Transcriptome and Metabolome Combined to Analyze Quinoa Grain Quality Differences of Different Colors Cultivars
Abstract
1. Introduction
2. Results
2.1. Grain Metabolism in Four Quinoa Cultivars
2.2. Metabolic Differences among Four Quinoa Cultivars
2.2.1. Analyses of Amino Acids and Their Derivatives
2.2.2. Analyses of Phenolic Acid-Related Metabolites
2.2.3. Analyses of Nucleotides and Their Derivatives
2.2.4. Analyses of Lipid-Related Metabolites
2.2.5. Analyses of Organic Acid-Related Metabolites
2.2.6. Analyses of Alkaloid-Related Metabolites
2.2.7. Analyses of Sugar- and Alcohol-Related Metabolites
2.2.8. Analyses of Lignin- and Coumarin-Related Metabolites
2.2.9. Analyses of Tannin-Related Metabolites
2.2.10. Analyses of Terpenoid-Related Metabolites
2.2.11. Analyses of Vitamin-Related Metabolites
2.2.12. Analyses of Other Metabolites
2.3. Analyses of Correlations between Metabolome and Transcriptome among Different Quinoa Cultivars
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Widely Targeted Metabolome Detection and Analysis
4.3. Transcriptome Sequencing and Data Analysis
4.4. Combined Transcriptome and Metabolome Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaikishun, S.; Li, W.; Yang, Z.; Song, S. Quinoa: In perspective of global challenges. Agronomy 2019, 9, 176. [Google Scholar] [CrossRef]
- Pirzadah, T.B.; Malik, B. Pseudocereals as super foods of 21st century: Recent technological interventions. J. Agric. Food Res. 2020, 2, 100052. [Google Scholar] [CrossRef]
- Reguera, M.; Conesa, C.M.; Gil-Gómez, A.; Haros, C.M.; Pérez-Casas, M.Á.; Briones-Labarca, L.; Bolaños, L.; Bonilla, I.; Álvarez, R.; Pinto, K.; et al. The impact of different agroecological conditions on the nutritional composition of quinoa seeds. PeerJ 2018, 14, e4442. [Google Scholar] [CrossRef] [PubMed]
- Adolf, V.I.; Shabala, S.; Andersen, M.N.; Razzaghi, F.; Jacobsen, S.E. Varietal differences of quinoa’s tolerance to saline conditions. Plant Soil 2012, 357, 117–129. [Google Scholar] [CrossRef]
- Hariadi, Y.; Marandon, K.; Tian, Y.; Jacobsen, S.-E.; Shabala, S. Ionic and osmotic relations in quinoa (Chenopodium quinoa willd.) plants grown at various salinity levels. J. Exp. Bot. 2011, 62, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Sezgin, A.C.; Sanlier, N. A new generation plant for the conventional cuisine: Quinoa (Chenopodium quinoa Willd.). Trends Food Sci. Technol. 2019, 86, 51–58. [Google Scholar] [CrossRef]
- Van de Vondel, J.; Lambrecht, M.A.; Delcour, J.A. Osborne extractability and chromatographic separation of protein from quinoa (Chenopodium quinoa Willd.) wholemeal. LWT 2020, 126, 109321. [Google Scholar] [CrossRef]
- Gawlik-Dziki, U.; Świeca, M.; Sułkowski, M.; Dziki, D.; Baraniak, B.; Czyż, J. Antioxidant and anticancer activities of Chenopodium quinoa leaves extracts–in vitro study. Food Chem. Toxicol. 2013, 57, 154–160. [Google Scholar] [CrossRef]
- Lápez-Cervantes, J.; Sánchez-Machado, D.; de la Mora-López, D.S.; Sanches-Silva, A. Quinoa (Chenopodium quinoa Willd.): Exploring a superfood from Andean indigenous cultures with potential to reduce cardiovascular disease (CVD) risk markers. Curr. Mol. Pharmacol. 2021, 14, 925–934. [Google Scholar] [CrossRef]
- Suárez-Estrella, D.; Torri, L.; Pagani, M.A.; Marti, A. Quinoa bitterness: Causes and solutions for improving product acceptability. J. Sci. Food. Agr 2018, 98, 4033–4041. [Google Scholar] [CrossRef]
- Maliro, M.F.A.; Guwela, V. Quinoa breeding in Africa: History, goals, and progress. In Quinoa: Improvement and Sustainable Production; Wiley-Blackwell: Hoboken, NJ, USA, 2015; pp. 161–172. [Google Scholar] [CrossRef]
- Garcia, M.; Condori, B.; Castillo, C.D. Agroecological and agronomic cultural practices of quinoa in South America. In Quinoa: Improvement and Sustainable Production; Wiley-Blackwell: Hoboken, NJ, USA, 2015; pp. 25–46. [Google Scholar] [CrossRef]
- Basantes-Morales, E.R.; Alconada, M.M.; Pantoja, J.L. Quinoa (Chenopodium quinoa Willd) production in the Andean Region: Challenges and potentials. J. Exp. Agric. Int. 2019, 36, 1–18. [Google Scholar] [CrossRef]
- Abugoch, J.; Lilian, E. Quinoa (Chenopodium quinoa Willd.): Composition, chemistry, nutritional, and functional properties. Adv. Food Nutr. Res. 2009, 58, 1–31. [Google Scholar] [CrossRef]
- Han, Y.; Chi, J.; Zhang, M.; Zhang, R.; Fan, S.; Huang, F.; Xue, K.; Liu, L. Characterization of saponins and phenolic compounds: Antioxidant activity and inhibitory effects on α-glucosidase in different varieties of colored quinoa (Chenopodium quinoa Willd). Biosci. Biotechnol. Biochem. 2019, 83, 2128–2139. [Google Scholar] [CrossRef] [PubMed]
- Pereira, E.; Encina-Zelada, C.; Barros, L.; Gonzales-Barron, U.; Cadavez, V.; Ferreira, I.C. Chemical and nutritional characterization of Chenopodium quinoa Willd (quinoa) grains: A good alternative to nutritious food. Food Chem. 2019, 280, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Z.; Wang, Z.; Hao, Y.; Wang, Y.; Yang, Z.; Li, W.; Wang, J. Physicochemical and functional properties of soluble dietary fiber from different colored quinoa varieties (Chenopodium quinoa Willd). J. Cereal Sci. 2020, 95, 103045. [Google Scholar] [CrossRef]
- Buzzini, P.; Arapitsas, P.; Goretti, M.; Branda, E.; Turchetti, B.; Pinelli, P.; Romani, A. Antimicrobial and antiviral activity of hydrolysable tannins. Mini-Rev. Med. Chem. 2008, 8, 1179. [Google Scholar] [CrossRef]
- Rauf, A.; Imran, M.; Abu-Izneid, T.; Ul-Haq, I.; Patel, S.; Pan, X.; Naz, S.; Silva, A.S.; Saeed, F.; Suleria, H.A.R.; et al. Proanthocyanidins: A comprehensive review. Biomed. Pharmacother. 2019, 116, 108999. [Google Scholar] [CrossRef]
- Qu, W.; Shi, S.; Li, P.; Pan, Z.; Venkitasamy, C. Extraction kinetics and properties of proanthocyanidins from pomegranate peel. Int. J. Food Eng. 2014, 10, 683–695. [Google Scholar] [CrossRef]
- Vega-Gálvez, A.; Miranda, M.; Vergara, J.; Uribe, E.; Puente, L.; Martínez, E.A. Nutrition facts and functional potential of quinoa (Chenopodium quinoa Willd.), an ancient Andean grain: A review. J. Sci. Food Agric. 2010, 90, 2541–2547. [Google Scholar] [CrossRef]
- Leinonen, I.; Iannetta, P.P.M.; Rees, R.M.; Russell, W.; Watson, C.; Barnes, A.P. Lysine supply is a critical factor in achieving sustainable global protein economy. Front. Sustain. Food Syst. 2019, 3, 27. [Google Scholar] [CrossRef]
- Quiroga, C.; Escalera, R.; Aroni, G.; Bonifacio, A.; González, J.A.; Villca, M.; Saravia, R.; Ruiz, A. Traditional processes and technological innovations in quinoa harvesting, processing and industrialization. In State of the Art Report on Quinoa around the World in 2013; FAO: Rome, Italy; CIRAD: Paris, France, 2015; pp. 258–296. ISBN 978-92-5-108558-5. [Google Scholar]
- Wang, S.; Zhu, F. Formulation and quality attributes of quinoa food products. Food Bioprocess Technol. 2016, 9, 49–68. [Google Scholar] [CrossRef]
- Ayseli, M.T.; Yilmaz, M.T.; Cebi, N.; Sagdic, O.; Ozmen, D.; Capanoglu, E. Physicochemical, rheological, molecular, thermal and sensory evaluation of newly developed complementary infant (6–24 months old) foods prepared with quinoa (Chenopodium quinoa Willd.) flour. Food Chem. 2020, 315, 126208. [Google Scholar] [CrossRef] [PubMed]
- Väkeväinen, K.; Ludena-Urquizo, F.; Korkala, E.; Lapveteläinen, A.; Peräniemi, S.; von Wright, A.; Plumed-Ferrer, C. Potential of quinoa in the development of fermented spoonable vegan products. LWT-Food Sci. Technol. 2020, 120, 108912. [Google Scholar] [CrossRef]
- Angeli, V.; Miguel Silva, P.; Crispim Massuela, D.; Khan, M.W.; Hamar, A.; Khajehei, F.; Graeff-Hönninger, S.; Piatti, C. Quinoa (Chenopodium quinoa Willd.): An overview of the potentials of the “Golden Grain” and socio-economic and environmental aspects of its cultivation and marketization. Foods 2020, 9, 216. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guoyo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Eriksson, L.; Kettanehwold, N.; Trygg, J.; Wikström, C.; Wold, S. Multi-and Megavariate Data Analysis: Part I: Basic Principles and Applications; Umetrics Inc.: San Jose, CA, USA, 2006. [Google Scholar]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Varet, H.; Loraine, B.; Jean-Yves, C.; Marie-Agnès, D. SARTools: A DESeq2- and EdgeR-Based R pipeline for comprehensive differential analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Gonzalez, I.; Dejean, S.; Martin, P.; Baccini, A. CCA: An R Package to Extend CanonicalCorrelation Analysis. J. Stat. Softw. 2008, 23, 1–14. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Kong, Z.; Huan, X.; Li, L.; Zhang, P.; Wang, Q.; Guo, Y.; Zhu, W.; Qin, P. Transcriptomics and metabolomics analyses of the mechanism of flavonoid synthesis in seeds of differently colored quinoa strains. Genomics 2022, 114, 138–148. [Google Scholar] [CrossRef]

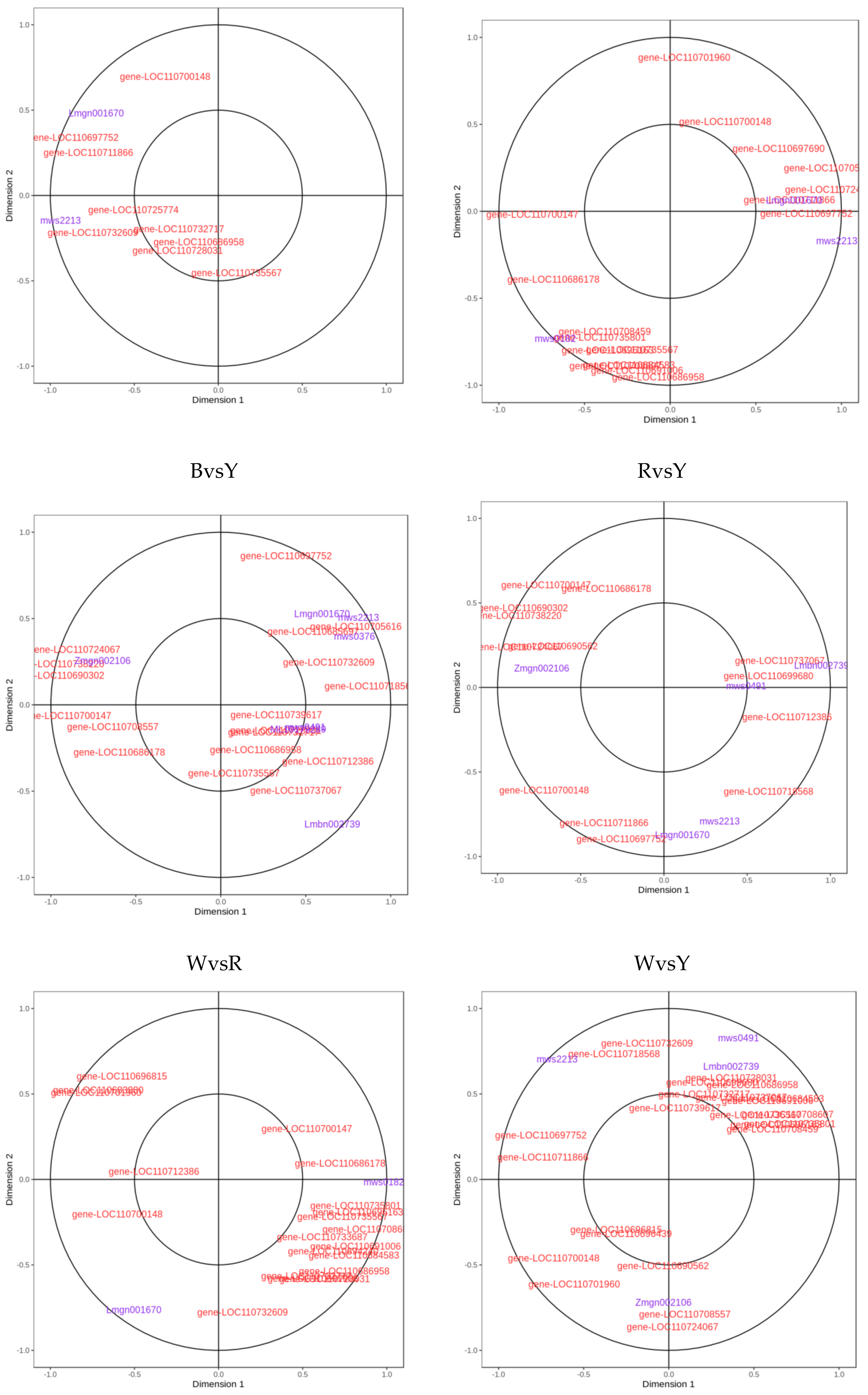
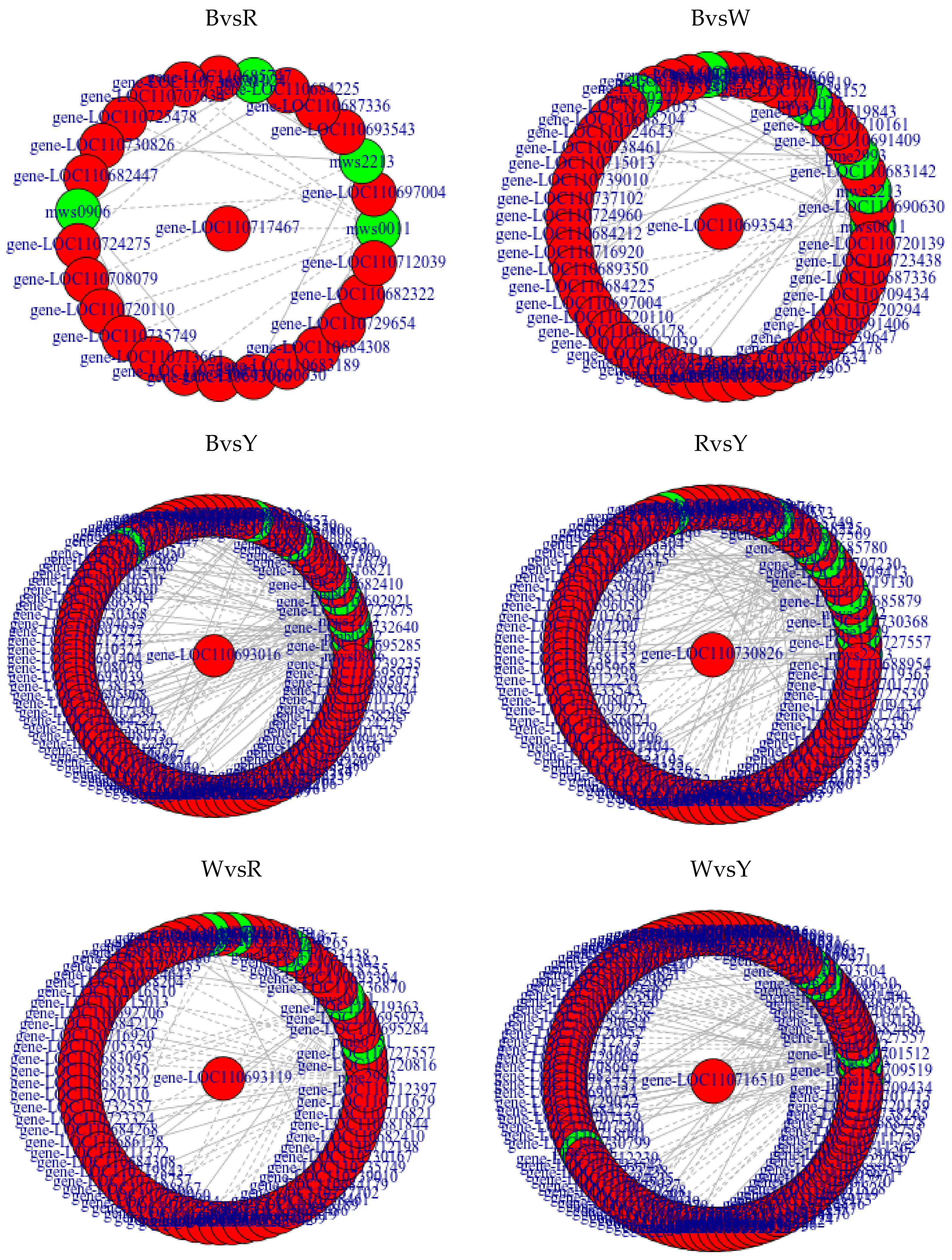

| Differential Amino Acids and Their Derivatives | Log2FC | |||||
|---|---|---|---|---|---|---|
| BvsR | BvsW | BvsY | RvsY | WvsR | WvsY | |
| L-threo-3-Methylaspartate | - | - | - | - | - | −1.02 |
| N-Acetyl-L-phenylalanine | - | - | 1.61 | 2.02 | - | 1.76 |
| Pipecolic acid | - | 1.68 | - | - | −1.89 | −1.40 |
| 3,4-Dihydroxy-L-phenylalanine | −7.05 | −9.92 | −10.07 | −3.02 | - | - |
| Homoarginine | - | - | 1.31 | 1.20 | - | 1.11 |
| N-Acetyl-L-glycine | - | - | −1.04 | - | −1.02 | −1.34 |
| 3-Hydroxy-3-methylpentane-1,5-dioic acid | - | −1.11 | - | - | - | |
| L-Homocystine | - | 1.09 | - | - | −1.67 | - |
| γ-Glu-Cys | - | 1.11 | - | - | −1.70 | |
| L-Ornithine | - | - | - | - | −1.40 | - |
| L-Alanine | - | - | - | - | −1.09 | −1.34 |
| L-Saccharopine | - | - | - | −1.03 | - | −1.16 |
| S-(5′-Adenosy)-L-homocysteine | - | - | - | - | - | −1.13 |
| L-Methionine | - | - | - | - | - | −1.06 |
| L-Tyramine | −1.69 | −1.31 | −1.77 | - | - | - |
| 2,6-Diaminooimelic acid | - | - | −1.57 | - | - | - |
| N-Acetyl-L-leucine | - | - | −1.07 | - | - | - |
| N-Acetyl-L-Glutamine | - | - | - | - | −1.06 | - |
| L-Alanyl-L-Alanine | - | - | −1.03 | −1.23 | - | −1.49 |
| N-Acetyl-L-glutamic acid | - | - | - | - | −1.14 | −1.50 |
| L-Lysine | - | 1.04 | - | - | −1.13 | - |
| L-Glutamic acid | - | - | −1.34 | - | - | - |
| L-Citrulline | - | 1.19 | - | - | −1.25 | −1.58 |
| L-Proline | - | - | −1.68 | - | - | −2.39 |
| L-Glutamine-O-glycoside | - | 1.76 | −1.89 | −1.76 | −1.89 | −3.65 |
| N-Acetyl-L-Tryptophan | - | - | - | - | 1.02 | 1.81 |
| L-Glutaminyl-L-valyl-L-valyl-L-cysteine | - | - | −1.65 | −1.90 | - | - |
| L-Aspartic acid-O-diglucoside | - | - | −2.02 | −1.73 | - | −2.11 |
| L-Glycyl-L-phenylalanine | - | 1.16 | - | - | −1.46 | −1.18 |
| L-Glycyl-L-isoleucine | - | 1.25 | - | - | −1.56 | −1.22 |
| L-Alanyl-L-Phenylalanine | - | 1.95 | - | - | −1.75 | −2.17 |
| Oxiglutatione | - | - | - | −1.37 | - | −1.95 |
| O-Acetylserine | - | - | −2.18 | −2.01 | −1.16 | −3.17 |
| N-Glycyl-L-leucine | - | 1.31 | -- | - | −1.60 | −1.26 |
| L-Phenylalanyl-L-phenylalanine | - | 1.78 | 1.61 | - | - | - |
| S-(Methyl)glutathione | - | - | −1.03 | - | - | - |
| L-Threonine | - | −1.63 | −1.10 | - | 1.03 | - |
| L-Asparagine | - | −1.16 | - | - | - | - |
| N-Methylglycine | - | - | −1.28 | - | - | −1.11 |
| Cycloleucine | - | 1.87 | - | - | −2.09 | −1.58 |
| L-Isoleucyl-L-Aspartate | - | 1.48 | - | - | −2.15 | −2.21 |
| L-Valyl-L-Phenylalanine | - | 1.01 | - | - | −1.12 | - |
| L-Prolyl-L-Phenylalanine | - | - | - | −1.10 | - | −1.22 |
| L-Homomethionine | - | 1.32 | - | - | - | −1.48 |
| Differential Tannin Species | Log2FC | ||||
|---|---|---|---|---|---|
| BvsR | BvsW | BvsY | RvsY | WvsR | |
| Gambiriin B3 | - | −11.00 | −11.00 | −10.70 | 10.70 |
| Arecatannin B1 | - | −11.84 | −11.84 | −11.13 | 11.13 |
| Procyanidin B4 | - | −17.73 | −17.73 | −16.95 | 16.95 |
| Cinnamtannin B2 | - | −10.93 | −10.93 | −9.97 | 9.97 |
| Cinnamtannin D1 | - | −12.80 | −12.80 | −12.78 | 12.78 |
| Procyanidin C1 | - | −17.43 | −17.43 | −17.31 | 17.31 |
| 1-O-Galloyl-D-glucose | - | - | - | 1.16 | - |
| Procyanidin B3 | - | −20.94 | −20.94 | −20.86 | 20.86 |
| Procyanidin B2 | - | −20.87 | −20.87 | −20.86 | 20.86 |
| 3-O-Methylgallic Acid | - | 1.52 | - | - | −1.44 |
| Procyanidin B1 | - | −12.42 | −12.42 | −12.07 | 12.07 |
| Epitheaflavic acid-3-O-Gallate | - | −19.58 | −19.58 | −19.72 | 19.72 |
| 2,3-Di-O-galloyl-D-glucose | 1.48 | - | - | −1.52 | 1.09 |
| Enzyme Point | Gene ID | Gene FPKM p Value | |||
|---|---|---|---|---|---|
| B | R | W | Y | ||
| K14454 aspartate aminotransferase, cytoplasmic [EC:2.6.1.1] | gene-LOC110705616 | 66.89 | 55.32 | 32.20 | 33.49 |
| K01692 enoyl-CoA hydratase [EC:4.2.1.17] | gene-LOC110738220 | 16.16 | 16.41 | 20.07 | 35.84 |
| K01593 aromatic-L-amino-acid/L-tryptophan decarboxylase [EC:4.1.1.28 4.1.1.105] | gene-LOC110728031 | 1.94 | 0.97 | 2.94 | 0.93 |
| K01426 amidase [EC:3.5.1.4] | gene-LOC110685697 | 0.10 | 0.02 | 0.02 | 0.00 |
| gene-LOC110718568 | 0.66 | 0.48 | 0.40 | 0.12 | |
| gene-LOC110735801 | 6.66 | 9.07 | 18.08 | 6.32 | |
| gene-LOC110737067 | 0.11 | 0.25 | 0.18 | 0.02 | |
| K00276 primary-amine oxidase [EC:1.4.3.21] | gene-LOC110732717 | 17.69 | 8.78 | 25.55 | 8.83 |
| ID of Metabolites | Name of Different Metabolites | Relative Content of Metabolites | |||
|---|---|---|---|---|---|
| B | R | W | Y | ||
| Zmgn002106 | N-Acetyl-L-phenylalanine | 443363.33 | 334590.00 | 399070.00 | 1355836.67 |
| pmb2620 | Cinnamic acid | 60389.00 | 23857.33 | 17504.33 | 4122.60 |
| mws0491 | Phenethylamine | 1252800.00 | 890490.00 | 1601766.67 | 409326.67 |
| mws0376 | Fumaric acid | 284246.67 | 155803.33 | 161770.00 | 83921.67 |
| mws0182 | p-Hydroxyphenyl acetic acid | 1349833.33 | 1604800.00 | 3897533.33 | 2410566.67 |
| ML10179289 | 2-Phenylethanol | 136783.33 | 112217.33 | 179213.33 | 57010.67 |
| Lmgn001670 | Salicylic acid-2-O-glucoside | 1586490.00 | 383650.00 | 22707.00 | 19206.00 |
| Lmbn002739 | 3-Hydroxycinnamic Acid | 392796.67 | 676676.67 | 640516.67 | 99752.67 |
| The Name of Metabolites | Gene Name | PCC |
|---|---|---|
| D-Erythrose-4-phosphate | gene-LOC110732956 | −0.9 |
| gene-LOC110694674 | −0.9 | |
| Isocitric Acid | gene-LOC110697015 | 0.92 |
| gene-LOC110735050 | −0.92 | |
| gene-LOC110683155 | −0.92 | |
| 3-Phospho-D-glyceric acid | gene-LOC110702638 | 0.97 |
| gene-LOC110735050 | −0.94 | |
| L-Methionine | gene-LOC110709420 | 0.93 |
| gene-LOC110708868 | −0.92 | |
| 2,6-Diaminooimelic acid | gene-LOC110719991 | 0.93 |
| gene-LOC110688007 | −0.92 | |
| N-Acetyl-L-glutamic acid | gene-LOC110726706 | 0.95 |
| gene-LOC110725557 | −0.97 | |
| gene-LOC110683155 | −0.97 | |
| L-Glutamic acid | gene-LOC110712605 | 0.93 |
| gene-LOC110688007 | −0.91 | |
| L-Citrulline | gene-LOC110725557 | −0.91 |
| gene-LOC110688707 | −0.94 | |
| L-Proline | gene-LOC110682443 | 0.91 |
| 2-Isopropylmalic Acid | gene-LOC110709420 | 0.94 |
| gene-LOC110683155 | −0.92 | |
| gene-LOC110725557 | −0.92 | |
| Phosphoenolpyruvate | gene-LOC110712605 | 0.99 |
| gene-LOC110688007 | −0.94 | |
| Anthranilic Acid | gene-LOC110732372 | 0.9 |
| gene-LOC110725557 | −0.9 | |
| gene-LOC110683155 | −0.9 | |
| O-Acetylserine | gene-LOC110704567 | 0.94 |
| gene-LOC110735050 | −0.92 | |
| 3-Methyl-2-Oxobutanoic acid | gene-LOC110683275 | 0.92 |
| gene-LOC110693518 | 0.92 | |
| gene-LOC110683155 | −0.95 | |
| L-Homoserine | gene-LOC110708482 | 0.91 |
| Citric Acid | gene-LOC110726706 | 0.9 |
| gene-LOC110683155 | −0.9 | |
| L-Asparagine | gene-LOC110705616 | 0.93 |
| 3-Isopropylmalic Acid | gene-LOC110726706 | 0.93 |
| gene-LOC110709420 | 0.93 | |
| gene-LOC110725557 | −0.93 |
| The Name of Metabolites | Gene Name | PCC |
|---|---|---|
| Lactobiose | gene-LOC110689873 | 0.92 |
| gene-LOC110729558 | −0.94 | |
| Melibiose | gene-LOC110683754 | −0.9 |
| gene-LOC110717984 | −0.9 | |
| gene-LOC110729558 | −0.94 | |
| Inositol | gene-LOC110710278 | 0.91 |
| gene-LOC110717984 | −0.91 | |
| L-Alanine | gene-LOC110681663 | 0.9 |
| D-Glucose | gene-LOC110700561 | 0.9 |
| D-Ribose | gene-LOC110700561 | 0.91 |
| D-(+)-Trehalose | gene-LOC110689873 | 0.96 |
| gene-LOC110729522 | 0.92 | |
| gene-LOC110729558 | −0.9 | |
| gene-LOC110722638 | −0.92 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Liu, J.; Li, L.; Zhang, P.; Wang, Q.; Qin, P. Transcriptome and Metabolome Combined to Analyze Quinoa Grain Quality Differences of Different Colors Cultivars. Int. J. Mol. Sci. 2022, 23, 12883. https://doi.org/10.3390/ijms232112883
Liu Y, Liu J, Li L, Zhang P, Wang Q, Qin P. Transcriptome and Metabolome Combined to Analyze Quinoa Grain Quality Differences of Different Colors Cultivars. International Journal of Molecular Sciences. 2022; 23(21):12883. https://doi.org/10.3390/ijms232112883
Chicago/Turabian StyleLiu, Yongjiang, Junna Liu, Li Li, Ping Zhang, Qianchao Wang, and Peng Qin. 2022. "Transcriptome and Metabolome Combined to Analyze Quinoa Grain Quality Differences of Different Colors Cultivars" International Journal of Molecular Sciences 23, no. 21: 12883. https://doi.org/10.3390/ijms232112883
APA StyleLiu, Y., Liu, J., Li, L., Zhang, P., Wang, Q., & Qin, P. (2022). Transcriptome and Metabolome Combined to Analyze Quinoa Grain Quality Differences of Different Colors Cultivars. International Journal of Molecular Sciences, 23(21), 12883. https://doi.org/10.3390/ijms232112883




