Molecular Characterization, Expression, Evolutionary Selection, and Biological Activity Analysis of CD68 Gene from Megalobrama amblycephala
Abstract
1. Introduction
2. Results
2.1. Sequence Analysis of MaCD68
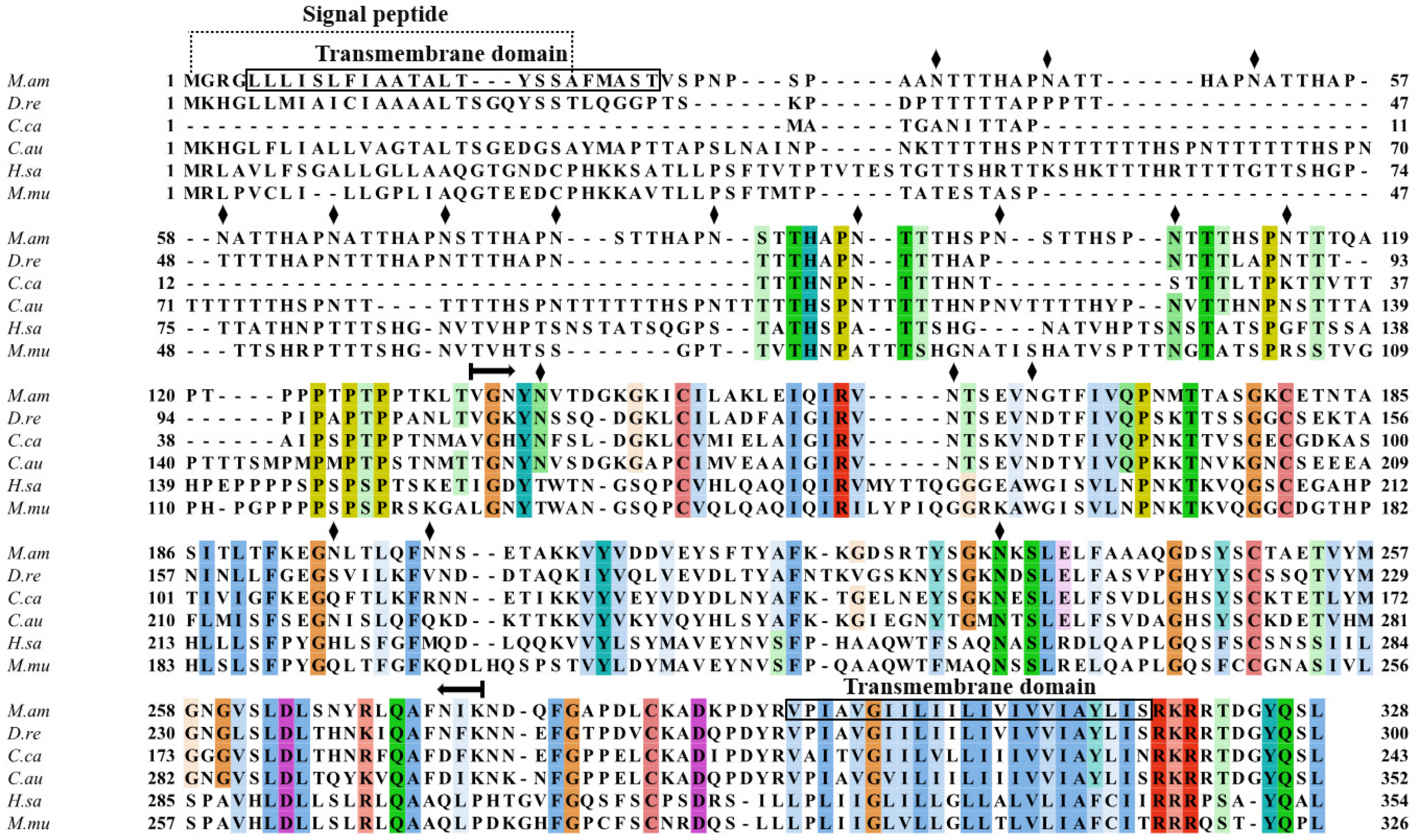
2.2. Multiple Sequence Alignment and Phylogenetic Analysis of MaCD68
2.3. Adaptive Evolutionary Analysis
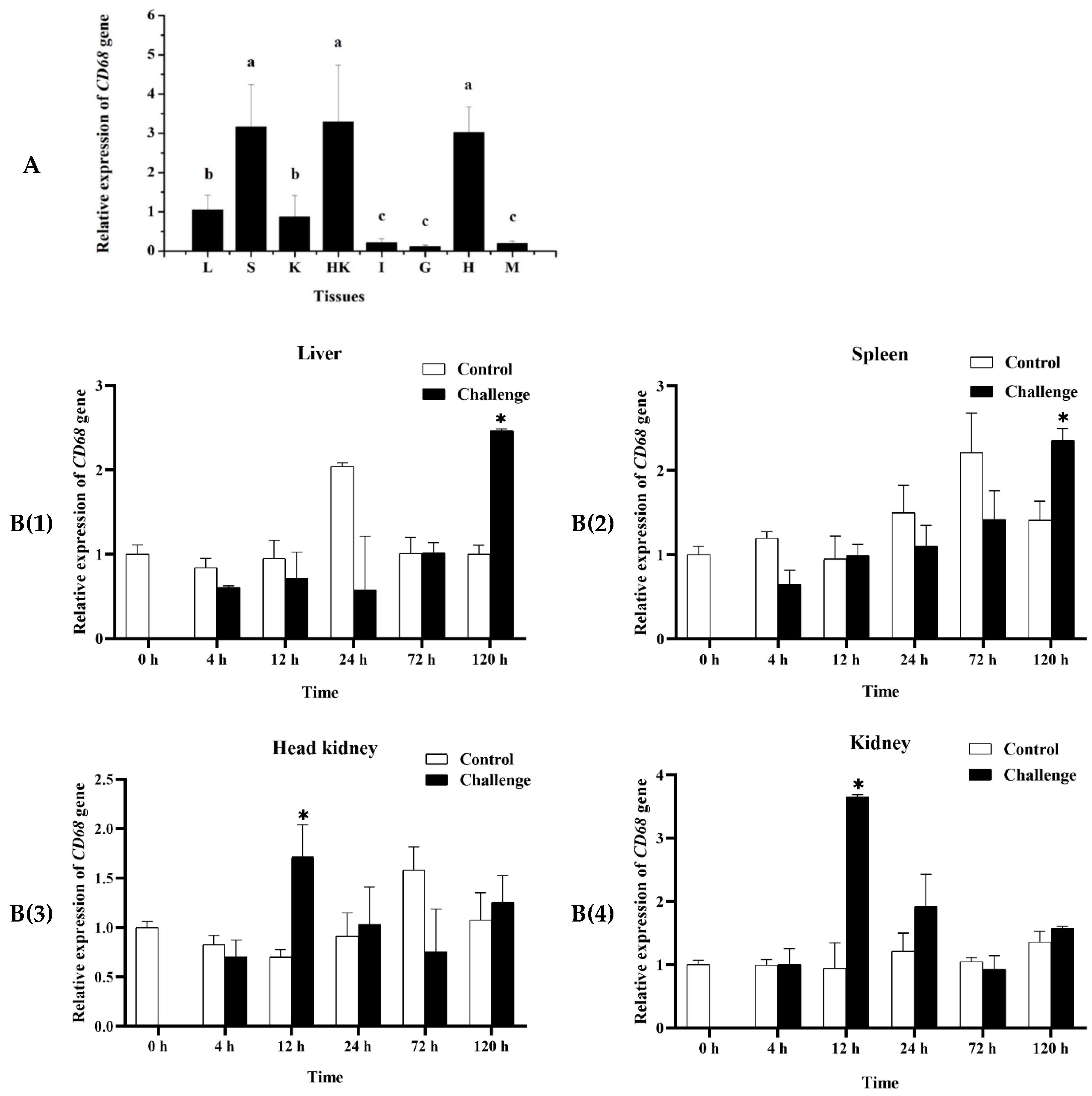
2.4. Expression of MaCD68 Gene in Healthy and Infected Tissues
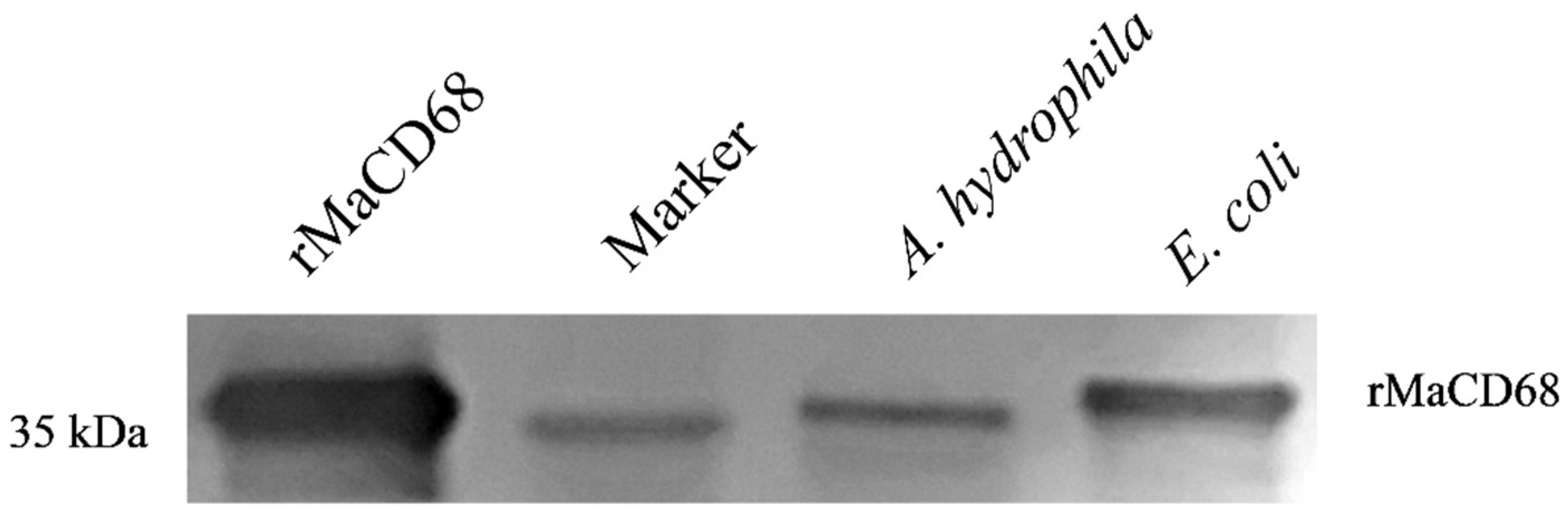
2.5. Preparation of Recombinant Protein and Polyclonal Antibody
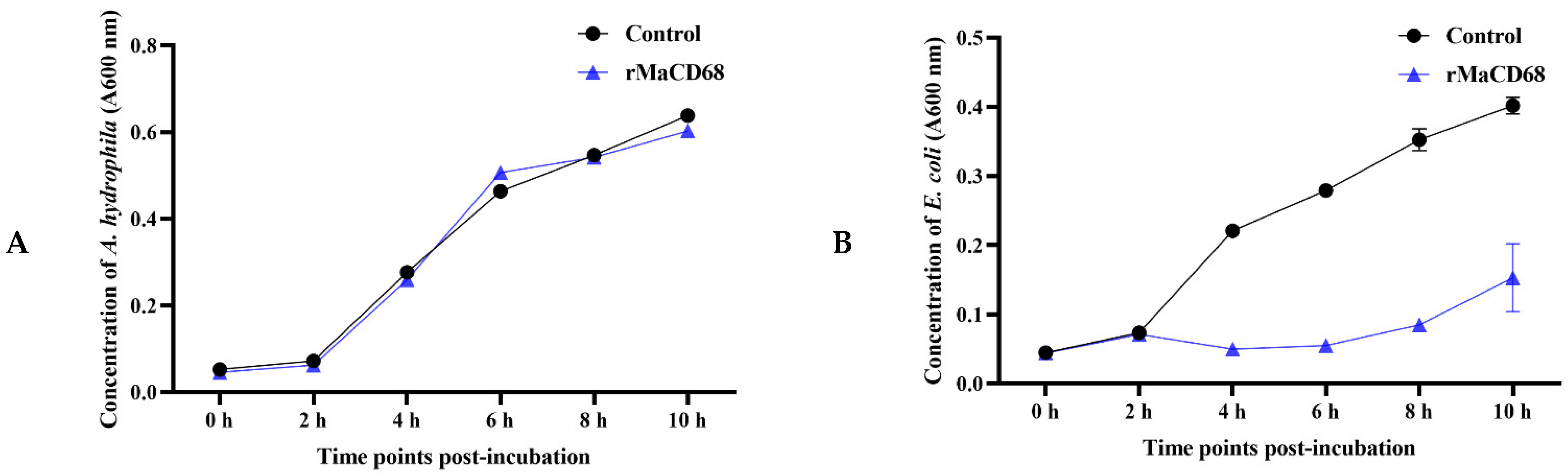
2.6. Biological Activities of rMaCD68 Protein
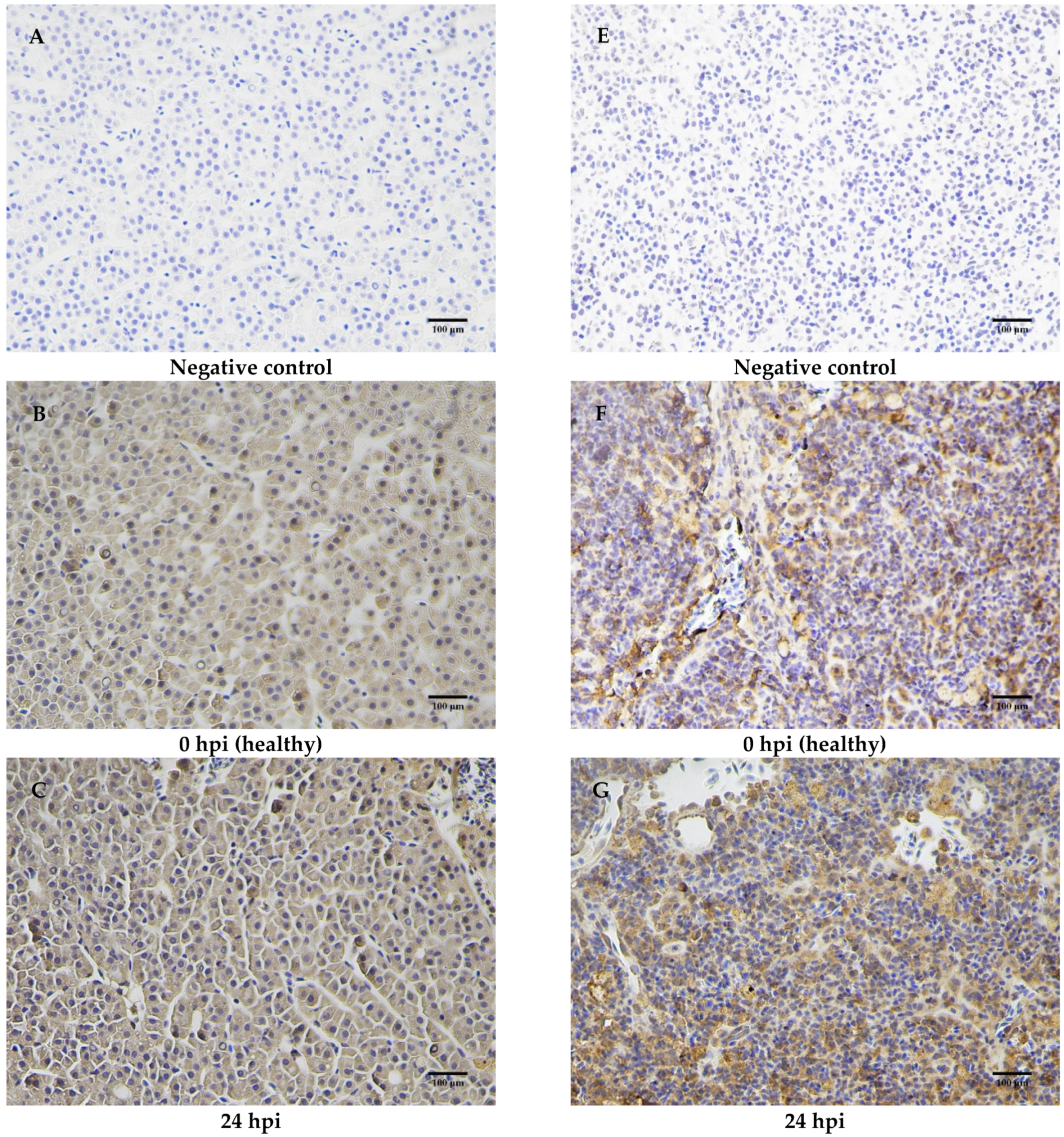
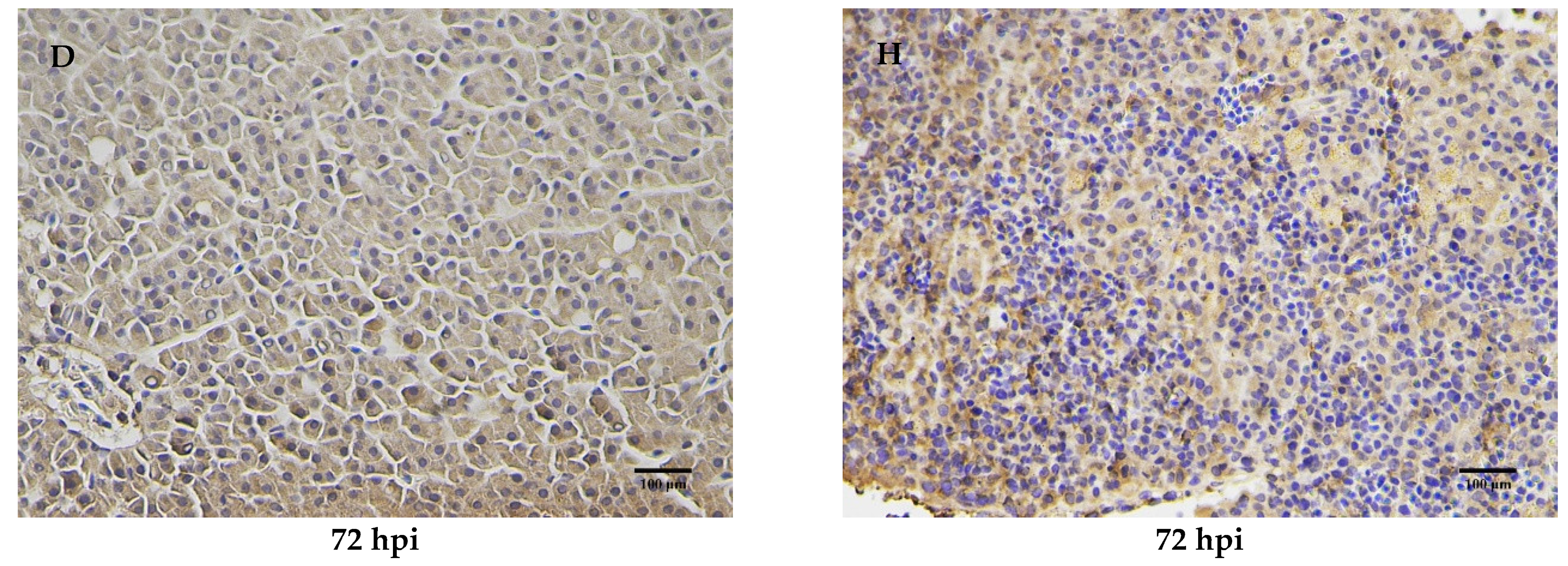
2.7. Immunohistochemical Analysis of MaCD68 Protein
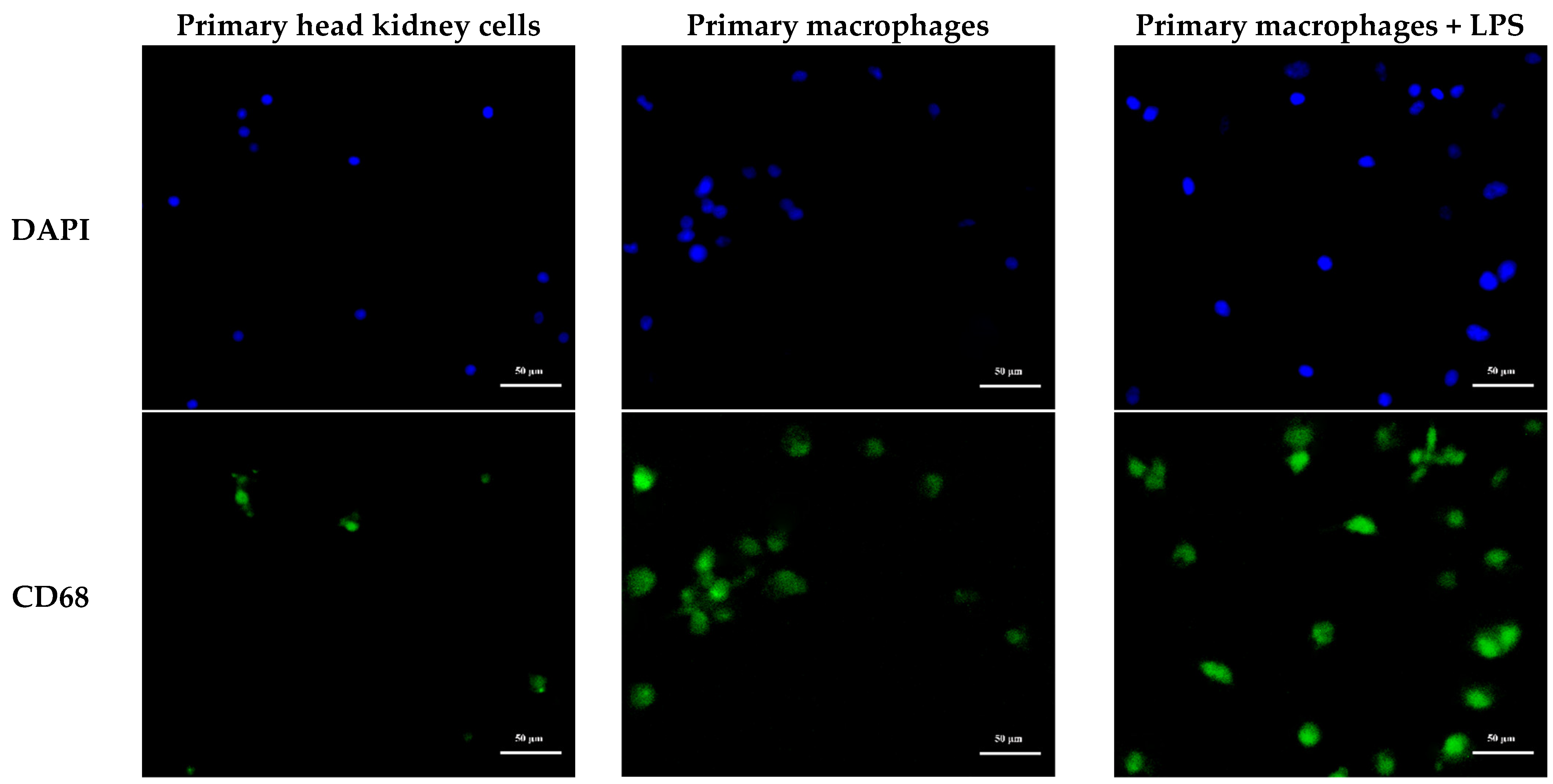
2.8. Subcellular Localization of MaCD68
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Experimental Fish and Samples Collection
4.3. cDNA Cloning of CD68
4.4. Bioinformatics Analysis
4.5. Adaptive Evolution Analysis
4.6. qRT-PCR Assay
4.7. Preparation of Recombinant Protein and Polyclonal Antibody
4.8. Western Blotting
4.9. Bacterial Agglutination Assay
4.10. Bacteriostatic Activity of rMaCD68
4.11. Bacterial Binding Assay
4.12. Immunohistochemistry Assay
4.13. Isolation of Macrophages
4.14. Immunofluorescence Assay
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Holness, C.L.; Simmons, D.L. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood 1993, 81, 1607–1613. [Google Scholar] [CrossRef] [PubMed]
- Holness, C.L.; da Silva, R.P.; Fawcett, J.; Gordon, S.; Simmons, D.L. Macrosialin, a mouse macrophage-restricted glycoprotein, is a member of the lamp/lgp family. J. Biol. Chem. 1993, 268, 9661–9666. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M. Lysosomal membrane glycoproteins. Structure, biosynthesis, and intracellular trafficking. J. Biol. Chem. 1991, 266, 21327–21330. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, J.; Yang, H.; Han, X.; Luo, X.; Liao, L.; Yang, B.; Zhu, T.; Huo, F.; Guo, W.; et al. Recruited CD68+CD206+ macrophages orchestrate graft immune tolerance to prompt xenogeneic-dentin matrix-based tooth root regeneration. Bioact. Mater. 2021, 6, 1051–1072. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, I.E.; Lewen, A.; Galow, L.V.; Cesetti, T.; Scheffel, J.; Regen, T.; Hanisch, U.K.; Kann, O. TLR4-activated microglia require IFN-γ to induce severe neuronal dysfunction and death in situ. Proc. Natl. Acad. Sci. USA 2016, 113, 212–217. [Google Scholar] [CrossRef]
- Wang, J.; Chen, H.; Chen, X.; Lin, H. Expression of Tumor-Related Macrophages and Cytokines After Surgery of Triple-Negative Breast Cancer Patients and its Implications. Med. Sci. Monit. 2016, 22, 115–120. [Google Scholar] [CrossRef]
- Ashley, J.W.; Shi, Z.; Zhao, H.; Li, X.; Kesterson, R.A.; Feng, X. Genetic ablation of CD68 results in mice with increased bone and dysfunctional osteoclasts. PLoS ONE 2011, 6, e25838. [Google Scholar] [CrossRef]
- Barros, M.H.; Hauck, F.; Dreyer, J.H.; Kempkes, B.; Niedobitek, G. Macrophage polarisation: An immunohistochemical approach for identifying M1 and M2 macrophages. PLoS ONE 2013, 8, e80908. [Google Scholar] [CrossRef]
- Greaves, D.R.; Gordon, S. Macrophage-specific gene expression: Current paradigms and future challenges. Int. J. Hematol. 2002, 76, 6–15. [Google Scholar] [CrossRef]
- Andreeva, E.R.; Pugach, I.M.; Orekhov, A.N. Subendothelial smooth muscle cells of human aorta express macrophage antigen in situ and in vitro. Atherosclerosis 1997, 135, 19–27. [Google Scholar] [CrossRef]
- Gottfried, E.; Kunz-Schughart, L.; Weber, A.; Rehli, M.; Peuker, A.; Müller, A.; Kastenberger, M.; Brockhoff, G.; Andreesen, R.; Kreutz, M. Expression of CD68 in non-myeloid cell types. Scand. J. Immunol. 2008, 67, 453–463. [Google Scholar] [CrossRef]
- La Rocca, G.; Anzalone, R.; Farina, F. The expression of CD68 in human umbilical cord mesenchymal stem cells: New evidences of presence in non-myeloid cell types. Scand. J. Immunol. 2009, 70, 161–162. [Google Scholar] [CrossRef]
- Pulford, K.A.; Sipos, A.; Cordell, J.L.; Stross, W.P.; Mason, D.Y. Distribution of the CD68 macrophage/myeloid associated antigen. Int. Immunol. 1990, 2, 973–980. [Google Scholar] [CrossRef]
- Strobl, H.; Scheinecker, C.; Csmarits, B.; Majdic, O.; Knapp, W. Flow cytometric analysis of intracellular CD68 molecule expression in normal and malignant haemopoiesis. Br. J. Haematol. 1995, 90, 774–782. [Google Scholar] [CrossRef]
- Barreda, D.R.; Hanington, P.C.; Walsh, C.K.; Wong, P.; Belosevic, M. Differentially expressed genes that encode potential markers of goldfish macrophage development in vitro. Dev. Comp. Immunol. 2004, 28, 727–746. [Google Scholar] [CrossRef]
- Santos, A.A.; Gutierre, R.C.; Antoniazzi, M.M.; Ranzani-Paiva, M.J.; Silva, M.R.; Oshima, C.T.; Egami, M.I. Morphocytochemical, immunohistochemical and ultrastructural characterization of the head kidney of fat snook Centropomus parallelus. J. Fish Biol. 2011, 79, 1685–1707. [Google Scholar] [CrossRef]
- Passantino, L.; Patruno, R.; Cianciotta, A.; Passantino, G.; Tafaro, A.; Gadaleta, C.; Ranieri, G. A phylogenetic comparison between acute monocytic leukemia cells and monocytes-macrophages in lower vertebrates. Immunopharmacol. Immunotoxicol. 2003, 25, 87–99. [Google Scholar] [CrossRef]
- Cui, K.; Li, Q.; Xu, D.; Zhang, J.; Gao, S.; Xu, W.; Mai, K.; Ai, Q. Establishment and characterization of two head kidney macrophage cell lines from large yellow croaker (Larimichthys crocea). Dev. Comp. Immunol. 2020, 102, 103477. [Google Scholar] [CrossRef]
- Mellquist, J.; Kasturi, L.; Spitalnik, S.; Shakin-Eshleman, S.H. The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 1998, 37, 6833–6837. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Killingsworth, M.C.; Myasoedova, V.A.; Orekhov, A.N.; Bobryshev, Y.V. CD68/macrosialin: Not just a histochemical marker. Lab Investig. 2017, 97, 4–13. [Google Scholar] [CrossRef]
- Carlsson, S.R.; Fukuda, M. Structure of Human Lysosomal Membrane Glycoprotein 1. J. Biol. Chem. 1989, 264, 20526–20531. [Google Scholar] [CrossRef]
- Arterburn, L.M.; Earles, B.J.; August, J.T. The disulfide structure of mouse lysosome-associated membrane protein 1. J. Biol. Chem. 1990, 265, 7419–7423. [Google Scholar] [CrossRef]
- Ferenbach, D.; Hughes, J. Macrophages and dendritic cells: What is the difference? Kidney Int. 2008, 74, 5–7. [Google Scholar] [CrossRef]
- Kumar, R.; Joy, K.P.; Singh, S.M. Morpho-histology of head kidney of female catfish Heteropneustes fossilis: Seasonal variations in melano-macrophage centers, melanin contents and effects of lipopolysaccharide and dexamethasone on melanins. Fish Physiol. Biochem. 2016, 42, 1287–1306. [Google Scholar] [CrossRef]
- Press, C.M.; Dannevig, B.; Landsverk, T. Immune and enzyme histochemical phenotypes of lymphoid and nonlymphoid cells within the spleen and head kidney of Atlantic salmon (Salmo salar L.). Fish Shellfish. Immunol. 1994, 4, 79–93. [Google Scholar] [CrossRef]
- Sahoo, P. Immunocompetent Organs in Teleosts; Narendra Publishing House: New Delhi, India, 2006; pp. 1–11. [Google Scholar]
- Li, R.; Qu, J.; Li, H.; Zhang, Q. Genome-wide identification and analysis of scavenger receptors and their expression profiling in response to Edwardsiella tarda infection in Japanese flounder (Paralichthys olivaceus). Dev. Comp. Immunol. 2022, 132, 104397. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Liu, H.; Yang, J.; Dong, X.; Wu, C. Abundant members of Scavenger receptors family and their identification, characterization and expression against Vibrio alginolyticus infection in juvenile Larimichthys crocea. Fish Shellfish Immunol. 2016, 50, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Geven, E.J.W.; Klaren, P.H.M. The teleost head kidney: Integrating thyroid and immune signalling. Dev. Comp. Immunol. 2017, 66, 73–83. [Google Scholar] [CrossRef]
- Lu, L.; Dai, S.; Liu, L.; Liu, J.; Zhang, X.; Huang, X.; Ouyang, P.; Geng, Y.; Li, Z.; Chen, D. Identification and characterization of high mobility group box 1 and high mobility group box 2 in Siberian sturgeon (Acipenser baerii). Gene 2022, 850, 146932. [Google Scholar] [CrossRef]
- Li, F.; Liu, X.; Ge, H.; Huang, J.; Zhang, Y.; Wang, Z. Transcriptome profiling and differential expression analysis of the immune-related genes during the early phase of acute infection with Aeromonas hydrophila in the Chinese sucker (Myxocyprinus asiaticus). Aquaculture 2021, 545, 737258. [Google Scholar] [CrossRef]
- Tan, P.; Dong, X.; Mai, K.; Xu, W.; Ai, Q. Vegetable oil induced inflammatory response by altering TLR-NF-κB signalling, macrophages infiltration and polarization in adipose tissue of large yellow croaker (Larimichthys crocea). Fish Shellfish Immunol. 2016, 59, 398–405. [Google Scholar] [CrossRef]
- Dai, Y.J.; Cao, X.F.; Zhang, D.D.; Li, X.F.; Liu, W.B.; Jiang, G.Z. Chronic inflammation is a key to inducing liver injury in blunt snout bream (Megalobrama amblycephala) fed with high-fat diet. Dev. Comp. Immunol. 2019, 97, 28–37. [Google Scholar] [CrossRef]
- Song, L.; Lee, C.; Schindler, C. Deletion of the murine scavenger receptor CD68. J. Lipid Res. 2011, 52, 1542–1550. [Google Scholar] [CrossRef]
- Hendrickx, D.A.E.; van Eden, C.G.; Schuurman, K.G.; Hamann, J.; Huitinga, I. Staining of HLA-DR, Iba1 and CD68 in human microglia reveals partially overlapping expression depending on cellular morphology and pathology. J. Neuroimmunol. 2017, 309, 12–22. [Google Scholar] [CrossRef]
- Wu, X.; Hollingshead, N.; Roberto, J.; Knupp, A.; Kenerson, H.; Chen, A.; Strickland, I.; Horton, H.; Yeung, R.; Soysa, R.; et al. Human Liver Macrophage Subsets Defined by CD32. Front. Immunol. 2020, 11, 2108. [Google Scholar] [CrossRef]
- Athanasou, N.; Puddle, B.; Quinn, J.; Woods, C.G. Use of monoclonal antibodies to recognise osteoclasts in routinely processed bone biopsy specimens. J. Clin. Pathol. 1991, 44, 664–666. [Google Scholar] [CrossRef]
- Jubb, A.M.; Soilleux, E.J.; Turley, H.; Steers, G.; Parker, A.; Low, I.; Blades, J.; Li, J.L.; Allen, P.; Leek, R.; et al. Expression of vascular notch ligand delta-like 4 and inflammatory markers in breast cancer. Am. J. Pathol. 2010, 176, 2019–2028. [Google Scholar] [CrossRef]
- Xia, Y.-T.; Cheng, E.H.-C.; Xia, Y.-J.; Wu, Q.-Y.; Zhang, L.H.-L.; Lin, S.-Y.; Dong, T.T.-X.; Qin, Q.-W.; Wang, W.-X.; Tsim, K.W.-K. Characterization of a macrophagic-like cell line derived from rabbit fish (Siganus fuscescens): An illustration of anti-inflammatory responses of the herbal extract of Scutellaria baicalensis. Fish Shellfish Immunol. Rep. 2021, 2, 100036. [Google Scholar] [CrossRef]
- Eto, S.F.; Fernandes, D.C.; Funnicelli, M.I.G.; Alecrim, J.V.C.; Souza, P.G.; Carvalho, F.C.A.; Belo, M.A.A.; Pizauro, J.M. Microglia extracellular traps in Oreochromis niloticus infected with Weissella cibaria. Fish Shellfish Immunol. 2021, 113, 148–153. [Google Scholar] [CrossRef]
- Eto, S.F.; Fernandes, D.C.; Baldassi, A.C.; Balbuena, T.S.; da Costa Alecrim, J.V.; Almeida de Carvalho, F.C.; Lima, C.; Lopes-Ferreira, M.; Pizauro, J.M. Proteomic analysis capsule synthesis and redox mechanisms in the intracellular survival of group B Streptococcus in fish microglia. Fish Shellfish Immunol. 2021, 118, 34–50. [Google Scholar] [CrossRef]
- Xu, D.; Li, Q.; Zhou, Y.; Shen, Y.; Lai, W.; Hao, T.; Ding, Y.; Mai, K.; Ai, Q. Functional analysis and regulation mechanism of interferon gamma in macrophages of large yellow croaker (Larimichthys crocea). Int. J. Biol. Macromol. 2022, 194, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Murdaca, G.; Greco, M.; Tonacci, A.; Negrini, S.; Borro, M.; Puppo, F.; Gangemi, S. IL-33/IL-31 Axis in Immune-Mediated and Allergic Diseases. Int. J. Mol. Sci. 2019, 20, 5856. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, S.; Moriyama, M.; Miyake, K.; Nakashima, H.; Tanaka, A.; Maehara, T.; Iizuka-Koga, M.; Tsuboi, H.; Hayashida, J.N.; Ishiguro, N.; et al. Interleukin-33 produced by M2 macrophages and other immune cells contributes to Th2 immune reaction of IgG4-related disease. Sci. Rep. 2017, 7, 42413. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Zhao, X.; Zhan, Q.; Cui, L.; Sun, Q.; Lin, L.; Wang, W.; Liu, H. Characterization and expression analysis of an intelectin gene from Megalobrama amblycephala with excellent bacterial binding and agglutination activity. Fish Shellfish Immunol. 2017, 61, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Zhao, X.; Wang, J.; Zhang, F.; Wang, W.; Liu, H. Intelectin mediated phagocytosis and killing activity of macrophages in blunt snout bream (Megalobrama amblycephala). Fish Shellfish Immunol. 2019, 87, 129–135. [Google Scholar] [CrossRef]
- Cui, H.; Shen, X.; Zheng, Y.; Guo, P.; Gu, Z.; Gao, Y.; Zhao, X.; Cheng, H.; Xu, J.; Chen, X.; et al. Identification, expression patterns, evolutionary characteristics and recombinant protein activities analysis of CD209 gene from Megalobrama amblycephala. Fish Shellfish Immunol. 2022, 126, 47–56. [Google Scholar] [CrossRef]
- Chen, S.X.; Ma, H.L.; Shi, Y.H.; Li, M.Y.; Chen, J. Molecular and functional characterization of a novel CD302 gene from ayu (Plecoglossus altivelis). Fish Shellfish Immunol. 2016, 55, 140–148. [Google Scholar] [CrossRef]
- Li, J.; Chen, Y.; Gu, W.; Xu, F.; Li, H.; Shan, S.; Sun, X.; Yin, M.; Yang, G.; Chen, L. Characterization of a common carp intelectin gene with bacterial binding and agglutination activity. Fish Shellfish Immunol. 2021, 108, 32–41. [Google Scholar] [CrossRef]
- Liu, H.; Chen, C.; Gao, Z.; Min, J.; Gu, Y.; Jian, J.; Jiang, X.; Cai, H.; Ebersberger, I.; Xu, M.; et al. The draft genome of blunt snout bream (Megalobrama amblycephala) reveals the development of intermuscular bone and adaptation to herbivorous diet. Gigascience 2017, 6, 1–13. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Guindon, S.; Delsuc, F.; Dufayard, J.F.; Gascuel, O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 2009, 537, 113–137. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Kimura, M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature 1977, 267, 275–276. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Yang, Z. Likelihood and Bayes estimation of ancestral population sizes in hominoids using data from multiple loci. Genetics 2002, 162, 1811–1823. [Google Scholar] [CrossRef]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




| Gene Name | LRT M1a versus M2a (Significance) | LRT M7 versus M8 (Significance) | dN/dS (Model) | Sites a under Positive Selection |
|---|---|---|---|---|
| CD68 | 27.44 (p < 0.01) | 42.02 (p < 0.01) | 2.21 (M2a), 1.50 (M8) | T46, H97, K252, A265, K305 |
| Foreground Branch(es) | LRT M1a versus MA (Significance) | LRT MA1 versus MA (Significance) | Sites a under Positive Selection |
|---|---|---|---|
| Characiformes | 28.19 (p < 0.01) | 23.20 (p < 0.01) | R225, G313 |
| Perciformes | 10.65 (p < 0.01) | 10.65 (p < 0.01) | N360 |
| Teleosts fish | 15.71 (p < 0.01) | 0 (p > 0.05) | Y297 |
| Foreground Branch(es) | Category | First Category of Sites (0) | Second Category of Sites (1) | Third Category of Sites (2a) | Fourth Category of Sites (2b) |
|---|---|---|---|---|---|
| Characiformes | Proportion | 0.42459 | 0.46201 | 0.05431 | 0.05909 |
| Foreground ω | 0.19077 | 1 | 998.84237 | 998.84237 | |
| Perciformes | Proportion | 0.45428 | 0.50616 | 0.01871 | 0.02084 |
| Foreground ω | 0.19187 | 1 | 999 | 999 | |
| Teleosts fish | Proportion | 0.27506 | 0.33848 | 0.17326 | 0.2132 |
| Foreground ω | 0.17821 | 1 | 1 | 1 |
| Names | Sequence (5′-3′) | Purpose |
|---|---|---|
| CD68-CDS-F | ATGGGACGCGGATTATTATTGATC | CDS amplification |
| CD68-CDS-R | CTATAGTGACTGGTACCCATCAG | |
| qCD68-F | CTATAGTGACTGGTACCCAT | qRT-PCR |
| qCD68-R | TGGGGAACGGTGTGAGTCTA | |
| qβ-actin-F | GCTCTTACAGGAAACGGGTC | qRT-PCR |
| qβ-actin-R | GCAGCAGCTCTGTAGGTCAT | |
| qGAPDH-F | TGCCGGCATCTCCCTCAA | qRT-PCR |
| qGAPDH-R | TCAGCAACACGGTGGCTGTAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, H.; Li, H.; Zhang, M.; Li, H.; Wang, X.; Wang, Z.; Zhai, W.; Chen, X.; Cheng, H.; Xu, J.; et al. Molecular Characterization, Expression, Evolutionary Selection, and Biological Activity Analysis of CD68 Gene from Megalobrama amblycephala. Int. J. Mol. Sci. 2022, 23, 13133. https://doi.org/10.3390/ijms232113133
Cui H, Li H, Zhang M, Li H, Wang X, Wang Z, Zhai W, Chen X, Cheng H, Xu J, et al. Molecular Characterization, Expression, Evolutionary Selection, and Biological Activity Analysis of CD68 Gene from Megalobrama amblycephala. International Journal of Molecular Sciences. 2022; 23(21):13133. https://doi.org/10.3390/ijms232113133
Chicago/Turabian StyleCui, Hujun, Hong Li, Minying Zhang, Hongping Li, Xu Wang, Zirui Wang, Wei Zhai, Xiangning Chen, Hanliang Cheng, Jianhe Xu, and et al. 2022. "Molecular Characterization, Expression, Evolutionary Selection, and Biological Activity Analysis of CD68 Gene from Megalobrama amblycephala" International Journal of Molecular Sciences 23, no. 21: 13133. https://doi.org/10.3390/ijms232113133
APA StyleCui, H., Li, H., Zhang, M., Li, H., Wang, X., Wang, Z., Zhai, W., Chen, X., Cheng, H., Xu, J., Zhao, X., & Ding, Z. (2022). Molecular Characterization, Expression, Evolutionary Selection, and Biological Activity Analysis of CD68 Gene from Megalobrama amblycephala. International Journal of Molecular Sciences, 23(21), 13133. https://doi.org/10.3390/ijms232113133





