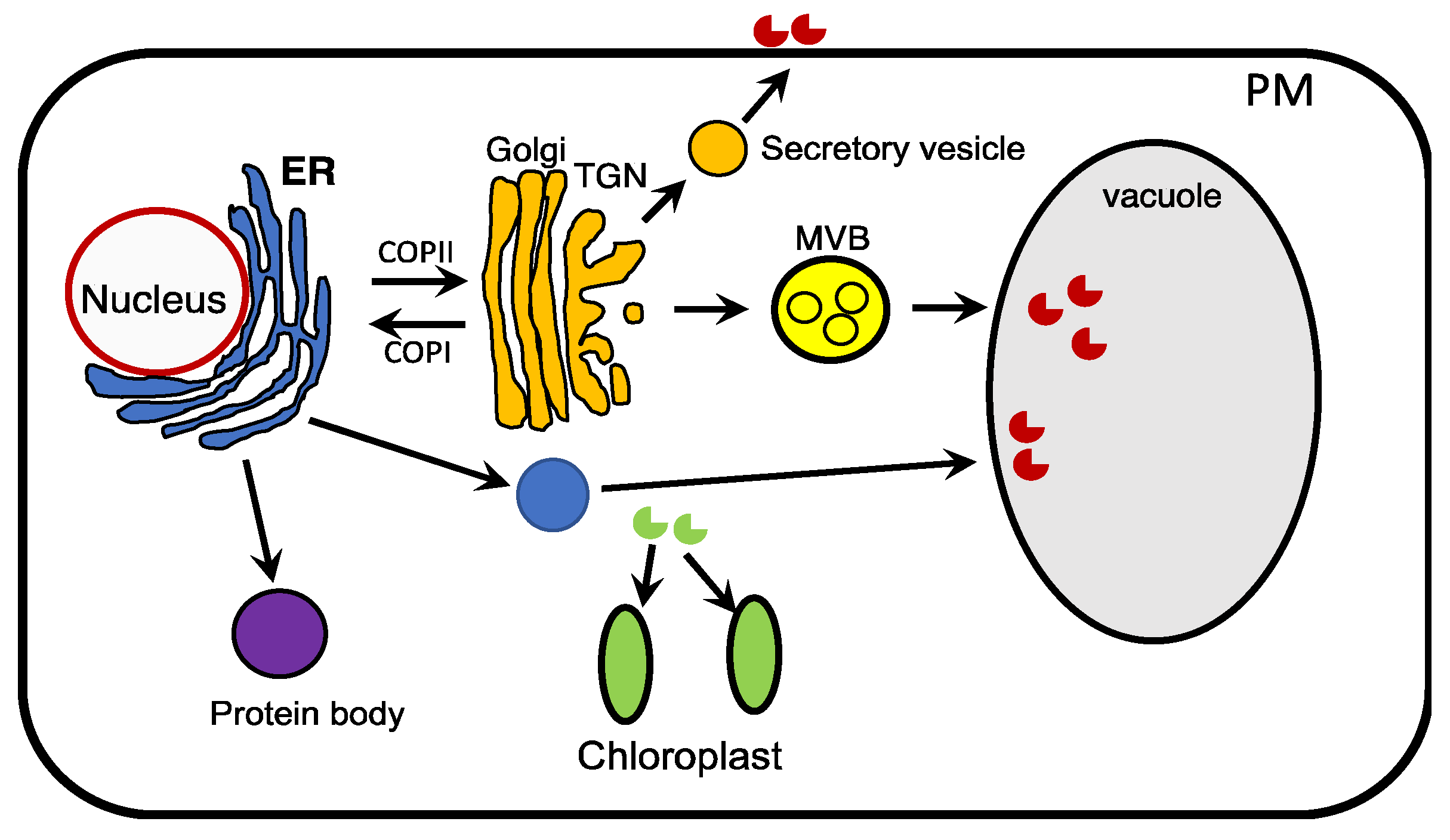
6. Subcellular Localization of Recombinant Proteins to Promote Their Accumulation
7. Summary and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Walsh, G. Biopharmaceutical benchmarks 2018. Nat. Biotechnol. 2018, 36, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Schillberg, S.; Spiegel, H. Recombinant Protein Production in Plants: A Brief Overview of Strengths and Challenges. Methods Mol. Biol. 2022, 2480, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Schillberg, S.; Raven, N.; Spiegel, H.; Rasche, S.; Buntru, M. Critical Analysis of the Commercial Potential of Plants for the Production of Recombinant Proteins. Front. Plant Sci. 2019, 10, 720. [Google Scholar] [CrossRef] [PubMed]
- De Block, M.; Herrera-Estrella, L.; Van Montagu, M.; Schell, J.; Zambryski, P. Expression of foreign genes in regenerated plants and in their progeny. EMBO J. 1984, 3, 1681–1689. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Estrella, L.; Block, M.D.; Messens, E.; Hernalsteens, J.P.; Montagu, M.V.; Schell, J. Chimeric genes as dominant selectable markers in plant cells. EMBO J. 1983, 2, 987–995. [Google Scholar] [CrossRef]
- Fischer, R.; Buyel, J.F. Molecular farming—The slope of enlightenment. Biotechnol. Adv. 2020, 40, 107519. [Google Scholar] [CrossRef]
- Kelada, K.D.; Tuse, D.; Gleba, Y.; McDonald, K.A.; Nandi, S. Process Simulation and Techno-Economic Analysis of Large-Scale Bioproduction of Sweet Protein Thaumatin II. Foods 2021, 10, 838. [Google Scholar] [CrossRef]
- McNulty, M.J.; Kelada, K.; Paul, D.; Nandi, S.; McDonald, K.A. Techno-economic process modelling and Monte Carlo simulation data of uncertainty quantification in field-grown plant-based manufacturing. Data Brief. 2021, 38, 107317. [Google Scholar] [CrossRef]
- Nandi, S.; Kwong, A.T.; Holtz, B.R.; Erwin, R.L.; Marcel, S.; McDonald, K.A. Techno-economic analysis of a transient plant-based platform for monoclonal antibody production. MAbs 2016, 8, 1456–1466. [Google Scholar] [CrossRef]
- McNulty, M.J.; Gleba, Y.; Tuse, D.; Hahn-Lobmann, S.; Giritch, A.; Nandi, S.; McDonald, K.A. Techno-economic analysis of a plant-based platform for manufacturing antimicrobial proteins for food safety. Biotechnol. Prog. 2020, 36, e2896. [Google Scholar] [CrossRef]
- Walwyn, D.R.; Huddy, S.M.; Rybicki, E.P. Techno-economic analysis of horseradish peroxidase production using a transient expression system in Nicotiana benthamiana. Appl. Biochem. Biotechnol. 2015, 175, 841–854. [Google Scholar] [CrossRef] [PubMed]
- Sethi, L.; Kumari, K.; Dey, N. Engineering of Plants for Efficient Production of Therapeutics. Mol. Biotechnol. 2021, 63, 1125–1137. [Google Scholar] [CrossRef]
- Burnett, M.J.B.; Burnett, A.C. Therapeutic recombinant protein production in plants: Challenges and opportunities. Plants People Planet 2020, 2, 121–132. [Google Scholar] [CrossRef]
- Kay, R.; Chan, A.; Daly, M.; Mcpherson, J. Duplication of Camv-35s Promoter Sequences Creates a Strong Enhancer for Plant Genes. Science 1987, 236, 1299–1302. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.G.; Liu, Q.; Smith, N.A.; Liang, G.L.; Wang, M.B. RNA Silencing in Plants: Mechanisms, Technologies and Applications in Horticultural Crops. Curr. Genom. 2016, 17, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Gomollon, S.; Baulcombe, D.C. Roles of RNA silencing in viral and non-viral plant immunity and in the crosstalk between disease resistance systems. Nat. Rev. Mol. Cell Biol. 2022, 23, 645–662. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.L.; Li, J.Y.; Wang, M.J.; Song, Z.T.; Liu, J.X. Protein Quality Control in Plant Organelles: Current Progress and Future Perspectives. Mol. Plant 2021, 14, 95–114. [Google Scholar] [CrossRef]
- Reuter, L.J.; Bailey, M.J.; Joensuu, J.J.; Ritala, A. Scale-up of hydrophobin-assisted recombinant protein production in tobacco BY-2 suspension cells. Plant Biotechnol. J. 2014, 12, 402–410. [Google Scholar] [CrossRef]
- Sadoch, J.; Pyc, M.; Urbanowicz, A.; Iglewski, A.; Pilarski, R. High-throughput evolutionary optimization of the induction medium towards recombinant protein production in BY-2 tobacco. Biotechnol. Bioeng. 2021, 118, 676–689. [Google Scholar] [CrossRef]
- Svobodnikova, L.; Kummerova, M.; Zezulka, S.; Babula, P. Possible use of a Nicotiana tabacum ‘Bright Yellow 2’ cell suspension as a model to assess phytotoxicity of pharmaceuticals (diclofenac). Ecotoxicol. Environ. Saf. 2019, 182, 109369. [Google Scholar] [CrossRef]
- Fox, J.L. First plant-made biologic approved. Nat. Biotechnol. 2012, 30, 472–473. [Google Scholar] [CrossRef]
- Hellwig, S.; Drossard, J.; Twyman, R.M.; Fischer, R. Plant cell cultures for the production of recombinant proteins. Nat. Biotechnol. 2004, 22, 1415–1422. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.K.; McDonald, K.A. Molecular Farming Using Bioreactor-Based Plant Cell Suspension Cultures for Recombinant Protein Production. In Molecular Farming in Plants: Recent Advances and Future Prospects; Springer: Berlin, Germany, 2012; pp. 37–67. [Google Scholar] [CrossRef]
- Santos, R.B.; Schiermeyer, A.; Abranches, R. Optimization of plant cell suspension cultures for the production of recombinant proteins. FEBS J. 2016, 283, 73–74. [Google Scholar]
- Khan, M.S.; Joyia, F.A.; Mustafa, G. Seeds as Economical Production Platform for Recombinant Proteins. Protein Pept. Lett. 2020, 27, 89–104. [Google Scholar] [CrossRef]
- Kurup, V.M.; Thomas, J. Edible Vaccines: Promises and Challenges. Mol. Biotechnol. 2020, 62, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Takaiwa, F.; Wakasa, Y.; Ozawa, K.; Sekikawa, K. Improvement of production yield and extraction efficacy of recombinant protein by high endosperm-specific expression along with simultaneous suppression of major seed storage proteins. Plant Sci. 2021, 302, 110692. [Google Scholar] [CrossRef] [PubMed]
- Cardi, T.; Lenzi, P.; Maliga, P. Chloroplasts as expression platforms for plant-produced vaccines. Expert Rev. Vaccines 2010, 9, 893–911. [Google Scholar] [CrossRef]
- Maliga, P. Engineering of Transgenes for High-level Protein Expression in Chloroplasts. In Vitro Cell. Dev. Biol. Anim. 2009, 45, S31. [Google Scholar]
- Maliga, P.; Kuroda, H.; Corneille, S.; Lutz, K.; Azhagiri, A.K.; Svab, Z.; Tregoning, J.; Nixon, P.; Dougan, G. Tobacco chloroplasts as a platform for vaccine production. In Plant Biotechnology 2002 and Beyond; Springer: Berlin, Germany, 2003; pp. 397–400. [Google Scholar]
- Mardanova, E.S.; Ravin, N.V. Transient expression of recombinant proteins in plants using potato virus X based vectors. Methods Enzymol. 2021, 660, 205–222. [Google Scholar] [CrossRef]
- Nosaki, S.; Miura, K. Transient expression of recombinant proteins in plants. Methods Enzymol. 2021, 660, 193–203. [Google Scholar] [CrossRef]
- Spiegel, H.; Schillberg, S.; Nolke, G. Production of Recombinant Proteins by Agrobacterium-Mediated Transient Expression. Methods Mol. Biol. 2022, 2480, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Kapila, J.; DeRycke, R.; VanMontagu, M.; Angenon, G. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 1997, 122, 101–108. [Google Scholar] [CrossRef]
- Janssen, B.J.; Gardner, R.C. Localized transient expression of GUS in leaf discs following cocultivation with Agrobacterium. Plant Mol. Biol. 1990, 14, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Vezina, L.P.; Faye, L.; Lerouge, P.; D’Aoust, M.A.; Marquet-Blouin, E.; Burel, C.; Lavoie, P.O.; Bardor, M.; Gomord, V. Transient co-expression for fast and high-yield production of antibodies with human-like N-glycans in plants. Plant Biotechnol. J. 2009, 7, 442–455. [Google Scholar] [CrossRef] [PubMed]
- Shih, S.M.H.; Doran, P.M. Foreign protein production using plant cell and organ cultures: Advantages and limitations. Biotechnol. Adv. 2009, 27, 1036–1042. [Google Scholar] [CrossRef]
- Naseri, Z.; Khezri, G.; Davarpanah, S.J.; Ofoghi, H. Virus-based vectors: A new approach for the production of recombinant proteins. J. Appl. Biotechnol. Rep. 2019, 6, 6–14. [Google Scholar] [CrossRef]
- Gleba, Y.; Klimyuk, V.; Marillonnet, S. Magnifection—A new platform for expressing recombinant vaccines in plants. Vaccine 2005, 23, 2042–2048. [Google Scholar] [CrossRef]
- Gleba, Y.; Klimyuk, V.; Marillonnet, S. Viral vectors for the expression of proteins in plants. Curr. Opin. Biotechnol. 2007, 18, 134–141. [Google Scholar] [CrossRef]
- Gleba, Y.; Marillonnet, S.; Klimyuk, V. Engineering viral expression vectors for plants: The ‘full virus’ and the ‘deconstructed virus’ strategies. Curr. Opin. Plant Biol. 2004, 7, 182–188. [Google Scholar] [CrossRef]
- Marillonnet, S.; Giritch, A.; Gils, M.; Kandzia, R.; Klimyuk, V.; Gleba, Y. In planta engineering of viral RNA replicons: Efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc. Natl. Acad. Sci. USA 2004, 101, 6852–6857. [Google Scholar] [CrossRef]
- Marillonnet, S.; Thoeringer, C.; Kandzia, R.; Klimyuk, V.; Gleba, Y. Systemic Agrobacterium tumefaciens-mediated transfection of viral replicons for efficient transient expression in plants. Nat. Biotechnol. 2005, 23, 718–723. [Google Scholar] [CrossRef] [PubMed]
- Benfey, P.N.; Chua, N.H. The Cauliflower Mosaic Virus 35S Promoter: Combinatorial Regulation of Transcription in Plants. Science 1990, 250, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Odell, J.T.; Nagy, F.; Chua, N.H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 1985, 313, 810–812. [Google Scholar] [CrossRef] [PubMed]
- Egelkrout, E.; Rajan, V.; Howard, J.A. Overproduction of recombinant proteins in plants. Plant Sci. 2012, 184, 83–101. [Google Scholar] [CrossRef]
- Makhzoum, A.; Benyammi, R.; Moustafa, K.; Tremouillaux-Guiller, J. Recent advances on host plants and expression cassettes’ structure and function in plant molecular pharming. BioDrugs 2014, 28, 145–159. [Google Scholar] [CrossRef]
- Smirnova, O.G.; Ibragimova, S.S.; Kochetov, A.V. Simple database to select promoters for plant transgenesis. Transgenic Res. 2012, 21, 429–437. [Google Scholar] [CrossRef]
- Christensen, A.H.; Quail, P.H. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996, 5, 213–218. [Google Scholar] [CrossRef]
- Grant, T.N.; De La Torre, C.M.; Zhang, N.; Finer, J.J. Synthetic introns help identify sequences in the 5′ UTR intron of the Glycine max polyubiquitin (Gmubi) promoter that give increased promoter activity. Planta 2017, 245, 849–860. [Google Scholar] [CrossRef]
- Hily, J.M.; Singer, S.D.; Yang, Y.; Liu, Z. A transformation booster sequence (TBS) from Petunia hybrida functions as an enhancer-blocking insulator in Arabidopsis thaliana. Plant Cell Rep. 2009, 28, 1095–1104. [Google Scholar] [CrossRef]
- Noad, R.J.; Turner, D.S.; Covey, S.N. Expression of functional elements inserted into the 35S promoter region of infectious cauliflower mosaic virus replicons. Nucleic Acids Res. 1997, 25, 1123–1129. [Google Scholar] [CrossRef][Green Version]
- Singer, S.D.; Cox, K.D.; Liu, Z. Both the constitutive cauliflower mosaic virus 35S and tissue-specific AGAMOUS enhancers activate transcription autonomously in Arabidopsis thaliana. Plant Mol. Biol. 2010, 74, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Boothe, J.; Nykiforuk, C.; Shen, Y.; Zaplachinski, S.; Szarka, S.; Kuhlman, P.; Murray, E.; Morck, D.; Moloney, M.M. Seed-based expression systems for plant molecular farming. Plant Biotechnol. J. 2010, 8, 588–606. [Google Scholar] [CrossRef] [PubMed]
- Sengupta-Gopalan, C.; Reichert, N.A.; Barker, R.F.; Hall, T.C.; Kemp, J.D. Developmentally regulated expression of the bean beta-phaseolin gene in tobacco seed. Proc. Natl. Acad. Sci. USA 1985, 82, 3320–3324. [Google Scholar] [CrossRef] [PubMed]
- Stoger, E.; Sack, M.; Perrin, Y.; Vaquero, C.; Torres, E.; Twyman, R.M.; Christou, P.; Fischer, R. Practical considerations for pharmaceutical antibody production in different crop systems. Mol. Breed. 2002, 9, 149–158. [Google Scholar] [CrossRef]
- Hood, E.E.; Bailey, M.R.; Beifuss, K.; Magallanes-Lundback, M.; Horn, M.E.; Callaway, E.; Drees, C.; Delaney, D.E.; Clough, R.; Howard, J.A. Criteria for high-level expression of a fungal laccase gene in transgenic maize. Plant Biotechnol. J. 2003, 1, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.C.; Guo, F.L.; Liu, B.; Huang, N.; Watkins, S.C. Expression and localization of human lysozyme in the endosperm of transgenic rice. Planta 2003, 216, 597–603. [Google Scholar] [CrossRef]
- Hennegan, K.; Yang, D.C.; Nguyen, D.; Wu, L.Y.; Goding, J.; Huang, J.M.; Guo, F.L.; Huang, N.; Watkins, S. Improvement of human lysozyme expression in transgenic rice grain by combining wheat (Triticum aestivum) puroindoline b and rice (Oryza sativa) Gt1 promoters and signal peptides. Transgenic Res. 2005, 14, 583–592. [Google Scholar] [CrossRef]
- Corrado, G.; Karali, M. Inducible gene expression systems and plant biotechnology. Biotechnol. Adv. 2009, 27, 733–743. [Google Scholar] [CrossRef]
- Caddick, M.X.; Greenland, A.J.; Jepson, I.; Krause, K.P.; Qu, N.; Riddell, K.V.; Salter, M.G.; Schuch, W.; Sonnewald, U.; Tomsett, A.B. An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nat. Biotechnol. 1998, 16, 177–180. [Google Scholar] [CrossRef]
- Roslan, H.A.; Salter, M.G.; Wood, C.D.; White, M.R.; Croft, K.P.; Robson, F.; Coupland, G.; Doonan, J.; Laufs, P.; Tomsett, A.B.; et al. Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 2001, 28, 225–235. [Google Scholar] [CrossRef]
- Tomsett, B.; Tregova, A.; Garoosi, A.; Caddick, M. Ethanol-inducible gene expression: First step towards a new green revolution? Trends Plant Sci. 2004, 9, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Garoosi, G.A.; Salter, M.G.; Caddick, M.X.; Tomsett, A.B. Characterization of the ethanol-inducible alc gene expression system in tomato. J. Exp. Bot. 2005, 56, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Werner, S.; Breus, O.; Symonenko, Y.; Marillonnet, S.; Gleba, Y. High-level recombinant protein expression in transgenic plants by using a double-inducible viral vector. Proc. Natl. Acad. Sci. USA 2011, 108, 14061–14066. [Google Scholar] [CrossRef]
- Dugdale, B.; Mortimer, C.L.; Kato, M.; James, T.A.; Harding, R.M.; Dale, J.L. In plant activation: An inducible, hyperexpression platform for recombinant protein production in plants. Plant Cell 2013, 25, 2429–2443. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, C.L.; Dugdale, B.; Dale, J.L. Updates in inducible transgene expression using viral vectors: From transient to stable expression. Curr. Opin. Biotechnol. 2015, 32, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Damaj, M.B.; Jifon, J.L.; Woodard, S.L.; Vargas-Bautista, C.; Barros, G.O.F.; Molina, J.; White, S.G.; Damaj, B.B.; Nikolov, Z.L.; Mandadi, K.K. Unprecedented enhancement of recombinant protein production in sugarcane culms using a combinatorial promoter stacking system. Sci. Rep. 2020, 10, 13713. [Google Scholar] [CrossRef]
- Sinaga, D.S.; Ho, S.L.; Lu, C.A.; Yu, S.M.; Huang, L.F. Knockdown expression of a MYB-related transcription factor gene, OsMYBS2, enhances production of recombinant proteins in rice suspension cells. Plant Methods 2021, 17, 99. [Google Scholar] [CrossRef]
- Kurbidaeva, A.; Purugganan, M. Insulators in Plants: Progress and Open Questions. Genes 2021, 12, 1422. [Google Scholar] [CrossRef]
- Nowak, W.; Gawlowska, M.; Jarmolowski, A.; Augustyniak, J. Effect of nuclear matrix attachment regions on transgene expression in tobacco plants. Acta Biochim. Pol. 2001, 48, 637–646. [Google Scholar] [CrossRef]
- Spiker, S.; Thompson, W.F. Nuclear Matrix Attachment Regions and Transgene Expression in Plants. Plant Physiol. 1996, 110, 15–21. [Google Scholar] [CrossRef][Green Version]
- Torney, F.; Partier, A.; Says-Lesage, V.; Nadaud, I.; Barret, P.; Beckert, M. Heritable transgene expression pattern imposed onto maize ubiquitin promoter by maize adh-1 matrix attachment regions: Tissue and developmental specificity in maize transgenic plants. Plant Cell Rep. 2004, 22, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, Y.; Ramos, P.L.; Soto, J.; Rodriguez, M.; Carlos, N.; Reyes, A.; Callard, D.; Sanchez, Y.; Pujol, M.; Fuentes, A. The M4 insulator, the TM2 matrix attachment region, and the double copy of the heavy chain gene contribute to the enhanced accumulation of the PHB-01 antibody in tobacco plants. Transgenic Res. 2020, 29, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Hoffer, P.; Ivashuta, S.; Pontes, O.; Vitins, A.; Pikaard, C.; Mroczka, A.; Wagner, N.; Voelker, T. Posttranscriptional gene silencing in nuclei. Proc. Natl. Acad. Sci. USA 2011, 108, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Pressman, S.; Bei, Y.; Carthew, R. Posttranscriptional gene silencing. Cell 2007, 130, 570. [Google Scholar] [CrossRef]
- Wesley, S.V.; Helliwell, C.; Wang, M.B.; Waterhouse, P. Posttranscriptional gene silencing in plants. Methods Mol. Biol. 2004, 265, 117–129. [Google Scholar] [CrossRef]
- Lechtenberg, B.; Schubert, D.; Forsbach, A.; Gils, M.; Schmidt, R. Neither inverted repeat T-DNA configurations nor arrangements of tandemly repeated transgenes are sufficient to trigger transgene silencing. Plant J. 2003, 34, 507–517. [Google Scholar] [CrossRef]
- Que, Q.; Wang, H.Y.; English, J.J.; Jorgensen, R.A. The Frequency and Degree of Cosuppression by Sense Chalcone Synthase Transgenes Are Dependent on Transgene Promoter Strength and Are Reduced by Premature Nonsense Codons in the Transgene Coding Sequence. Plant Cell 1997, 9, 1357–1368. [Google Scholar] [CrossRef]
- Schubert, D.; Lechtenberg, B.; Forsbach, A.; Gils, M.; Bahadur, S.; Schmidt, R. Silencing in Arabidopsis T-DNA transformants: The predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell 2004, 16, 2561–2572. [Google Scholar] [CrossRef]
- Vaucheret, H.; Beclin, C.; Elmayan, T.; Feuerbach, F.; Godon, C.; Morel, J.B.; Mourrain, P.; Palauqui, J.C.; Vernhettes, S. Transgene-induced gene silencing in plants. Plant J. 1998, 16, 651–659. [Google Scholar] [CrossRef]
- Beclin, C.; Boutet, S.; Waterhouse, P.; Vaucheret, H. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 2002, 12, 684–688. [Google Scholar] [CrossRef]
- Luo, Z.; Chen, Z. Improperly Terminated, Unpolyadenylated mRNA of Sense Transgenes Is Targeted by RDR6-Mediated RNA Silencing in Arabidopsis. Plant Cell 2007, 19, 943–958. [Google Scholar] [CrossRef] [PubMed]
- Baeg, K.; Iwakawa, H.; Tomari, Y. The poly(A) tail blocks RDR6 from converting self mRNAs into substrates for gene silencing. Nat. Plants 2017, 3, 17036. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.B.; Last, R.L. Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J. 1997, 11, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Beyene, G.; Buenrostro-Nava, M.T.; Damaj, M.B.; Gao, S.J.; Molina, J.; Mirkov, T.E. Unprecedented enhancement of transient gene expression from minimal cassettes using a double terminator. Plant Cell Rep. 2011, 30, 13–25. [Google Scholar] [CrossRef]
- de Felippes, F.F.; McHale, M.; Doran, R.L.; Roden, S.; Eamens, A.L.; Finnegan, E.J.; Waterhouse, P.M. The key role of terminators on the expression and post-transcriptional gene silencing of transgenes. Plant J. 2020, 104, 96–112. [Google Scholar] [CrossRef]
- de Felippes, F.F.; Shand, K.; Waterhouse, P.M. Identification of a Transferrable Terminator Element That Inhibits Small RNA Production and Improves Transgene Expression Levels. Front. Plant Sci. 2022, 13, 1602. [Google Scholar] [CrossRef] [PubMed]
- Schunmann, P.H.D.; Llewellyn, D.J.; Surin, B.; Boevink, P.; De Feyter, R.C.; Waterhouse, P.M. A suite of novel promoters and terminators for plant biotechnology. Funct. Plant Biol. 2003, 30, 443–452. [Google Scholar] [CrossRef]
- Suzaki, T.; Tsudat, M.; Ezura, H.; Day, B.; Miura, K. Agroinfiltration-based efficient transient protein expression in leguminous plants. Plant Biotechnol. 2019, 36, 119–123. [Google Scholar] [CrossRef]
- Wang, P.H.; Kumar, S.; Zeng, J.; McEwan, R.; Wright, T.R.; Gupta, M. Transcription Terminator-Mediated Enhancement in Transgene Expression in Maize: Preponderance of the AUGAAU Motif Overlapping with Poly(A) Signals. Front. Plant Sci. 2020, 11, 570778. [Google Scholar] [CrossRef]
- Yamamoto, T.; Hoshikawa, K.; Ezura, K.; Okazawa, R.; Fujita, S.; Takaoka, M.; Mason, H.S.; Ezura, H.; Miura, K. Improvement of the transient expression system for production of recombinant proteins in plants. Sci. Rep. 2018, 8, 4755. [Google Scholar] [CrossRef]
- Diamos, A.G.; Mason, H.S. Chimeric 3′ flanking regions strongly enhance gene expression in plants. Plant Biotechnol. J. 2018, 16, 1971–1982. [Google Scholar] [CrossRef] [PubMed]
- Diamos, A.G.; Hunter, J.G.L.; Pardhe, M.D.; Rosenthal, S.H.; Sun, H.Y.; Foster, B.C.; DiPalma, M.P.; Chen, Q.; Mason, H.S. High Level Production of Monoclonal Antibodies Using an Optimized Plant Expression System. Front. Bioeng. Biotechnol. 2020, 7, 472. [Google Scholar] [CrossRef] [PubMed]
- Arzola, L.; Chen, J.X.; Rattanaporn, K.; Maclean, J.M.; McDonald, K.A. Transient Co-Expression of Post-Transcriptional Gene Silencing Suppressors for Increased in Planta Expression of a Recombinant Anthrax Receptor Fusion Protein. Int. J. Mol. Sci. 2011, 12, 4975–4990. [Google Scholar] [CrossRef]
- Huang, T.K.; Falk, B.W.; Dandekar, A.M.; McDonald, K.A. Enhancement of Recombinant Protein Production in Transgenic Nicotiana benthamiana Plant Cell Suspension Cultures with Co-Cultivation of Agrobacterium Containing Silencing Suppressors. Int. J. Mol. Sci. 2018, 19, 1561. [Google Scholar] [CrossRef] [PubMed]
- Garabagi, F.; Gilbert, E.; Loos, A.; McLean, M.D.; Hall, J.C. Utility of the P19 suppressor of gene-silencing protein for production of therapeutic antibodies in Nicotiana expression hosts. Plant Biotechnol. J. 2012, 10, 1118–1128. [Google Scholar] [CrossRef] [PubMed]
- Habibi, P.; Soccol, C.R.; O’Keefe, B.R.; Krumpe, L.R.H.; Wilson, J.; de Macedo, L.L.P.; Faheem, M.; Dos Santos, V.O.; Prado, G.S.; Botelho, M.A.; et al. Gene-silencing suppressors for high-level production of the HIV-1 entry inhibitor griffithsin in Nicotiana benthamiana. Process Biochem. 2018, 70, 45–54. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014, 83, 779–812. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. Structural Insights into the Mechanism of Scanning and Start Codon Recognition in Eukaryotic Translation Initiation. Trends Biochem. Sci. 2017, 42, 589–611. [Google Scholar] [CrossRef]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, G.; Jeon, E.; Sohn, E.J.; Lee, Y.; Kang, H.; Lee, D.W.; Kim, D.H.; Hwang, I. The immediate upstream region of the 5′-UTR from the AUG start codon has a pronounced effect on the translational efficiency in Arabidopsis thaliana. Nucleic Acids Res. 2014, 42, 485–498. [Google Scholar] [CrossRef]
- Fan, Q.; Treder, K.; Miller, W.A. Untranslated regions of diverse plant viral RNAs vary greatly in translation enhancement efficiency. BMC Biotechnol. 2012, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Asrani, K.H.; Farelli, J.D.; Stahley, M.R.; Miller, R.L.; Cheng, C.J.; Subramanian, R.R.; Brown, J.M. Optimization of mRNA untranslated regions for improved expression of therapeutic mRNA. RNA Biol. 2018, 15, 756–762. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R. The 5′-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Res. 2002, 30, 3401–3411. [Google Scholar] [CrossRef]
- Gallie, D.R.; Walbot, V. Identification of the motifs within the tobacco mosaic virus 5′-leader responsible for enhancing translation. Nucleic Acids Res. 1992, 20, 4631–4638. [Google Scholar] [CrossRef] [PubMed]
- Mauro, V.P.; Chappell, S.A. A critical analysis of codon optimization in human therapeutics. Trends Mol. Med. 2014, 20, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Mauro, V.P.; Chappell, S.A. Considerations in the Use of Codon Optimization for Recombinant Protein Expression. Methods Mol. Biol. 2018, 1850, 275–288. [Google Scholar] [CrossRef]
- Mauro, V.P. Codon Optimization in the Production of Recombinant Biotherapeutics: Potential Risks and Considerations. BioDrugs 2018, 32, 69–81. [Google Scholar] [CrossRef]
- Agarwal, P.; Gautam, T.; Singh, A.K.; Burma, P.K. Evaluating the effect of codon optimization on expression of bar gene in transgenic tobacco plants. J. Plant Biochem. Biot. 2019, 28, 189–202. [Google Scholar] [CrossRef]
- Kwon, K.C.; Chan, H.T.; Leon, I.R.; Williams-Carrier, R.; Barkan, A.; Daniell, H. Codon Optimization to Enhance Expression Yields Insights into Chloroplast Translation. Plant Physiol. 2016, 172, 62–77. [Google Scholar] [CrossRef]
- Benchabane, M.; Goulet, C.; Rivard, D.; Faye, L.; Gomord, V.; Michaud, D. Preventing unintended proteolysis in plant protein biofactories. Plant Biotechnol. J. 2008, 6, 633–648. [Google Scholar] [CrossRef]
- Gils, M.; Kandzia, R.; Marillonnet, S.; Klimyuk, V.; Gleba, Y. High-yield production of authentic human growth hormone using a plant virus-based expression system. Plant Biotechnol. J. 2005, 3, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Wang, Y.; Zhang, Y.; Pan, T.; Duan, E.; Bao, X.; Zhu, J.; Teng, X.; Zhang, P.; Gu, C.; et al. Endomembrane-mediated storage protein trafficking in plants: Golgi-dependent or Golgi-independent? FEBS Lett. 2022, 596, 2215–2230. [Google Scholar] [CrossRef] [PubMed]
- Tsao, H.E.; Lui, S.N.; Lo, A.H.; Chen, S.; Wong, H.Y.; Wong, C.K.; Jiang, L.; Wong, K.B. Structural insights into how vacuolar sorting receptors recognize the sorting determinants of seed storage proteins. Proc. Natl. Acad. Sci. USA 2022, 119, e2111281119. [Google Scholar] [CrossRef] [PubMed]
- Marin Viegas, V.S.; Ocampo, C.G.; Petruccelli, S. Vacuolar deposition of recombinant proteins in plant vegetative organs as a strategy to increase yields. Bioengineered 2017, 8, 203–211. [Google Scholar] [CrossRef]
- Li, X.; Li, X.; Fan, B.; Zhu, C.; Chen, Z. Specialized endoplasmic reticulum-derived vesicles in plants: Functional diversity, evolution, and biotechnological exploitation. J. Integr. Plant Biol. 2022, 64, 821–835. [Google Scholar] [CrossRef]
- Kopertekh, L.; Schiemann, J. Transient Production of Recombinant Pharmaceutical Proteins in Plants: Evolution and Perspectives. Curr. Med. Chem. 2019, 26, 365–380. [Google Scholar] [CrossRef]
- de Virgilio, M.; De Marchis, F.; Bellucci, M.; Mainieri, D.; Rossi, M.; Benvenuto, E.; Arcioni, S.; Vitale, A. The human immunodeficiency virus antigen Nef forms protein bodies in leaves of transgenic tobacco when fused to zeolin. J. Exp. Bot. 2008, 59, 2815–2829. [Google Scholar] [CrossRef]
- Kogan, M.J.; Dalcol, I.; Gorostiza, P.; Lopez-Iglesias, C.; Pons, M.; Sanz, F.; Ludevid, D.; Giralt, E. Self-assembly of the amphipathic helix (VHLPPP)8. A mechanism for zein protein body formation. J. Mol. Biol. 2001, 312, 907–913. [Google Scholar] [CrossRef]
- Llop-Tous, I.; Madurga, S.; Giralt, E.; Marzabal, P.; Torrent, M.; Ludevid, M.D. Relevant elements of a maize gamma-zein domain involved in protein body biogenesis. J. Biol. Chem. 2010, 285, 35633–35644. [Google Scholar] [CrossRef]
- Mainieri, D.; Rossi, M.; Archinti, M.; Bellucci, M.; De Marchis, F.; Vavassori, S.; Pompa, A.; Arcioni, S.; Vitale, A. Zeolin. A new recombinant storage protein constructed using maize gamma-zein and bean phaseolin. Plant Physiol. 2004, 136, 3447–3456. [Google Scholar] [CrossRef]
- Llop-Tous, I.; Ortiz, M.; Torrent, M.; Ludevid, M.D. The expression of a xylanase targeted to ER-protein bodies provides a simple strategy to produce active insoluble enzyme polymers in tobacco plants. PLoS ONE 2011, 6, e19474. [Google Scholar] [CrossRef] [PubMed]
- Urry, D.W. Entropic elastic processes in protein mechanisms. II. Simple (passive) and coupled (active) development of elastic forces. J. Protein Chem. 1988, 7, 81–114. [Google Scholar] [CrossRef] [PubMed]
- Urry, D.W. Entropic elastic processes in protein mechanisms. I. Elastic structure due to an inverse temperature transition and elasticity due to internal chain dynamics. J. Protein Chem. 1988, 7, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.; Dzuricky, M.; Chilkoti, A. Elastin-like polypeptides as models of intrinsically disordered proteins. FEBS Lett. 2015, 589, 2477–2486. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Zhu, H.; Menassa, R.; Gyenis, L.; Richman, A.; Brandle, J. Elastin-like polypeptide fusions enhance the accumulation of recombinant proteins in tobacco leaves. Transgenic Res. 2007, 16, 239–249. [Google Scholar] [CrossRef]
- Kaldis, A.; Ahmad, A.; Reid, A.; McGarvey, B.; Brandle, J.; Ma, S.; Jevnikar, A.; Kohalmi, S.E.; Menassa, R. High-level production of human interleukin-10 fusions in tobacco cell suspension cultures. Plant Biotechnol. J. 2013, 11, 535–545. [Google Scholar] [CrossRef]
- Floss, D.M.; Sack, M.; Stadlmann, J.; Rademacher, T.; Scheller, J.; Stoger, E.; Fischer, R.; Conrad, U. Biochemical and functional characterization of anti-HIV antibody-ELP fusion proteins from transgenic plants. Plant Biotechnol. J. 2008, 6, 379–391. [Google Scholar] [CrossRef]
- Phan, H.T.; Pohl, J.; Floss, D.M.; Rabenstein, F.; Veits, J.; Le, B.T.; Chu, H.H.; Hause, G.; Mettenleiter, T.; Conrad, U. ELPylated haemagglutinins produced in tobacco plants induce potentially neutralizing antibodies against H5N1 viruses in mice. Plant Biotechnol. J. 2013, 11, 582–593. [Google Scholar] [CrossRef]
- Saberianfar, R.; Joensuu, J.J.; Conley, A.J.; Menassa, R. Protein body formation in leaves of Nicotiana benthamiana: A concentration-dependent mechanism influenced by the presence of fusion tags. Plant Biotechnol. J. 2015, 13, 927–937. [Google Scholar] [CrossRef]
- Linder, M.B.; Szilvay, G.R.; Nakari-Setala, T.; Penttila, M.E. Hydrophobins: The protein-amphiphiles of filamentous fungi. FEMS Microbiol. Rev. 2005, 29, 877–896. [Google Scholar] [CrossRef]
- Linder, M.B.; Qiao, M.; Laumen, F.; Selber, K.; Hyytia, T.; Nakari-Setala, T.; Penttila, M.E. Efficient purification of recombinant proteins using hydrophobins as tags in surfactant-based two-phase systems. Biochemistry 2004, 43, 11873–11882. [Google Scholar] [CrossRef] [PubMed]
- Bankar, S.B.; Bule, M.V.; Singhal, R.S.; Ananthanarayan, L. Glucose oxidase—An overview. Biotechnol. Adv. 2009, 27, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Zoschke, R.; Bock, R. Chloroplast Translation: Structural and Functional Organization, Operational Control, and Regulation. Plant Cell 2018, 30, 745–770. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Li, X.; Fan, B.; Zhu, C.; Chen, Z. Chloroplasts Protein Quality Control and Turnover: A Multitude of Mechanisms. Int. J. Mol. Sci. 2022, 23, 7760. [Google Scholar] [CrossRef]
- Lehtimaki, N.; Koskela, M.M.; Mulo, P. Posttranslational Modifications of Chloroplast Proteins: An Emerging Field. Plant Physiol. 2015, 168, 768–775. [Google Scholar] [CrossRef]
- Chen, M.H.; Huang, L.F.; Li, H.M.; Chen, Y.R.; Yu, S.M. Signal peptide-dependent targeting of a rice alpha-amylase and cargo proteins to plastids and extracellular compartments of plant cells. Plant Physiol. 2004, 135, 1367–1377. [Google Scholar] [CrossRef]
- Villarejo, A.; Buren, S.; Larsson, S.; Dejardin, A.; Monne, M.; Rudhe, C.; Karlsson, J.; Jansson, S.; Lerouge, P.; Rolland, N.; et al. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 2005, 7, 1224–1231. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Z.; Li, X.; Fan, B.; Zhu, C.; Chen, Z. Maximizing the Production of Recombinant Proteins in Plants: From Transcription to Protein Stability. Int. J. Mol. Sci. 2022, 23, 13516. https://doi.org/10.3390/ijms232113516
Feng Z, Li X, Fan B, Zhu C, Chen Z. Maximizing the Production of Recombinant Proteins in Plants: From Transcription to Protein Stability. International Journal of Molecular Sciences. 2022; 23(21):13516. https://doi.org/10.3390/ijms232113516
Chicago/Turabian StyleFeng, Ziru, Xifeng Li, Baofang Fan, Cheng Zhu, and Zhixiang Chen. 2022. "Maximizing the Production of Recombinant Proteins in Plants: From Transcription to Protein Stability" International Journal of Molecular Sciences 23, no. 21: 13516. https://doi.org/10.3390/ijms232113516
APA StyleFeng, Z., Li, X., Fan, B., Zhu, C., & Chen, Z. (2022). Maximizing the Production of Recombinant Proteins in Plants: From Transcription to Protein Stability. International Journal of Molecular Sciences, 23(21), 13516. https://doi.org/10.3390/ijms232113516






