Exploring Next Generation Probiotics for Metabolic and Microbiota Dysbiosis Linked to Xenobiotic Exposure: Holistic Approach
Abstract
1. Introduction
2. Results
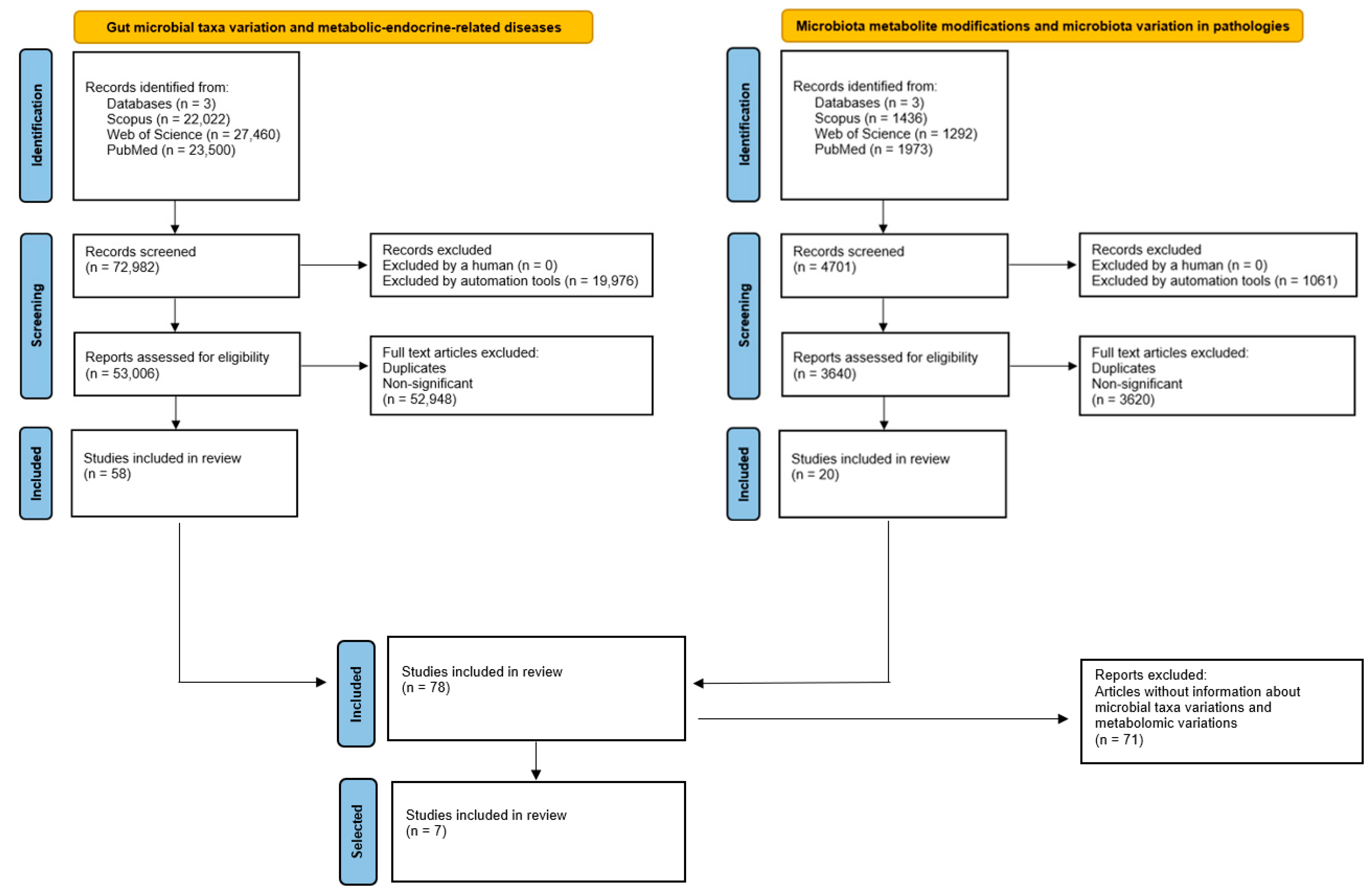
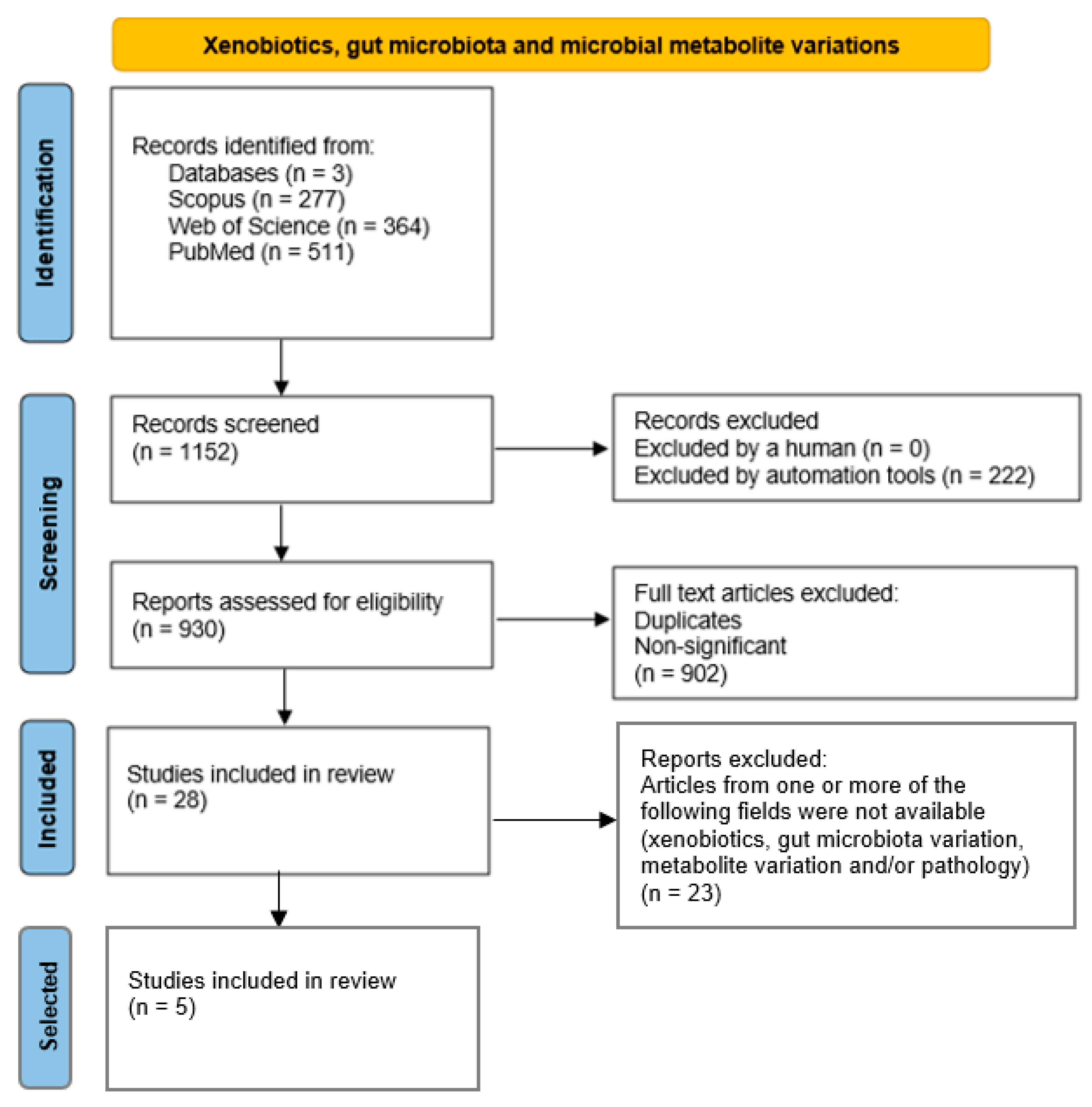
2.1. Extraction of Data and Analysis
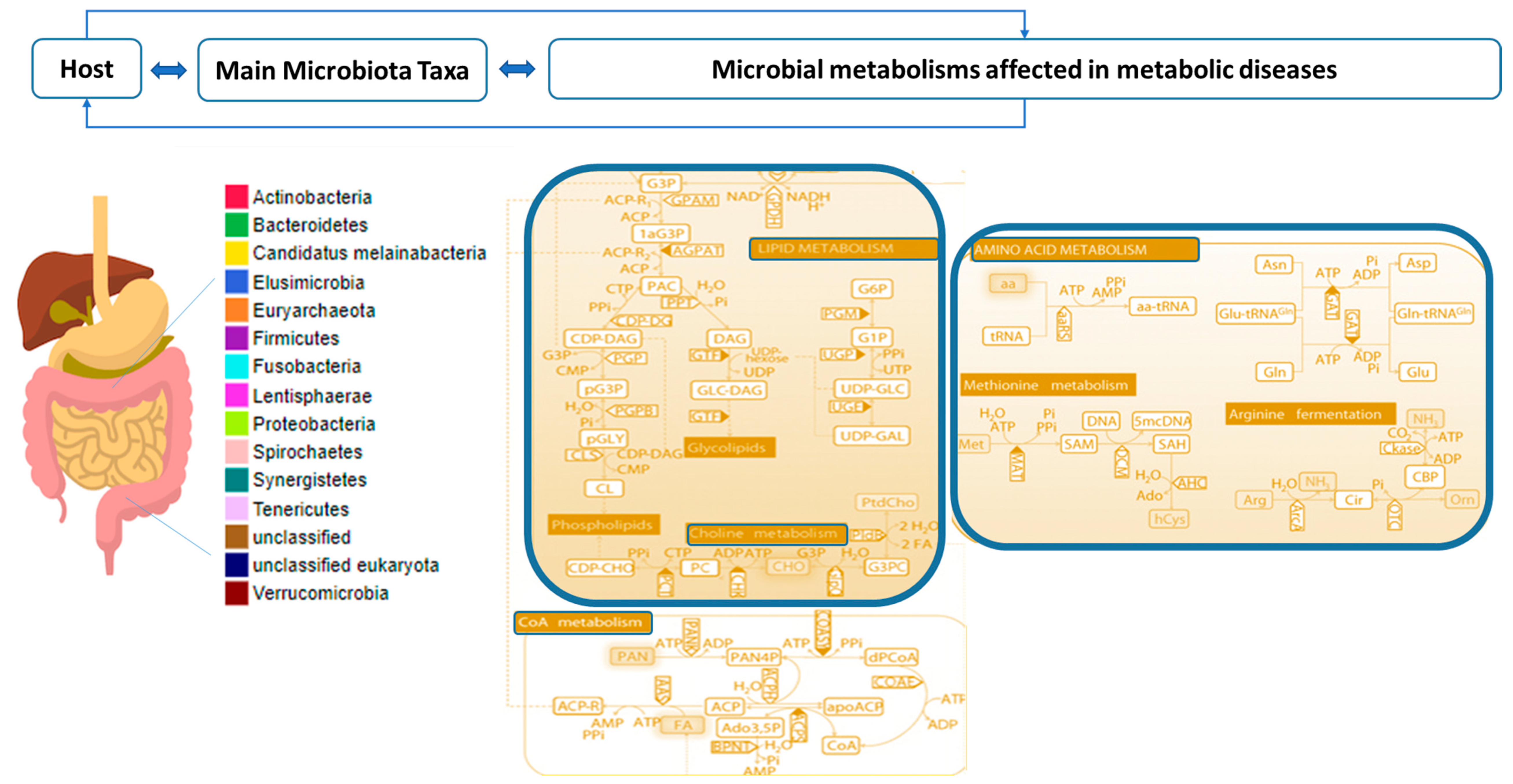
2.2. Combined Analysis of Microbiota Taxa and Metabolites in Metabolic Diseases
2.3. Microbiota Taxa and Metabolite Profiles Linked to Xenobiotic Exposure
2.4. NGP Studies for Interventional Metabolic Dysbiosis
3. Discussion
3.1. Limitations of the Study
3.2. Future Perspectives
4. Materials and Methods
5. Conclusions
- To increase scientific data availability on the interplay between metabolic and molecular pathways involving xenobiotic exposure and their biodegradation, gut microbiota taxa and metabolite modification needs to be studied continuously, using improved methods. It will allow for the development of new biological-based treatments for mitigating metabolic disorders and diseases.
- Relevant modifications of potential signature metabolites mediated by targeted microbiota taxa belong to lipid, bile acid, acetyl-CoA, and amino acid metabolisms.
- The selection and application of appropriate NGPs from healthy microbiota, after elucidating their abundance, functionality, and key molecular mechanisms, seems to be a promising strategy to potentially restore the homeostasis of the intestinal microbiota, taking into account food safety and risk assessment studies and their clinical impact in murine models and subsequently validation in human studies.
- Exploring the uses of NGPs in animals, plants, and/or bioremediation following the preliminary steps of the One Health approach before clinical administration can overcome many safety issues posed by the use of new beneficial microbes in humans. Moreover, it could demonstrate the metabolic potential of NGPs to help refine doses and formulations.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- García-Córcoles, M.T.; Cipa, M.; Rodríguez-Gómez, R.; Rivas, A.; Olea-Serrano, F.; Vílchez, J.L.; Zafra-Gómez, A. Determination of Bisphenols with Estrogenic Activity in Plastic Packaged Baby Food Samples Using Solid-Liquid Extraction and Clean-up with Dispersive Sorbents Followed by Gas Chromatography Tandem Mass Spectrometry Analysis. Talanta 2018, 178, 441–448. [Google Scholar] [CrossRef]
- Zhai, Q.; Feng, S.; Arjan, N.; Chen, W. A next Generation Probiotic, Akkermansia Muciniphila. Crit. Rev. Food Sci. Nutr. 2019, 59, 3227–3236. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.; Machado, D.; Andrade, J.C.; Mendo, S.; Gomes, A.M.; Freitas, A.C. Evolving Trends in Next-Generation Probiotics: A 5W1H Perspective. Crit. Rev. Food Sci. Nutr. 2020, 60, 1783–1796. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, D.K.; Renuka; Dangi, A.K.; Shandilya, U.K.; Puniya, A.K.; Shukla, P. Chapter 44 New-Generation Probiotics Perspectives and Applications. In Microbiome and Metabolome in Diagnosis, Therapy, and Other Strategic Applications; Academic Press: Cambridge, MA, USA, 2019; pp. 417–424. [Google Scholar]
- Satokari, R. Modulation of Gut Microbiota for Health by Current and Next-Generation Probiotics. Nutrients 2019, 11, 1921. [Google Scholar] [CrossRef]
- Chang, C.-J.; Lin, T.-L.; Tsai, Y.-L.; Wu, T.-R.; Lai, W.-F.; Lu, C.-C.; Lai, H.-C. Next Generation Probiotics in Disease Amelioration. J. Food Drug Anal. 2019, 27, 615–622. [Google Scholar] [CrossRef]
- Cani, P.D.; de Vos, W.M. Next-Generation Beneficial Microbes: The Case of Akkermansia Muciniphila. Front. Microbiol. 2017, 8, 1765. [Google Scholar] [CrossRef] [PubMed]
- Saarela, M.H. Safety Aspects of next Generation Probiotics. Curr. Opin. Food Sci. 2019, 30, 8–13. [Google Scholar] [CrossRef]
- Tan, H.; Zhai, Q.; Chen, W. Investigations of Bacteroides Spp. towards next-Generation Probiotics. Food Res. Int. 2019, 116, 637–644. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, P.W.; Marchesi, J.R.; Hill, C. Next-Generation Probiotics: The Spectrum from Probiotics to Live Biotherapeutics. Nat. Microbiol. 2017, 2, 17057. [Google Scholar] [CrossRef] [PubMed]
- Koppel, N.; Maini Rekdal, V.; Balskus, E.P. Chemical Transformation of Xenobiotics by the Human Gut Microbiota. Science 2017, 356, eaag2770. [Google Scholar] [CrossRef]
- Abdelsalam, N.A.; Ramadan, A.T.; ElRakaiby, M.T.; Aziz, R.K. Toxicomicrobiomics: The Human Microbiome vs. Pharmaceutical, Dietary, and Environmental Xenobiotics. Front. Pharmacol. 2020, 11, 390. [Google Scholar] [CrossRef]
- Del Chierico, F.; Nobili, V.; Vernocchi, P.; Russo, A.; De Stefanis, C.; Gnani, D.; Furlanello, C.; Zandonà, A.; Paci, P.; Capuani, G.; et al. Gut Microbiota Profiling of Pediatric Nonalcoholic Fatty Liver Disease and Obese Patients Unveiled by an Integrated Meta-Omics-Based Approach. Hepatology 2017, 65, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Thingholm, L.B.; Rühlemann, M.C.; Koch, M.; Fuqua, B.; Laucke, G.; Boehm, R.; Bang, C.; Franzosa, E.A.; Hübenthal, M.; Rahnavard, A.; et al. Obese Individuals with and without Type 2 Diabetes Show Different Gut Microbial Functional Capacity and Composition. Cell Host Microbe 2019, 26, 252–264.e10. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lou, H.; Peng, Y.; Chen, S.; Zhang, Y.; Li, X. Comprehensive Relationships between Gut Microbiome and Faecal Metabolome in Individuals with Type 2 Diabetes and Its Complications. Endocrine 2019, 66, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Budinska, E.; Gojda, J.; Heczkova, M.; Bratova, M.; Dankova, H.; Wohl, P.; Bastova, H.; Lanska, V.; Kostovcik, M.; Dastych, M.; et al. Microbiome and Metabolome Profiles Associated with Different Types of Short Bowel Syndrome: Implications for Treatment. JPEN J. Parenter. Enter. Nutr. 2020, 44, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Franzosa, E.A.; Sirota-Madi, A.; Avila-Pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; McIver, L.J.; et al. Gut Microbiome Structure and Metabolic Activity in Inflammatory Bowel Disease. Nat. Microbiol. 2019, 4, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut Microbiota Dysbiosis Contributes to the Development of Hypertension. Microbiome 2017, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, X.; Hu, X.; Niu, H.; Tian, R.; Wang, H.; Pang, H.; Jiang, L.; Qiu, B.; Chen, X.; et al. Alterations in the Gut Microbiome and Metabolism with Coronary Artery Disease Severity. Microbiome 2019, 7, 68. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, F.; Wang, T.; Xu, Y.; Qiu, J.; Qian, Y. Host Metabolic Disorders Induced by Alterations in Intestinal Flora under Dietary Pesticide Exposure. J. Agric. Food Chem. 2021, 69, 6303–6317. [Google Scholar] [CrossRef]
- Wang, D.; Jin, Y.; Teng, M.; Sen, Y.; Zhou, Z.; Zhu, W. In Utero and Lactational Exposure to BDE-47 Promotes Obesity Development in Mouse Offspring Fed a High-Fat Diet: Impaired Lipid Metabolism and Intestinal Dysbiosis. Arch. Toxicol. Arch. Für Toxikol. 2018, 92, 1847–1860. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Sun, W.; Liu, W.; Wang, Y.; Jia, M.; Tian, S.; Chen, X.; Zhu, W.; Zhou, Z. A Common Fungicide Tebuconazole Promotes Colitis in Mice via Regulating Gut Microbiota. Environ. Pollut. 2022, 292, 118477. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Yuan, P.; Lei, H.; Zhang, L.; Deng, D.; Zhang, L.; Chen, X. Long-Term Chronic Exposure to Di-(2-Ethylhexyl)-Phthalate Induces Obesity via Disruption of Host Lipid Metabolism and Gut Microbiota in Mice. Chemosphere 2022, 287, 132414. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Zeng, Z.; Wang, C.; Luo, T.; Wang, S.; Zhou, J.; Ni, Y.; Fu, Z.; Jin, Y. Insights into a Possible Mechanism Underlying the Connection of Carbendazim-Induced Lipid Metabolism Disorder and Gut Microbiota Dysbiosis in Mice. Toxicol. Sci. 2018, 166, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Guo, Y.; Hu, C.; Lam, P.K.S.; Lam, J.C.W.; Zhou, B. Dysbiosis of Gut Microbiota by Chronic Coexposure to Titanium Dioxide Nanoparticles and Bisphenol A: Implications for Host Health in Zebrafish. Environ. Pollut. 2018, 234, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Qi, S.; Mu, X.; Yuan, L.; Li, Y.; Qiu, J. Bisphenol F Induces Liver-Gut Alteration in Zebrafish. Sci. Total Environ. 2022, 851, 157974. [Google Scholar] [CrossRef]
- Gu, J.; Zhu, Y.; Guo, M.; Yin, X.; Liang, M.; Lou, X.; Chen, J.; Zhou, L.; Fan, D.; Shi, L.; et al. The Potential Mechanism of BPF-Induced Neurotoxicity in Adult Zebrafish: Correlation between Untargeted Metabolomics and Gut Microbiota. Sci. Total Environ. 2022, 839, 156221. [Google Scholar] [CrossRef]
- Wang, X.; Shen, M.; Zhou, J.; Jin, Y. Chlorpyrifos Disturbs Hepatic Metabolism Associated with Oxidative Stress and Gut Microbiota Dysbiosis in Adult Zebrafish. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2019, 216, 19–28. [Google Scholar] [CrossRef]
- Huang, Z.; Xiao, X.; Wang, D.; Zhong, Y.; Ding, Q.; You, J. Joint Effects of Micro-Sized Polystyrene and Chlorpyrifos on Zebrafish Based on Multiple Endpoints and Gut Microbial Effects. J. Environ. Sci. 2023, 126, 184–197. [Google Scholar] [CrossRef]
- Zhang, R.; Pan, Z.; Wang, X.; Shen, M.; Zhou, J.; Fu, Z.; Jin, Y. Short-Term Propamocarb Exposure Induces Hepatic Metabolism Disorder Associated with Gut Microbiota Dysbiosis in Adult Male Zebrafish. Acta Biochim. Biophys. Sin. 2019, 51, 88–96. [Google Scholar] [CrossRef]
- Bao, Z.; Zhao, Y.; Wu, A.; Lou, Z.; Lu, H.; Yu, Q.; Fu, Z.; Jin, Y. Sub-Chronic Carbendazim Exposure Induces Hepatic Glycolipid Metabolism Disorder Accompanied by Gut Microbiota Dysbiosis in Adult Zebrafish (Daino Rerio). Sci. Total Environ. 2020, 739, 140081. [Google Scholar] [CrossRef]
- Jiang, J.; Chen, L.; Wu, S.; Lv, L.; Liu, X.; Wang, Q.; Zhao, X. Effects of Difenoconazole on Hepatotoxicity, Lipid Metabolism and Gut Microbiota in Zebrafish (Danio rerio). Environ. Pollut. 2020, 265, 114844. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Luo, T.; Zhu, Z.; Pan, Z.; Yang, J.; Wang, W.; Fu, Z.; Jin, Y. Imazalil Exposure Induces Gut Microbiota Dysbiosis and Hepatic Metabolism Disorder in Zebrafish. Comp. Biochem. Physiol. C Toxicol. Pharm. 2017, 202, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.-P.; Junaid, M.; Xin, G.-Y.; Wang, Y.; Ma, Y.-B.; Pei, D.-S. Disruption of Intestinal Homeostasis Through Altered Responses of the Microbial Community, Energy Metabolites, and Immune System in Zebrafish after Chronic Exposure to DEHP. Front. Microbiol. 2021, 12, 729530. [Google Scholar] [CrossRef] [PubMed]
- Adamovsky, O.; Buerger, A.N.; Vespalcova, H.; Sohag, S.R.; Hanlon, A.T.; Ginn, P.E.; Craft, S.L.; Smatana, S.; Budinska, E.; Persico, M.; et al. Evaluation of Microbiome-Host Relationships in the Zebrafish Gastrointestinal System Reveals Adaptive Immunity Is a Target of Bis(2-Ethylhexyl) Phthalate (DEHP) Exposure. Environ. Sci. Technol. 2020, 54, 5719–5728. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hu, C.; Lok-Shun Lai, N.; Zhang, W.; Hua, J.; Lam, P.K.S.; Lam, J.C.W.; Zhou, B. Acute Exposure to PBDEs at an Environmentally Realistic Concentration Causes Abrupt Changes in the Gut Microbiota and Host Health of Zebrafish. Environ. Pollut. 2018, 240, 17–26. [Google Scholar] [CrossRef]
- Hu, C.; Bai, Y.; Sun, B.; Tang, L.; Chen, L. Significant Impairment of Intestinal Health in Zebrafish after Subchronic Exposure to Methylparaben. Sci. Total Environ. 2022, 838, 156389. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Liu, W.; Wang, J.; Shi, J.; Sun, Y.; Wang, W.; Ning, G.; Liu, R.; Hong, J. Akkermansia muciniphila Improves Metabolic Profiles by Reducing Inflammation in Chow Diet-Fed Mice. J. Mol. Endocrinol. 2017, 58, 1–14. [Google Scholar] [CrossRef]
- Grajeda-Iglesias, C.; Durand, S.; Daillère, R.; Iribarren, K.; Lemaitre, F.; Derosa, L.; Aprahamian, F.; Bossut, N.; Nirmalathasan, N.; Madeo, F.; et al. Oral Administration of Akkermansia Muciniphila Elevates Systemic Antiaging and Anticancer Metabolites. Aging 2021, 13, 6375–6405. [Google Scholar] [CrossRef]
- Munukka, E.; Rintala, A.; Toivonen, R.; Nylund, M.; Yang, B.; Takanen, A.; Hänninen, A.; Vuopio, J.; Huovinen, P.; Jalkanen, S.; et al. Faecalibacterium Prausnitzii Treatment Improves Hepatic Health and Reduces Adipose Tissue Inflammation in High-Fat Fed Mice. ISME J. 2017, 11, 1667–1679. [Google Scholar] [CrossRef] [PubMed]
- López-Almela, I.; Romaní-Pérez, M.; Bullich-Vilarrubias, C.; Benítez-Páez, A.; Gómez Del Pulgar, E.M.; Francés, R.; Liebisch, G.; Sanz, Y. Bacteroides Uniformis Combined with Fiber Amplifies Metabolic and Immune Benefits in Obese Mice. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef]
- Yang, J.-Y.; Lee, Y.-S.; Kim, Y.; Lee, S.-H.; Ryu, S.; Fukuda, S.; Hase, K.; Yang, C.-S.; Lim, H.S.; Kim, M.-S.; et al. Gut Commensal Bacteroides Acidifaciens Prevents Obesity and Improves Insulin Sensitivity in Mice. Mucosal Immunol. 2017, 10, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Shan, K.; Pan, L.-L.; Feng, N.; Lv, Z.; Sun, Y.; Li, J.; Wu, C.; Zhang, H.; Chen, W.; et al. Clostridium Butyricum CGMCC0313.1 Protects against Autoimmune Diabetes by Modulating Intestinal Immune Homeostasis and Inducing Pancreatic Regulatory T Cells. Front. Immunol. 2017, 8, 1345. [Google Scholar] [CrossRef]
- Péan, N.; Le Lay, A.; Brial, F.; Wasserscheid, J.; Rouch, C.; Vincent, M.; Myridakis, A.; Hedjazi, L.; Dumas, M.-E.; Grundberg, E.; et al. Dominant Gut Prevotella Copri in Gastrectomised Non-Obese Diabetic Goto–Kakizaki Rats Improves Glucose Homeostasis through Enhanced FXR Signalling. Diabetologia 2020, 63, 1223–1235. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, Y.; Kim, Y.; Seo, Y.; Lee, H.; Ha, J.; Lee, J.; Choi, Y.; Oh, H.; Yoon, Y. Akkermansia Muciniphila Prevents Fatty Liver Disease, Decreases Serum Triglycerides, and Maintains Gut Homeostasis. Appl. Environ. Microbiol. 2020, 86, e03004-19. [Google Scholar] [CrossRef] [PubMed]
- van der Lugt, B.; van Beek, A.A.; Aalvink, S.; Meijer, B.; Sovran, B.; Vermeij, W.P.; Brandt, R.M.C.; de Vos, W.M.; Savelkoul, H.F.J.; Steegenga, W.T.; et al. Akkermansia Muciniphila Ameliorates the Age-Related Decline in Colonic Mucus Thickness and Attenuates Immune Activation in Accelerated Aging Ercc1−/Δ7 Mice. Immun. Ageing 2019, 16, 6. [Google Scholar] [CrossRef] [PubMed]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A Purified Membrane Protein from Akkermansia Muciniphila or the Pasteurized Bacterium Improves Metabolism in Obese and Diabetic Mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Liu, Y.; Huang, Y.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Influence of Oral Administration of Akkermansia Muciniphila on the Tissue Distribution and Gut Microbiota Composition of Acute and Chronic Cadmium Exposure Mice. FEMS Microbiol. Lett. 2019, 366, fnz160. [Google Scholar] [CrossRef] [PubMed]
- Zhai, R.; Xue, X.; Zhang, L.; Yang, X.; Zhao, L.; Zhang, C. Strain-Specific Anti-Inflammatory Properties of Two Akkermansia Muciniphila Strains on Chronic Colitis in Mice. Front. Cell. Infect. Microbiol. 2019, 9, 239. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Guo, X.; Zhang, M.; Ou, Z.; Wu, D.; Deng, L.; Lu, Z.; Zhang, J.; Deng, G.; Chen, S.; et al. An Akkermansia Muciniphila Subtype Alleviates High-Fat Diet-Induced Metabolic Disorders and Inhibits the Neurodegenerative Process in Mice. Anaerobe 2020, 61, 102138. [Google Scholar] [CrossRef] [PubMed]
- Grander, C.; Adolph, T.E.; Wieser, V.; Lowe, P.; Wrzosek, L.; Gyongyosi, B.; Ward, D.V.; Grabherr, F.; Gerner, R.R.; Pfister, A.; et al. Recovery of Ethanol-Induced Akkermansia Muciniphila Depletion Ameliorates Alcoholic Liver Disease. Gut 2018, 67, 891–901. [Google Scholar] [CrossRef]
- Zhang, L.; Qin, Q.; Liu, M.; Zhang, X.; He, F.; Wang, G. Akkermansia Muciniphila Can Reduce the Damage of Gluco/Lipotoxicity, Oxidative Stress and Inflammation, and Normalize Intestine Microbiota in Streptozotocin-induced Diabetic Rats. Pathog. Dis. 2018, 76, fty028. [Google Scholar] [CrossRef] [PubMed]
- Ou, Z.; Deng, L.; Lu, Z.; Wu, F.; Liu, W.; Huang, D.; Peng, Y. Protective Effects of Akkermansia Muciniphila on Cognitive Deficits and Amyloid Pathology in a Mouse Model of Alzheimer’s Disease. Nutr. Diabetes 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Fabersani, E.; Portune, K.; Campillo, I.; López-Almela, I.; la Paz, S.M.; Romaní-Pérez, M.; Benítez-Páez, A.; Sanz, Y. Bacteroides Uniformis CECT 7771 Alleviates Inflammation within the Gut-Adipose Tissue Axis Involving TLR5 Signaling in Obese Mice. Sci. Rep. 2021, 11, 11788. [Google Scholar] [CrossRef] [PubMed]
- Gómez del Pulgar, E.M.; Benítez-Páez, A.; Sanz, Y. Safety Assessment of Bacteroides Uniformis CECT 7771, a Symbiont of the Gut Microbiota in Infants. Nutrients 2020, 12, 551. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.X.; Wang, S.-Y.; Kuo, C.-H.; Tsai, I.-L. Metabolome Analysis for Investigating Host-Gut Microbiota Interactions. J. Formos. Med. Assoc. 2019, 118 (Suppl. 1), S10–S22. [Google Scholar] [CrossRef] [PubMed]
- Visconti, A.; Le Roy, C.I.; Rosa, F.; Rossi, N.; Martin, T.C.; Mohney, R.P.; Li, W.; de Rinaldis, E.; Bell, J.T.; Venter, J.C.; et al. Interplay between the Human Gut Microbiome and Host Metabolism. Nat. Commun. 2019, 10, 4505. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Xu, C.; Zhang, D.; Ju, F.; Ni, Y. MetOrigin: Discriminating the Origins of Microbial Metabolites for Integrative Analysis of the Gut Microbiome and Metabolome. iMeta 2022, 1, e10. [Google Scholar] [CrossRef]
- Motta, B.M.; Grander, C.; Gögele, M.; Foco, L.; Vukovic, V.; Melotti, R.; Fuchsberger, C.; De Grandi, A.; Cantaloni, C.; Picard, A.; et al. Microbiota, Type 2 Diabetes and Non-Alcoholic Fatty Liver Disease: Protocol of an Observational Study. J. Transl. Med. 2019, 17, 408. [Google Scholar] [CrossRef]
- Aron-Wisnewsky, J.; Vigliotti, C.; Witjes, J.; Le, P.; Holleboom, A.G.; Verheij, J.; Nieuwdorp, M.; Clément, K. Gut Microbiota and Human NAFLD: Disentangling Microbial Signatures from Metabolic Disorders. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 279–297. [Google Scholar] [CrossRef]
- Tang, W.H.W.; Kitai, T.; Hazen, S.L. Gut Microbiota in Cardiovascular Health and Disease. Circ. Res. 2017, 120, 1183–1196. [Google Scholar] [CrossRef]
- Collins, S.L.; Patterson, A.D. The Gut Microbiome: An Orchestrator of Xenobiotic Metabolism. Acta Pharm. Sin. B 2020, 10, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut Microbiota in Human Metabolic Health and Disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in Health and Diseases. Signal Transduct. Target. Ther. 2022, 7, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, M.; Galvez-Ontiveros, Y.; Rivas, A. Endobolome, a New Concept for Determining the Influence of Microbiota Disrupting Chemicals (MDC) in Relation to Specific Endocrine Pathogenesis. Front. Microbiol. 2020, 11, 578007. [Google Scholar] [CrossRef]
- Bidkhori, G.; Lee, S.; Edwards, L.A.; Chatelier, E.L.; Almeida, M.; Ezzamouri, B.; Onate, F.P.; Ponte, N.; Shawcross, D.L.; Proctor, G.; et al. The Reactobiome Unravels a New Paradigm in Human Gut Microbiome Metabolism. bioRxiv 2021. [Google Scholar] [CrossRef]
- López-Moreno, A.; Ruiz-Moreno, Á.; Pardo-Cacho, J.; Cerk, K.; Torres-Sánchez, A.; Ortiz, P.; Úbeda, M.; Aguilera, M. Culturing and Molecular Approaches for Identifying Microbiota Taxa Impacting Children’s Obesogenic Phenotypes Related to Xenobiotic Dietary Exposure. Nutrients 2022, 14, 241. [Google Scholar] [CrossRef] [PubMed]
- van der Hee, B.; Wells, J.M. Microbial Regulation of Host Physiology by Short-Chain Fatty Acids. Trends Microbiol. 2021, 29, 700–712. [Google Scholar] [CrossRef]
- Chen, J.; Vitetta, L. Gut Microbiota Metabolites in NAFLD Pathogenesis and Therapeutic Implications. Int. J. Mol. Sci. 2020, 21, 5214. [Google Scholar] [CrossRef]
- Tam, B.T.; Murphy, J.; Khor, N.; Morais, J.A.; Santosa, S. Acetyl-CoA Regulation, OXPHOS Integrity and Leptin Levels Are Different in Females with Childhood vs Adulthood Onset of Obesity. Endocrinology 2020, 161, bqaa142. [Google Scholar] [CrossRef]
- Ejtahed, H.-S.; Angoorani, P.; Soroush, A.-R.; Hasani-Ranjbar, S.; Siadat, S.-D.; Larijani, B. Gut Microbiota-Derived Metabolites in Obesity: A Systematic Review. Biosci. Microbiota Food Health 2020, 39, 65–76. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia Muciniphila in Overweight and Obese Human Volunteers: A Proof-of-Concept Exploratory Study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ni, Y.; Qian, L.; Fang, Q.; Zheng, T.; Zhang, M.; Gao, Q.; Zhang, Y.; Ni, J.; Hou, X.; et al. Decreased Abundance of Akkermansia Muciniphila Leads to the Impairment of Insulin Secretion and Glucose Homeostasis in Lean Type 2 Diabetes. Adv. Sci. 2021, 8, 2100536. [Google Scholar] [CrossRef]
- Ortiz, P.; Torres-Sánchez, A.; López-Moreno, A.; Cerk, K.; Ruiz-Moreno, Á.; Monteoliva-Sánchez, M.; Ampatzoglou, A.; Aguilera, M.; Gruszecka-Kosowska, A. Impact of Cumulative Environmental and Dietary Xenobiotics on Human Microbiota: Risk Assessment for One Health. J. Xenobiot. 2022, 12, 56–63. [Google Scholar] [CrossRef] [PubMed]
- González-Domínguez, R.; Jáuregui, O.; Queipo-Ortuño, M.I.; Andrés-Lacueva, C. Characterization of the Human Exposome by a Comprehensive and Quantitative Large-Scale Multianalyte Metabolomics Platform. Anal. Chem. 2020, 92, 13767–13775. [Google Scholar] [CrossRef] [PubMed]
- Lindell, A.E.; Zimmermann-Kogadeeva, M.; Patil, K.R. Multimodal Interactions of Drugs, Natural Compounds and Pollutants with the Gut Microbiota. Nat. Rev. Microbiol. 2022, 20, 431–443. [Google Scholar] [CrossRef]
- Ampatzoglou, A.; Gruszecka-Kosowska, A.; Torres-Sánchez, A.; López-Moreno, A.; Cerk, K.; Ortiz, P.; Monteoliva-Sánchez, M.; Aguilera, M. Incorporating the Gut Microbiome in the Risk Assessment of Xenobiotics and Identifying Beneficial Components for One Health. Front. Microbiol. 2022, 13, 872583. [Google Scholar] [CrossRef]
- Gao, P. The Exposome in the Era of One Health. Environ. Sci. Technol. 2021, 55, 2790–2799. [Google Scholar] [CrossRef]
- Perraudeau, F.; McMurdie, P.; Bullard, J.; Cheng, A.; Cutcliffe, C.; Deo, A.; Eid, J.; Gines, J.; Iyer, M.; Justice, N.; et al. Improvements to Postprandial Glucose Control in Subjects with Type 2 Diabetes: A Multicenter, Double Blind, Randomized Placebo-Controlled Trial of a Novel Probiotic Formulation. BMJ Open Diabetes Res. Care 2020, 8, e001319. [Google Scholar] [CrossRef]
- El Hage, R.; Hernandez-Sanabria, E.; Van de Wiele, T. Emerging Trends in “Smart Probiotics”: Functional Consideration for the Development of Novel Health and Industrial Applications. Front. Microbiol. 2017, 8, 1889. [Google Scholar] [CrossRef]
- Cunningham, M.; Azcarate-Peril, M.A.; Barnard, A.; Benoit, V.; Grimaldi, R.; Guyonnet, D.; Holscher, H.D.; Hunter, K.; Manurung, S.; Obis, D.; et al. Shaping the Future of Probiotics and Prebiotics. Trends Microbiol. 2021, 29, 667–685. [Google Scholar] [CrossRef]
- McDonough, C.M.; Xu, H.S.; Guo, T.L. Toxicity of Bisphenol Analogues on the Reproductive, Nervous, and Immune Systems, and Their Relationships to Gut Microbiome and Metabolism: Insights from a Multi-Species Comparison. Crit. Rev. Toxicol. 2021, 51, 283–300. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]


| Ref. | Clinical Traits | Microbiota Taxa Modification | Metabolite Modifications—Pathways |
|---|---|---|---|
| [13] | n = 115; HC n = 54; OB n = 8; NAFLD n = 27; NASH n = 26 | ↑ Bradyrhizobium, Anaerococcus, Peptoniphilus, Propionibacterium acnes, Dorea, and Ruminococcus ↓ Oscillospira in NAFLD, NASH and OB vs. HC | 2-Butanone, 4-Methyl-2-pentanone Ketone pathways |
| [14] | n = 1280; LN-NonT2D n = 633; OB-NonT2D n = 494; OBT2D n = 153 | ↓ Akkermansia, Faecalibacterium prausnitzii, Oscillibacter, and Alistipes in OB | Indolepropionate, 2-Methylbutyrylcarnitine, Valine, Isovalerate, Glutamine, Tyrosine, 3-Phenylpropionate, Phenylalanine, Oxalate, N1-Methyl-2-pyridone-5-carboxamide, Docosapentaenoate, 1-Stearoyl-GPE (18:0), 10-Heptadecenoate, 1-Arachidonoyl-GPI (20:4), Inosine, Glycylvaline, Citrulline, Gamma-CEHC, 1-Linoleoyl-GPC (18:2), Adrenate (22:4n6), Epiandrosterone sulphate, 2-Linoleoyl-GPC (18:2), 1-Oleyl-GPC (18:1), 1-Dihomo-linoleoyl-GPC (20:2), Cinnamoylglycine Amino acid and phospholipid metabolism pathways |
| [15] | n = 100; HC n = 35; T2D+ n = 49; T2D- n = 16 | ↑ Coprococcus 1 ↓ Bacteroides and Prevotella in T2D+ and T2D- vs. HC; ↑ Parasutterella in T2D+ vs. HC; ↑ Blautia and Eubacterium hallii group in T2D- vs. HC | HDL cholesterol, LDL cholesterol, Acetate, Butyrate, Linoleic acid, Palmitoylcarnitine, Lysophosphatidylcholine (18:2), Phosphatidylcholine (16:0/17:0), Diacylglycerol (15:0/18:3), Diacylglycerol (15:0/20:3), Glycoursodeoxycholic acid, Chenodeoxyglycocholate, Glycocholic acid, Cholic acid Lipid metabolism, bile acid metabolism and cholesterol pathways |
| [16] | n = 69; HC n = 40; Non-PN SBS n = 5; SBS I n = 10; SBS II n = 14 | ↑ Lactobacillus and Klebsiella ↓ Coprococcus, Faecalibacterium, Lachnospira, and Ruminococcus in SBS patients; ↓ Blautia, Bacteroides, Odoribacter, Oscillospira, Prevotella, Roseburia, and Sutterella in SBS I and SBS II; ↑ Streptococcus and Staphylococcus in SBS I | Butanoic acid, Pentanoic acid, 1-Nonanol, p-Cresol, Geranil acetone, γ-Undecalactone, Indole, Phenol, Decanoic acid, Dodecanoic acid, Nonanal, Octanal, Hexanal, 2-pentyl furan, Lythocholic acid, Taurocholic acid, Chenodeoxycholic acid Deoxycholic acid, Glycodeoxycholic acid, Cholic acid, Glycocholic acid, Glycochenodeoxycholic acid Volatile organic compounds and bile acid metabolism pathways |
| [17] | n = 155; Non-IBD n = 34; CD n = 68; UC n = 53 | ↑ Eubacterium ventriosum, Coprococcus catus, Roseburia hominis, Dorea longicatena, Eubacterium hallii, Eubacterium siraeum, Alistipes shaii, Alistipes putredinis, Alistipes finegoldii, Roseburia inulinivorans, Roseburia intestinalis, Faecalibacterium prausnitzii, Eubacterium eligens, Bacteroidales bacterium ph8, Alistipes indistinctus, Alistipes senegalensis, Ruminococcus callidus, Holdemania filiformis, Fordonibacter pamelaeae, Lachnospiraceae bacterium 1, Adlercreutzia equolifaciens, and Alistipes onderdonkii in Non-IBD controls; ↑ Unclassified Roseburia species in CD and UC; ↑ Bifidobacterium breve and Clostridium symbiosum in UC; ↑ Blautia producta, Lactobacillus gasseri, Enterococcus faecium, Lachnospiraceae bacterium 2, Clostridium clostridioforme, Ruminococcus gnavus, and Escherichia coli in CD | Caprylic acid, Carnosol, Urobilin, Pipecolic acid, 4-Methylcatechol, 2-Hydroxyhexadecanoate, Cholestenone, 5ɑ-Cholestanol, Dodecanedioic acid, Caproic acid, Hydrocinnamic acid, 3-Methyladipate-pimelate, Undecanedionate, Azelaic acid, 2-Hydroxyphenethylamine, Linoleoyl ethanolamide, Palmitoylethanolamide, Docosapentaenoic acid, Eicosatrienoic acid, Taurine, N-Acetylputrescine, ADMA, Cholate, Chenodeoxycholate, Phytosphingosine, C 18:0 CE, C14 carnitine, C3-DC-CH3 carnitine Bile acid metabolism pathways |
| [18] | n = 196; HC n = 41; pHT n = 56; HT n = 99 | ↑ Prevotella and Klebsiella in pHT or HT; ↑ Porphyromonas and Actinomyces in HT; ↓ Faecalibacterium, Oscillibacter, Roseburia, Subdoligranulum, Blautia, Bifidobacterium, Coprococcus, Butyrivibrio, Eggerthella, Streptococcus, and Akkermansia in pHT and HT | Hippurin-1, Trichloroethanol glucuronide, PS(O-18:0/0:0), LysoPC(18:2), S-Carboxymethyl-L-cysteine, Pyridine, LysoPC (22:5), 3-Keto stearic acid, Petunidin 3-rhamnoside 5-glucoside, Nɑ-Acetyl-L-arginine, 9,10-Dichloro-octadecanoic acid, PA(12:0/0:0) Glucuronide detoxification and antioxidant pathways |
| [19] | n = 201; HC n = 40; CAD n = 161 | ↑ Actinomyces, Haemophilus, Granulicatella, Weissella, Veillonella, Streptococcus, Klebsiella, Rothia, Enterococcus (CAG17); ↓ Faecalibacterium, Lachnospiraceae, Roseburia, Oscilibacter (CAG4); Lachnospiracea incertae sedis, Ruminococcus 2, Dorea, Blautia, Clostridium XVIII (CAG14); Anaerostipes, Blautia, Lactobacillus, Fusicatenibacter, Clostridium XIVa, Gemella, Bifidobacterium, Saccharibacteria genera incertae sedis (CAG15); Roseburia, Lachnospiracea incertae sedis, Clostridium XIVb, Parasutterella, Butyricicoccus in CAD | Steroids, Sphingolipids, Phosphatidylethanolamine, Phosphatidylcholine, Ceramides, Glycerophospholipid, Fatty acyls, Carboxylic acids, Benzene/derivatives, Fatty acyl carnitines, Prenol lipids, Glycerolipids, Potassium chloride, Addictives/ingredients, Taurine, Aminoacids (L-Leucine) Amino acid and lipid metabolism pathways |
| Ref., Xenobiotic, Biosample | Microbiota Taxa Modification | Metabolite Modification | Health Effects |
|---|---|---|---|
| [20] Chlorfenapyr; acetamiprid; and chlorfenapyr + acetamiprid Kunming mice n = 60; CK n = 20; C n = 10; A n = 10; AC n = 10; N = 10 Faeces and serum | ↑ Helicobacter, Desulfovibrio, Oscillibacter, Intestinimonas, Roseburia, Lachnoclostridium, Ruminiclostridium, and Butyricimonas in chlorfenapyr ↓ Lactobacillus, Bacteroides, Parasutterella, Erysipelatoclostridium, Enterorhabdus, Alloprevotella, and Enterococcus in chlorfenapyr ↑ Lactobacillus and Marvinbryantia in acetamiprid ↓ Muribaculum, Parabacteroides, and Unidentified Clostridiales in acetamiprid | ↑ Trimethylamine-N-oxide, cholic acid derivative, 5-β-cholanoic acid, 3-β-hydroxy-5-cholenoic acid, 7-ketodeoxycholic acid, avicholate, methylcholate, and uric acid in C, A, and AC (Faeces) ↓ Free fatty acid in C, A and AC (Serum) ↑ Betaine in A and AC (Faeces) ↑ Long-chain free fatty acids and esters in A and C (Faeces) ↓ Phosphatidylcholine and phosphatidylethanolamine in A and C (Serum) ↑ 5-Hydroxyinoleacetic acid and indole-2-carboxylic acid in A (Faeces) ↑ Free fatty acid, N-acetyl-tryptophan, and N-acetyl-phenylalanine in A (Serum) ↓ 3-(Aminomethyl)-indole, indoline, and indolemethanamine in C (Faeces) ↑ Tryptophan in C (Serum) | Glucose homeostasis |
| [21] 2,2′,4,4′-Tetrabromodiphenyl ether ICR mice n = 36; ND+V n = 6; ND+L-BDE n = 6; ND+H-BDE n = 6; HFD + V n = 6; HFD+L-BDE n = 6; HFD+H-BDE n = 6 Faeces and serum | ↑ Parasutterella and Gemella in ND+L-BDE ↓ Christensenellaceae R-7 group, Atopostipes, Family XIII UCG-001, and Bacillus in ND+L-BDE ↑ Candidatus Saccharimonas, Ruminococcaceae UCG-013, Staphylococcus, Eubacterium nodatum group, Gemella, Corynebacterium 1, and Paenalcaligenes in ND+H-BDE↑ Staphylococcus in HFD+L-BDE ↓ Bacteroides, Ruminiclostridium 9, Helicobacter, Alloprevotella, Oscillibacter, Christensenellaceae R-7 group, Ruminiclostridium 5, Odoribacter, Ruminococcaceae UCG-010, and Rikenella in HFD+L-BDE ↓ Turicibacter, and Anaerotruncus in HFD+L-BDE and HFD+H-BDE ↑ Dorea, Lactococcus, and Eubacterium nodatum group in HFD+H-BDE ↓ Ruminococcaceae UCG-014, Ruminococcaceae UCG-009, Candidatus Saccharimonas, Ruminiclostridium 5, and Family XIII UCG-001 in HFD+H-BDE | ↑ Bile acids, succinate, taurine, glycine, α-glucosa, β-glucose, arabinose, and galactose in ND-BDE (Faeces) ↓ Methionine in ND-BDE (Faeces) ↑ Bile acids, choline, α-ketoglutarate, and α-glucose in HFD-BDE (Faeces) ↓ Propionate and β-glucose in HFD-BDE (Faeces) ↑ Pyruvate, lactate, phosphoric acid, glutamine, ornithine, 3-hydoxybutyric acid, isoleucine, and octadecanoic acid in HFD-BDE (Serum) ↓ Palmitelaidic acid and uric acid in HFD-BDE (Serum) | Obesity Steatosis Glucose homeostasis |
| [22] Tebuconazole ICR mice n = 24; Control n = 8; L-TEB n = 8; H-TEB n = 8 C57BL/6 mice n = 16; Control n = 8; TEB+DSS n = 8 Serum | ↑ S24-7, Coprococcus, and Akkermansia in H-TEB ↓ Clostridiales, Ruminococcaceae, Ruminococcus, Oscillospira, Mucispirillum, Rikenellaceae, and Dehalobacterium in H-TEB ↑ Rikenellaceae, Akkermansia, and Bilophila in TEB+DSS ↓ S24-7 in TEB+DSS | ↑ α-Glucose, β-glucose, taurine, leucine, lysine, alanine, creatine, glutamine, and glutamate in H-TEB ↓ Lipid, lactate, acetate, and choline in H-TEB ↑ α-Glucose, β-glucose, taurine, leucine, lysine, alanine, and creatine in TEB+DSS ↓ Lipid, lactate, acetate, and choline in TEB+DSS | Colitis |
| [23] Di(2-ethylhexyl) phthalate (DEHP) C57BL/6J mice n = 24; Control n = 8; L-DEHP n = 8; H-DEHP n = 8 Liver | ↑ Streptococcus and Butyrivibrio ↓ Lactobacillus | ↑ Stearic acid (18:0), linoleic acid (18:2n6), α-linolenic acid (18:3n3), γ-linolenic acid (18:3n6), arachidonic acid (20:4n6), eicosapentaenoic acid (20:5n3), docosaexaenoic acid (22:6n3), glycerophosphoserine, and glycerophosphoglycerol in DEHP ↓ Glycerophosphocholine, glycerophosphoinositol, lysophosphosphatidylethanolamine, lysophosphatidylcholine, phosphatidylethanolamine, and sphingomyelin in DEHP | Obesity |
| [24] Carbendazim C57BL/6 mice n = 32; Control n = 8; L-CBZ n = 8; M-CBZ n = 8; H-CBZ n = 8 Faeces | ↑ Actinobacteria ↓ Bacteroidetes and Verrucomicrobia | ↑ Propionate and butyrate in CBZ ↓ Acetate in CBZ | Hyperlipidaemia |
| Ref., Xenobiotic, Doses | Metabolite Modifications | Gut Microbiota Taxa Modification | Health Status |
|---|---|---|---|
| [25] Bisphenol A BPA (2 and 20 µg/L) | ↑ Serotonin in BPA-female ↓ Serotonin in BPA-male | ↑ Hyphomicrobium in BPA | Intestinal health and oxidative stress |
| [26] Bisphenol F BPF (0.5, 5, and 50 µg/L) | ↑ Glutamate, arginine, succinate, D-serine, L-tyrosine, adenine, inosine, hypoxanthine, xanthine, and guanine in BPF | ↑ Ralstonia in BPF ↓ Gemmobacter in BPF | Hepatic fibrosis and steatosis |
| [27] Bisphenol F BPF (2, 20, and 200 μg/L) | L-glutamine, L-tyrosine, L-tryptophan, L-glutamate, L-leucine, L-isoleucine, and L-proline in BPF | ↑ Microbacterium, Mycobacterium, Pseudomonas, and uncultured bacteria in BPF ↓ Burkholderia–Caballeronia–Paraburkholderia, Bifidobacterium, Cetobacterium, and Halomonas in BPF | Neurotoxicity |
| [28] Chlorpyrifos CPF (30, 100, and 300 µg/L) | Celobiose, α-tocopherol, gentiobiose, β-mannosylglycerate, glucose-6-phosphate, gluconic acid, isomaltose, 3-hydroxyflavone, L-malic acid, glucose, mannose, 3-hydroxypropionic acid, maltose, lactic acid, 4-aminobutyric acid, phenyl β-D glucopyranoside, N-acetyl- β-D-mannosamine, fructose, heptadecanoic acid, neohesperidin, 2-monopalmitin, adrenosterone, 7-α-hydroxycholesterol, ethanolamine, glycerol, D-glyceric acid, 2-hydroxyvaleric acid, 4-cholesten-3 one 4, ergosterol, myristic acid, L-4-hydroxyphenylglycine, 3-hydroxy-L-proline, O-methylthreonine, cycloleucine, picolinic acid, shikimic acid, glutamic acid, β-alanine, oxoproline, serine, urail, phenanthrene, abietic acid, pantothenic acid, and cis-gondoic acid in CPF | ↑ β-Proteobacteria in CPF ↓ α-Proteobacteria and γ-Proteobacteria in CPF | Hepatic metabolism and oxidative stress |
| [29] Chlorpyrifos Micro-Siced Polystyrene CPF (0.02, 0.2, 2, 20, and 200 μg/g) mPS (50 and 500 μg/g) | Chlorpyrifos-oxon and mPS-adsorbed chlorpyrifos (MIX1 and MIX2) | ↑ Xanthobacter and Methylobacterium-Methylorubrum in CPF ↓ ZOR0006, Chitinibacter, Paucibacter, Rhodococcus, and Cetobacterium in CPF ↑ Vibrio, Rhodococcus, and unclassified_f_Rhizobiaceae in chlorpyrifos-loaded mPS ↓ Aeromonas, Cetobacterium, Chitinibacter, and Flavobacterium in chlorpyrifos-loaded mPS | Hepatic metabolism, intestinal health, oxidative stress and locomotivity |
| [30] Propamocarb PM (100 and 1000 μg/L) | ↑ Sucrose-6-phosphate, 1-kestose, glucose-6-phosphate, glycerol, lactic acid, thymine, ribitol, ribulose-5-phosphate, oxoproline, orotic acid, pyridoxine, glutamic acid, and succinic acid in PM ↓ 6-methylmercaptopurine, 3-aminoisobutyric acid, glutamine, lysine, isoleucine, L-allothreonine, glycine, serine, isocitric acid, fumaric acid, L-malic acid, aspartic acid, phenylalanine, valine, threonine, and methionine in PM | Deefgea, Flavobacterium, Cupriavidus, Megamonas, Sediminibacterium, Acinetobacter, Cetobacterium, and Shewanella in PM | Hepatic metabolism |
| [31] Carbendazim CBZ (30 and 100 μg/L) | ↓ Glucose and pyruvate in CBZ | ↑ Phascolarctobacterium, Macellibacteroides, Shewanella, Faecalibaculum, Turicibacter, [Eubacterium]_xylanophilum_group, and Crenobacter in CBZ ↓ Erysipelatoclostridium, Chryseobacterium, Bryobacter, Gemmobacter, Caulobacter, Nicotiana_otophora, Pelomonas, and Alistipes in CBZ | Hepatic metabolism |
| [32] Difenoconazole DFZ (0.4, 1, and 2 mg/L) | ↑ Triglycerides and malondialdehyde in DFZ | ↑ Plesiomonas, Aeromonas, Firmicutes, Ochrobactrum, Rhodobacteraceae, Enterobacteriaceae, Comamonadaceae, Gemmobacter, Shewanella, and Bacteroides in DFZ ↓ Cetobacterium in DFZ | Hepatic metabolism and intestinal health |
| [33] Imazalil IMZ (100 and 1000 μg/L) | ↑ Cellobiose, maltose, maltotriose, L-threose, sucrose-6-phosphate, trehalose-6-phosphate, 3-aminoisobutyric acid, ribose-5-phosphate, 6-phosphogluconic acid, pyrubic acid, citramalic acid, cholesterol, palmitic acid, phytanic acid, heptadecanoic acid, stearic acid, arachidonic acid, and myristic acid in IMZ ↓ AMP, dTMP, glutamine, alanine, serine, threonine, isoleucine, proline, valine, malate, pantothenic acid, taurine, orotic acid, and lauric acid in IMZ | ↑ Fusobacteria and Firmicutes in IMZ ↓ Bacteroidetes and Proteobacteria in IMZ | Hepatic metabolism and intestinal health |
| [34] Di-2-(ethylhexyl) phthalate DEHP (10, 33, and 100 μg/L) | ↑ Triglycerides, pyruvate, and glucose in DEHP-female ↑ Triglycerides, pyruvate, and non-esterified fatty acids in DEHP-male | ↑ Proteobacteria and Firmicutes in DEHP-female ↓ Fusobacteria, Bacteroidetes, and Actinobacteria in DEHP-female ↑ Proteobacteria and Bacteroidetes in DEHP-male ↓ Fusobacteria, Firmicutes, and Actinobacteria in DEHP-male | Intestinal health and obesity |
| [35] Di-2-(ethylhexyl) phthalate DEHP (3 mg/kg) | ↑ Thioguanine in DEHP-female ↓ D-fructose-6-phosphate in DEHP-female ↑ Choline, ethanolamine, and thioredoxin in DEHP-male ↓ L-Glutamine, L-citruline, and folic acid in DEHP-male | ↑ Fusobacteria, Bacteroidetes, and Verrucomicrobia in DEHP | Intestinal health |
| [36] Polybrominated Diphenyl Ethers PBDE mixture (DE-71) (5 ng/L) | ↓ Serotonin in DE-71 | ↑ Streptococcus, Bacillus, Helicobacter, Moraxella, Fischerella, Xanthomarina, and Tannerella in DE-71 male ↓ Lactobacillus, Chlamydia, Glutamicibacter, Paenibacillus, Olsenella, Ralstonia, Mycoplasma, Mucilaginibacter, Ruminiclostridium, unclassified Firmicutes sensu stricto, Eubacterium, Prevotella, and Fusobacterium in DE-71 male ↑ Streptococcus, Lactobacillus, Haemophilus, Leptospira, Paenibacillus, Staphylococcus, Helicobacter, Mucilaginibacter, Neisseria, Pseudomonas, Aeromonas, and Listeria in DE-71 female ↓ Acinetobacter, Bacillus, Glutamicibacter, Mycoplasma, Ruminiclostridium, unclassified Lachnospiraceae, unclassified Firmicutes sensu stricto, Eubacterium, Moraxella, Fischerella, Fusobacterium, Plesiomonas, Burkholderia, Xanthomarina, Xenorhabdus, Nonomuraea, Alicyclobacillus, and Mannheimia in DE-71 female | Intestinal health and oxidative stress |
| [37] Methylparaben MeP (1, 3, 10 μg/L) | ↑ Serotonin in MeP-male ↓ Serotonin in MeP-female | ↑ Mycoplasma and Cetobacterium in MeP | Intestinal health and oxidative stress |
| Ref., NGP Strain, Doses, Target | Metabolite Modifications | Health Effects |
|---|---|---|
| [38] Akkermansia muciniphila (ATCC BAA-835), 2 × 108 CFU/200 µL, C57BL/6 mice | ↑ α-Tocopherol and β-sitosterol ↓ Citrulline and ornithine Vitamin and Amino acid metabolites | ↑ Glucose tolerance ↓ Weight gain ↓ Fat mass |
| [39] Akkermansia muciniphila, 1 × 108 to 109 CFU/100 µL, C57BL/6 mice | ↑ N1, N12-Diacetylspermine, N1-acetylspermine, N1-acetylspermidine, N1, N8-diacetylspermidine, spermidine, ornithine, putrescine, acetate, propionate, butyrate, 2-hydroxybutyrate, ketoisovaleric acid, ketoisocaproic acid, ferulic acid, 2-hydroxy-3-methylbutyric acid, deoxycholic acid, hyodeoxycholic acid, murideoxycholic acid, hyocholic acid, lithocholic acid, Ω-muricholic acid, taurodeoxycholic acid, tauro-muricholic acid, taurohyodeoxycholic acid, tauroursodeoxycholic acid, chenodeoxycholic acid, β-muricholic acid, and ursodeoxycholic acid Polyamine metabolites, short-chain fatty acids and bile acid metabolites | ↑ Pleiotropic metabolic effects supporting gut homeostasis and host health. ↑ Antiaging and anticancer effects |
| [40] Faecalibacterium prausnitzii (ATCC 27766), 2 × 108 CFU/220 µL, C57BL/6N mice | ↑ Dihomo-γ-linolenic acid (20:3n6) ↓ Stearic acid (18:0), arachidonic acid (20:4n6), eicosapentaenoic acid (20:5n3), and docosahexanoic acid (22:6n3) ↓ Palmitic acid (16:00) ↓ Linoleic acid (18:2n-6), α-linoleic acid (18:3n3), and eicosapentaenoic acid (20:5n3) Fatty acid and lipid metabolites | ↑ Weight gain ↓ Hepatic injury |
| [41] Bacteroides uniformis (CECT 7771), 5 × 107 CFU/day, C57BL/6J mice | ↑ Butyrate, stearic acid (18:0), and arachidic acid (20:0)↓ Monounsaturated fatty acids, diunsaturated fatty acids, and polyunsaturated fatty acids Short-chain fatty acids and fatty acid lipid metabolites | ↑ Glucose tolerance ↓ Weight gain ↓ Serum cholesterol |
| [42] Bacteroides acidifaciens (JCM10556), 5 × 109 CFU/100 µL, C57BL/6 mice | ↑ Cholate and taurine ↓ Butyrate Short-chain fatty acids and bile acid metabolites | ↓ Weight gain ↓ Fat mass ↓ Insuline resistance |
| [43] Clostridium butyricum (CGMCC0313.1), 2.5 × 108 CFU/kg/day, NOD mice | ↑ Butyric acid Short-chain fatty acids metabolites | ↓ Diabetes ↓ Diabetes-induced energy metabolic dysfunction |
| [44] Prevotella copri (DSM 18205), 5 × 108 CFU, GK/Ox rats | ↑ Cholic acid, allolithocholic acid, chenodeoxycholic acid, and ω-muricholic acid Total and primary bile acids metabolites | ↑ Glucose tolerance |
| Ref., NGP Strain, Doses, Target | Metabolite Modifications | Health Effects |
| [45] Akkermansia muciniphila (ATCC BAA-835), 1 × 108–109 CFU/mL, C57BL/6N mice | Not determined | ↓ Fatty liver disease |
| [46] Akkermansia muciniphila MucT (ATTC BAA-835), 2 × 108 CFU/200 µL, Ercc1−/Δ7 mice | Not determined | ↑ Restoration of mucus layer |
| [47] Akkermansia muciniphila MucT (ATTC BAA-835), 2 × 108 CFU/150 µL, C57BL/6J mice | Not determined | ↑ Glucose tolerance ↓ Body weight ↓ Fat mass gain ↓ Insuline resistance |
| [48] Akkermansia muciniphila MucT (ATTC BAA-835), 1 × 108 CFU/200 µL, C57BL/6 mice | Not determined | ↓ [Cd] in kidney |
| [49] Akkermansia muciniphila strain (139) and (ATCCT), 2 × 108 CFU/200 µL, C57BL/6 mice | Not determined | ↓ Colitis |
| [50] Akkermansia muciniphilasub, 1 × 109 CFU/200 µL, C57BL/6 mice | Not determined | ↑ Blood glucose control ↓ Weight gain ↓ Liver steatosis ↓ Memory decay |
| [51] Akkermansia muciniphila MucT (CCUG 64013), 1.5 × 109 CFU/200 µL, C57BL/6 mice | Not determined | ↑ Restoration of mucus layer ↓ Hepatic injury, steatosis |
| [52] Akkermansia muciniphila (DSM 22959), 5 × 106–5×108/500 µL, SD rats | Not determined | ↑ Liver function |
| [53] Akkermansia muciniphila (GP01), 5 × 109 CFU/200 µL, APP/PS1 mice | Not determined | ↑ Glucose tolerance ↓ Hyperlipidemia ↓ Hepatic steatosis ↓ Intestinal barrier dysfunction |
| [54] Bacteroides uniformis (CECT 7771), 1 × 108 CFU, C57BL/6 mice | Not determined | ↓ Weight gain ↓ Cholesterol, triglycerides, glucose |
| [55] Bacteroides uniformis (CECT 7771), 1 × 108–1 × 1010 CFU/day, Wistar rats | Not determined | ↓ Hepatic alanine aminotransferase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres-Sánchez, A.; Ruiz-Rodríguez, A.; Ortiz, P.; Moreno, M.A.; Ampatzoglou, A.; Gruszecka-Kosowska, A.; Monteoliva-Sánchez, M.; Aguilera, M. Exploring Next Generation Probiotics for Metabolic and Microbiota Dysbiosis Linked to Xenobiotic Exposure: Holistic Approach. Int. J. Mol. Sci. 2022, 23, 12917. https://doi.org/10.3390/ijms232112917
Torres-Sánchez A, Ruiz-Rodríguez A, Ortiz P, Moreno MA, Ampatzoglou A, Gruszecka-Kosowska A, Monteoliva-Sánchez M, Aguilera M. Exploring Next Generation Probiotics for Metabolic and Microbiota Dysbiosis Linked to Xenobiotic Exposure: Holistic Approach. International Journal of Molecular Sciences. 2022; 23(21):12917. https://doi.org/10.3390/ijms232112917
Chicago/Turabian StyleTorres-Sánchez, Alfonso, Alicia Ruiz-Rodríguez, Pilar Ortiz, María Alejandra Moreno, Antonis Ampatzoglou, Agnieszka Gruszecka-Kosowska, Mercedes Monteoliva-Sánchez, and Margarita Aguilera. 2022. "Exploring Next Generation Probiotics for Metabolic and Microbiota Dysbiosis Linked to Xenobiotic Exposure: Holistic Approach" International Journal of Molecular Sciences 23, no. 21: 12917. https://doi.org/10.3390/ijms232112917
APA StyleTorres-Sánchez, A., Ruiz-Rodríguez, A., Ortiz, P., Moreno, M. A., Ampatzoglou, A., Gruszecka-Kosowska, A., Monteoliva-Sánchez, M., & Aguilera, M. (2022). Exploring Next Generation Probiotics for Metabolic and Microbiota Dysbiosis Linked to Xenobiotic Exposure: Holistic Approach. International Journal of Molecular Sciences, 23(21), 12917. https://doi.org/10.3390/ijms232112917








