Cheminformatics Study on Structural and Bactericidal Activity of Latest Generation β-Lactams on Widespread Pathogens
Abstract
1. Introduction
2. Results and Discussion
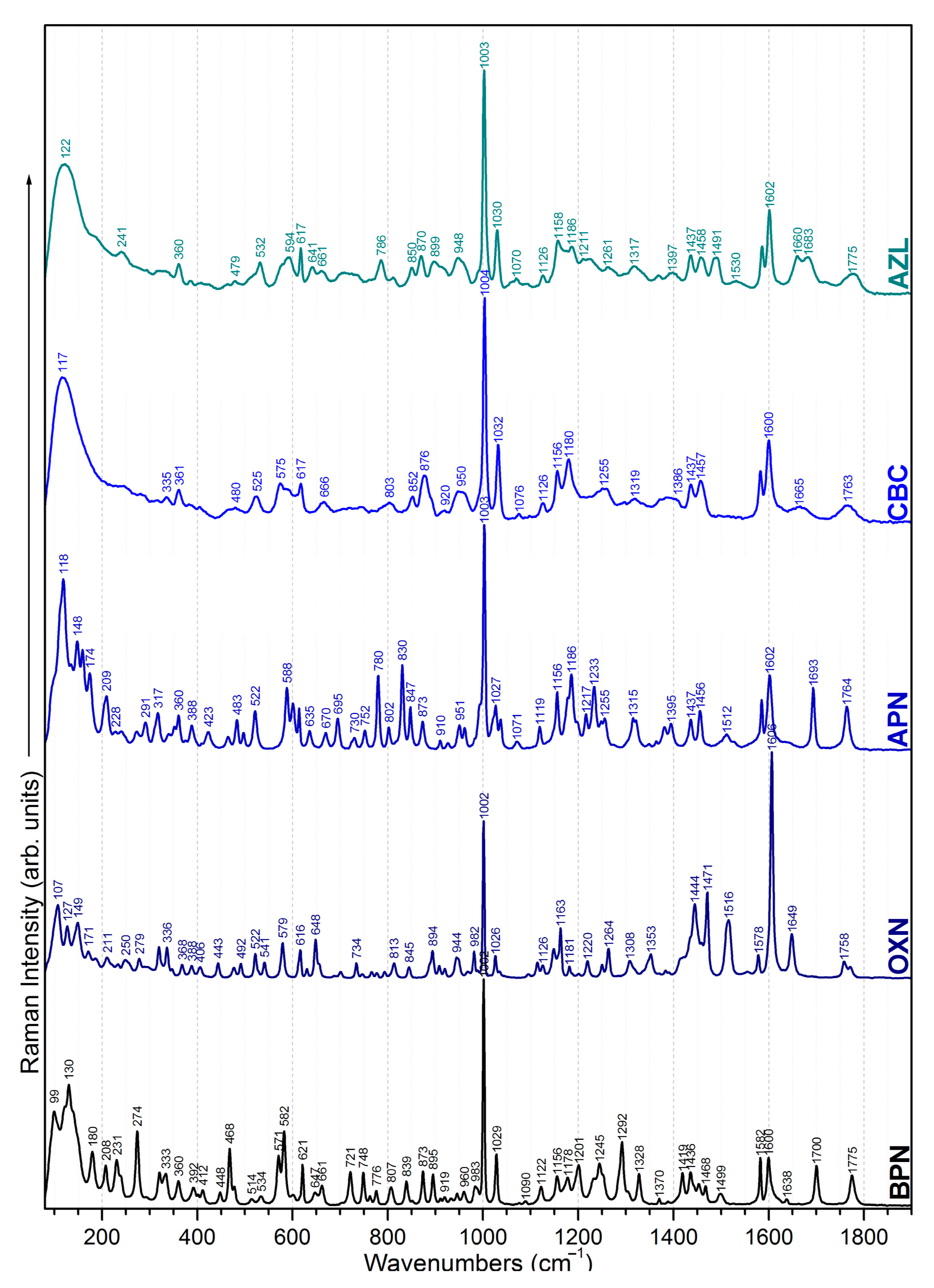
2.1. Chemical Structure (Dis)Similarities Observed in the Vibrational Spectra
2.1.1. Common Bands
2.1.2. Specific Bands
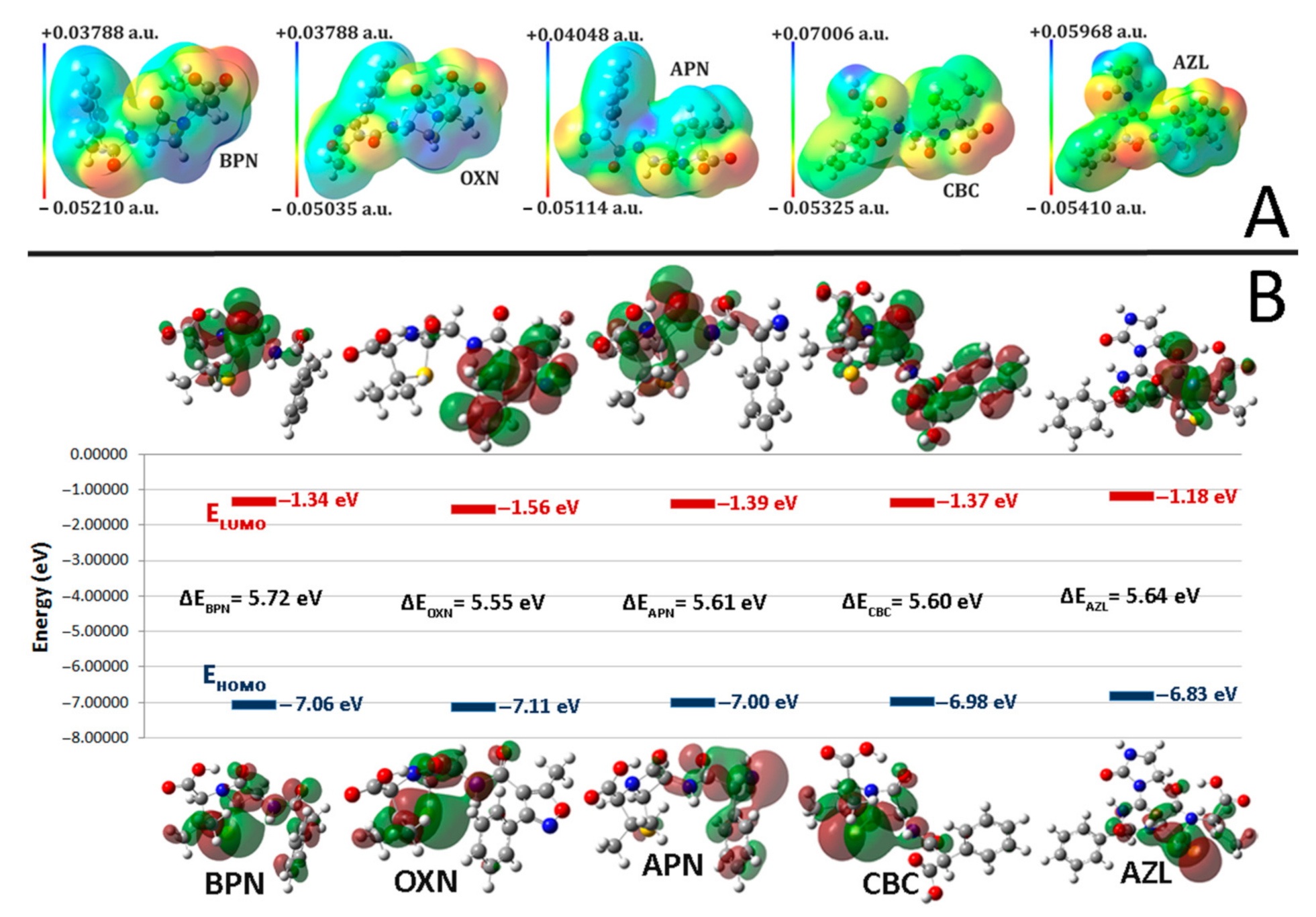
2.2. Molecular Electrostatic Potentials (MEPs)
2.3. Frontier Molecular Orbitals (FMO)
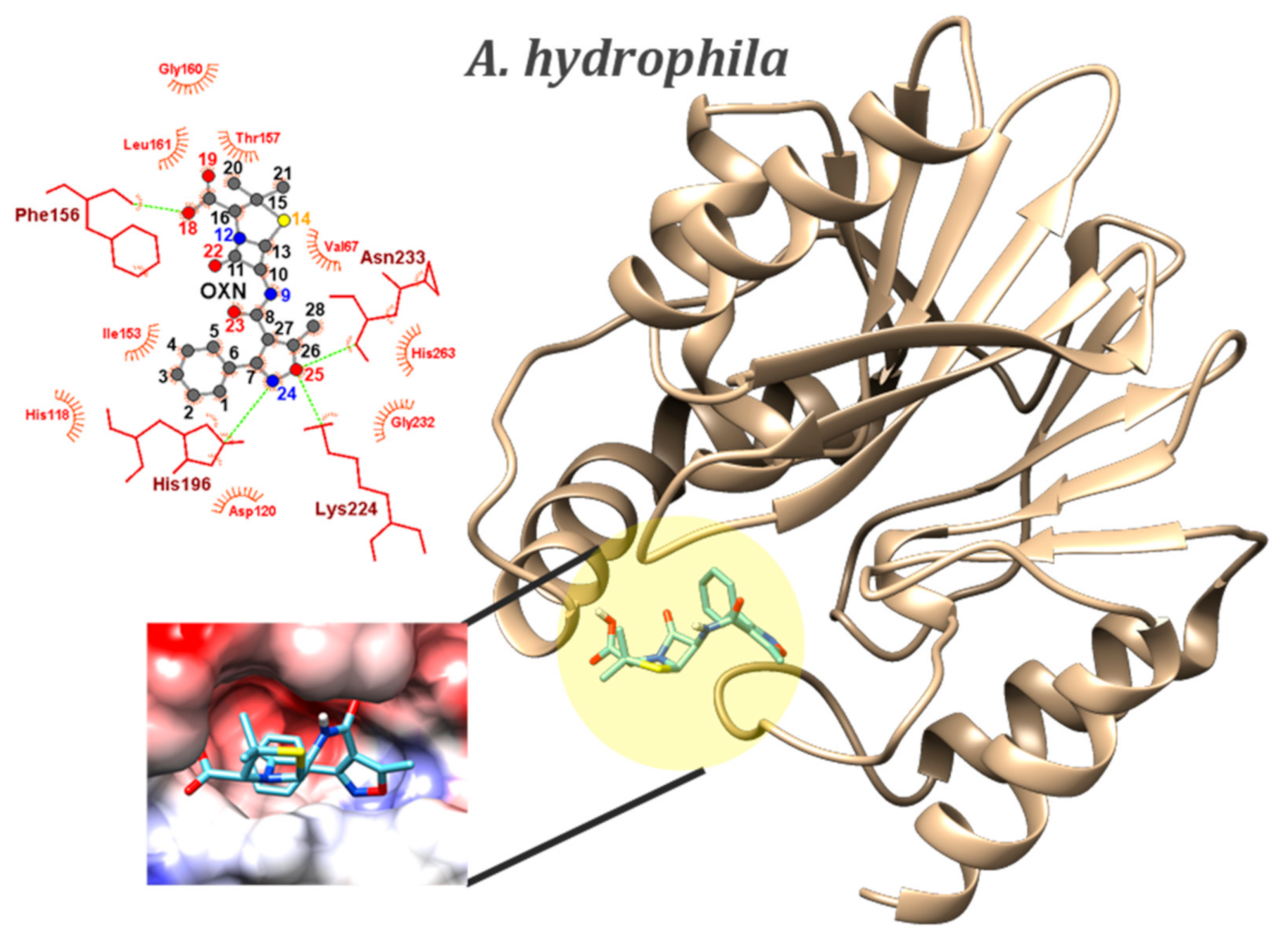
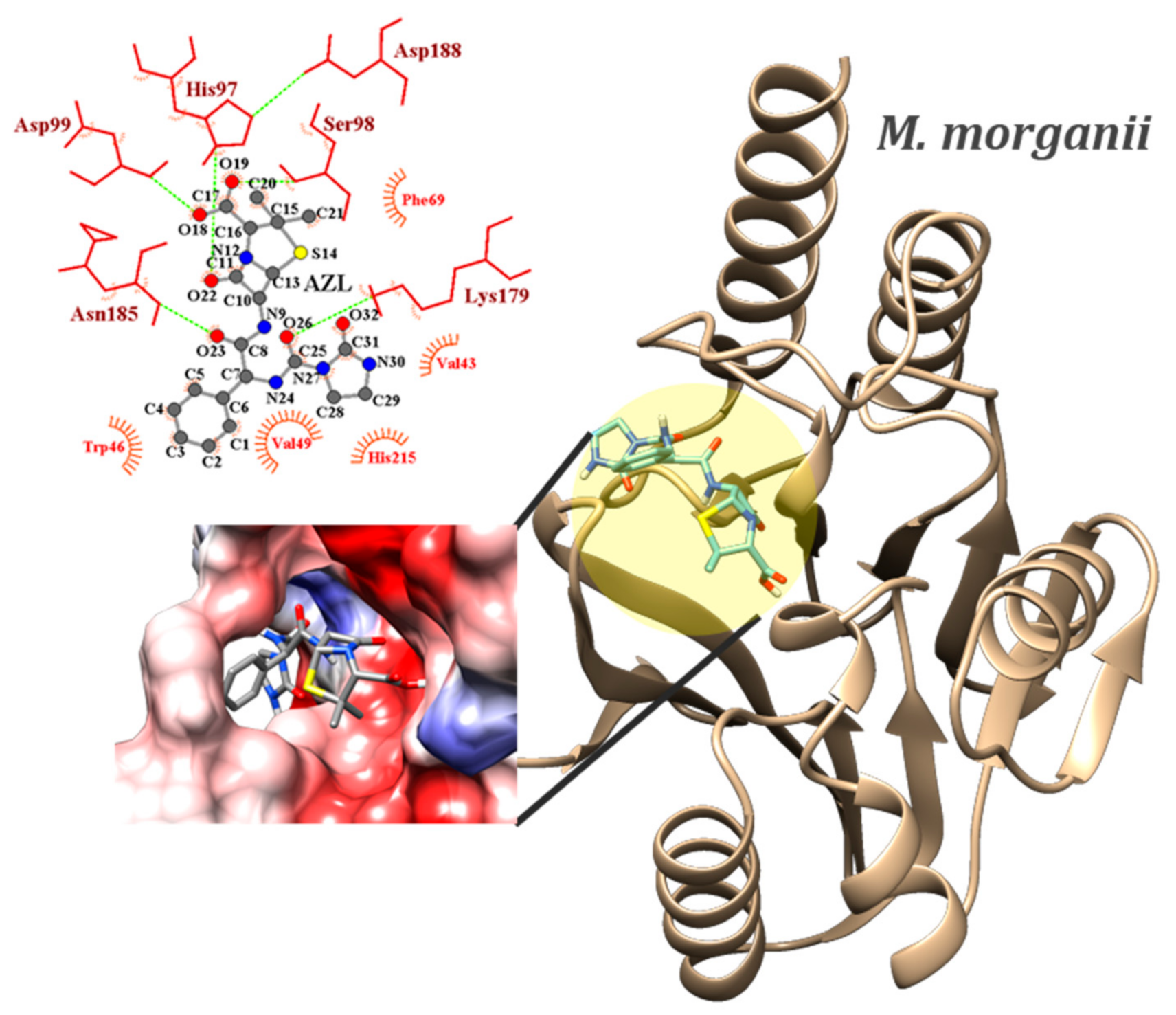
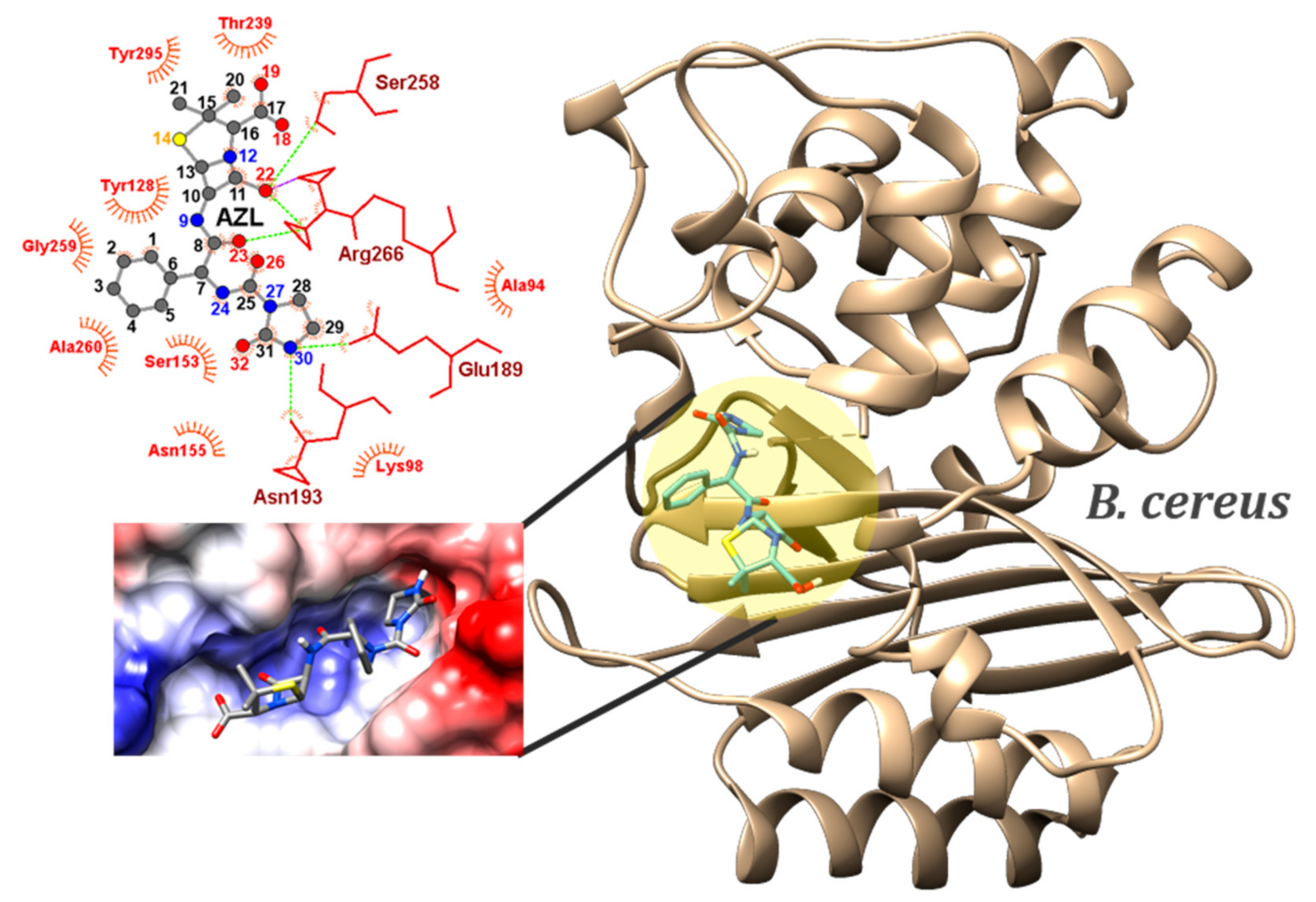
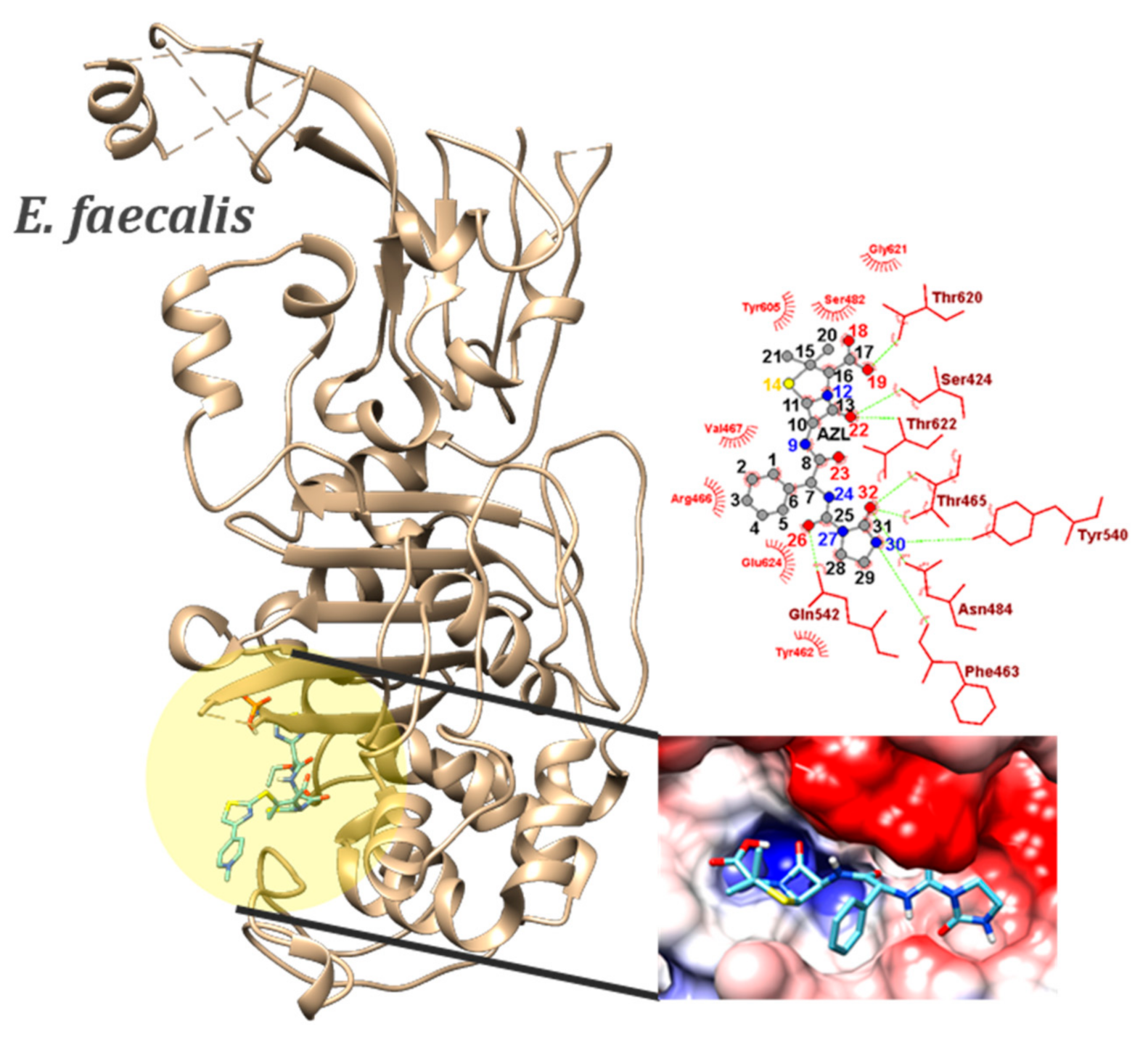
2.4. Molecular Docking
2.5. Disk Diffusion Tests
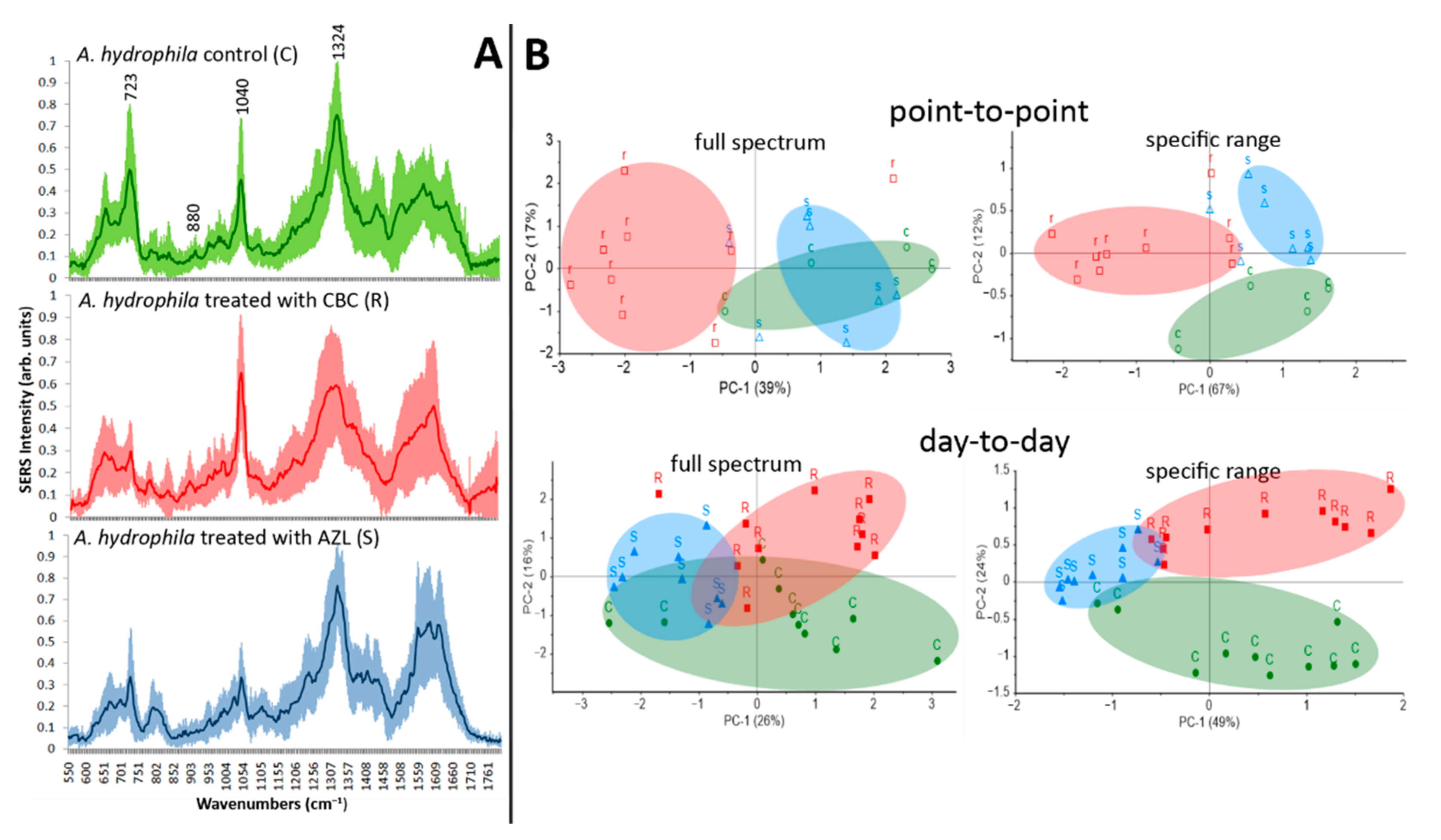
2.6. Bacterial Resistogram Based on SERS and PCA
3. Materials and Methods
3.1. Sample Preparation
3.1.1. Chemicals
3.1.2. Bacterial Strains
3.1.3. Disk Diffusion Tests
3.2. Vibrational Analysis
3.2.1. FT-Raman Analysis
3.2.2. In Situ Silver Nanoparticles (AgNPs) Synthesis
3.2.3. SERS Spectra Measurements
3.3. Computational Details
3.3.1. Input Files for DFT Calculations
3.3.2. Selecting the Ligands and Receptors
3.3.3. Building the Ligand-Receptor Systems
3.3.4. Setting the Search Box
3.3.5. PCA Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sweetman-Martindale, S.C. The Complete Drug Reference; 3-Pharmaceutical Press: London, UK, 2009. [Google Scholar]
- Iliescu, T.; Baia, M.; Pavel, I. Raman and SERS investigations of potassium benzylpenicillin. J. Raman Spectrosc. 2006, 37, 318–325. [Google Scholar] [CrossRef]
- Talebi Tari, M.; Ahmadinejad, N.; Salari, A.A. Investigation of the electronic structure and the structural stability of selected penicillins by Density Functional calculations of 14N nuclear quadrupole resonance parameters. J. Iran. Chem. Soc. 2014, 11, 759–767. [Google Scholar] [CrossRef][Green Version]
- Reipa, V.; Horvath, J.J. Surface-enhanced Raman study of benzylpenicillin. Appl. Spectrosc. 1992, 46, 1009–1013. [Google Scholar] [CrossRef]
- Filgueiras, A.L.; Paschoal, D.; Dos Santos, H.F.; Sant’Ana, A.C. Adsorption study of antibiotics on silver nanoparticle surfaces by surface-enhanced Raman scattering spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 136 Pt B, 979–985. [Google Scholar] [CrossRef]
- Jiang, X.; Qin, X.; Yin, D.; Gong, M.; Yang, L.; Zhao, B.; Ruan, W. Rapid monitoring of benzylpenicillin sodium using Raman and surface enhanced Raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 140, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Bellows, J.C.; Chen, F.P.; Prasad, P.N. Determination of drug polymorphs by laser Raman spectroscopy. I. Ampicillin and Griseofulvin. Drug Dev. Ind. Pharm. 1977, 3, 451–458. [Google Scholar] [CrossRef]
- Baraldi, C.; Tinti, A.; Ottani, S.; Gamberini, M.C. Characterization of polymorphic ampicillin forms. J. Pharm. Biomed. Anal. 2014, 100, 329–340. [Google Scholar] [CrossRef]
- Khan, E.; Shukla, A.; Srivastava, A.; Shweta; Tandon, P. Molecular structure, spectral analysis and hydrogen bonding analysis of ampicillin trihydrate: A combined DFT and AIM approach. New J. Chem. 2015, 39, 9800–9812. [Google Scholar] [CrossRef]
- Shukla, A.; Khan, E.; Srivastava, A.; Tandon, P.; Sinha, K. A computational study on molecular structure, multiple interactions, chemical reactivity and molecular docking studies on 6[D (−) α-amino-phenyl-acetamido] penicillanic acid (ampicillin). Mol. Simul. 2016, 42, 863–873. [Google Scholar] [CrossRef]
- Shukla, A.; Khan, E.; Tandon, P.; Sinha, K. Study of vibrational spectra and hydrogen bonding network in dimeric and tetrameric model of ampicillin using DFT and AIM approach. J. Mol. Struct. 2017, 1131, 225–235. [Google Scholar] [CrossRef]
- Soriano-Correa, C.; Sánchez Ruiz, J.F.; Raya, A.; Esquivel, R.O. Electronic structure and physicochemical properties of selected penicillins. Int. J. Quantum Chem. 2007, 107, 628–636. [Google Scholar] [CrossRef]
- Jiang, X.; Chen, Y.; Du, J.; Yang, M.; Shen, Y.; Li, X.; Han, X.; Yang, L.; Zhao, B. SERS investigation and high sensitive detection of carbenicillin disodium drug on the Ag substrate. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 204, 241–247. [Google Scholar] [CrossRef]
- Tarrat, N.; Benoit, M.; Giraud, M.; Ponchet, A.; Casanove, M.J. The gold/ampicillin interface at the atomic scale. Nanoscale 2015, 7, 14515–14524. [Google Scholar] [CrossRef]
- El-Zahry, M.R.; Refaat, I.H.; Mohamed, H.A.; Rosenberg, E.; Lendl, B. Utility of surface enhanced Raman spectroscopy (SERS) for elucidation and simultaneous determination of some penicillins and penicilloic acid using hydroxylamine silver nanoparticles. Talanta 2015, 144, 710–716. [Google Scholar] [CrossRef]
- Rishi, P.; Vij, S.; Maurya, I.K.; Kaur, U.J.; Bharati, S.; Tewari, R. Peptides as adjuvants for ampicillin and oxacillin against methicillin-resistant Staphylococcus aureus (MRSA). Microb. Pathog. 2018, 124, 11–20. [Google Scholar] [CrossRef]
- He, X.; Li, S.; Yin, Y.; Xu, J.; Gong, W.; Li, G.; Qian, L.; Yin, Y.; He, X.; Guo, T.; et al. Membrane vesicles are the dominant structural components of ceftazidime-induced biofilm formation in an oxacillin-sensitive MRSA. Front. Microbiol. 2019, 10, 571. [Google Scholar] [CrossRef]
- Agnes, M.; Thanassoulas, A.; Stavropoulos, P.; Nounesis, G.; Miliotis, G.; Miriagou, V.; Athanasiou, E.; Benkovics, G.; Malanga, M.; Yannakopoulou, K. Designed positively charged cyclodextrin hosts with enhanced binding of penicillins as carriers for the delivery of antibiotics: The case of oxacillin. Int. J. Pharm. 2017, 531, 480–491. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Xiao, S.; Lu, J.; Sun, L.; An, S. A simple and sensitive AuNPs-based colorimetric aptasensor for specific detection of azlocillin. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 271, 120924. [Google Scholar] [CrossRef]
- Colniță, A.; Marconi, D.; Dina, N.E.; Brezeștean, I.; Bogdan, D.; Turcu, I. 3D silver metallized nanotrenches fabricated by nanoimprint lithography as flexible SERS detection platform. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 276, 121232. [Google Scholar] [CrossRef]
- Liu, T.-T.; Lin, Y.-H.; Hung, C.-S.; Liu, T.-J.; Chen, Y.; Huang, Y.-C.; Tsai, T.-H.; Wang, H.-H.; Wang, D.-W.; Wang, J.-K.; et al. A high speed detection platform based on surface-enhanced Raman scattering for monitoring antibiotic-induced chemical changes in bacteria cell wall. PLoS ONE 2009, 4, e5470. [Google Scholar] [CrossRef]
- Han, Y.-Y.; Lin, Y.-C.; Cheng, W.-C.; Lin, Y.-T.; Teng, L.-J.; Wang, J.-K.; Wang, Y.-L. Rapid antibiotic susceptibility testing of bacteria from patients’ blood via assaying bacterial metabolic response with surface-enhanced Raman spectroscopy. Sci. Rep. 2020, 10, 12538. [Google Scholar] [CrossRef]
- Jia, H.; Zeng, J.; Song, W.; An, J.; Zhao, B. Preparation of silver nanoparticles by photo-reduction for surface-enhanced Raman scattering. Thin Solid Film. 2006, 496, 281–287. [Google Scholar] [CrossRef]
- Mircescu, N.E.; Zhou, H.; Leopold, N.; Chiş, V.; Ivleva, N.P.; Niessner, R.; Wieser, A.; Haisch, C. Towards a receptor-free immobilization and SERS detection of urinary tract infections causative pathogens. Anal. Bioanal. Chem. 2014, 406, 3051–3058. [Google Scholar] [CrossRef]
- Zhou, H.; Yang, D.; Ivleva, N.P.; Mircescu, N.E.; Niessner, R.; Haisch, C. SERS detection of bacteria in water by in situ coating with Ag nanoparticles. Anal. Chem. 2014, 86, 1525–1533. [Google Scholar] [CrossRef]
- Zhou, H.; Yang, D.; Ivleva, N.P.; Mircescu, N.E.; Schubert, S.; Niessner, R.; Wieser, A.; Haisch, C. Label-free in situ discrimination of live and dead bacteria by surface-enhanced Raman scattering. Anal. Chem. 2015, 87, 6553–6561. [Google Scholar] [CrossRef]
- Chen, K.-H.; Lee, S.-H.; Kok, L.-C.; Ishdorj, T.-O.; Chang, H.-Y.; Tseng, F.-G. A 3D-ACEK/SERS system for highly efficient and selectable electrokinetic bacteria concentration/detection/ antibiotic-susceptibility-test on whole blood. Biosens. Bioelectron. 2022, 197, 113740. [Google Scholar] [CrossRef]
- Zhang, P.; Fu, Y.; Zhao, H.; Liu, X.; Wu, X.; Lin, T.; Wang, H.; Song, L.; Fang, Y.; Lu, W.; et al. Dynamic insights into increasing antibiotic resistance in Staphylococcus aureus by label-free SERS using a portable Raman spectrometer. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 273, 121070. [Google Scholar] [CrossRef]
- Skvortsova, A.; Trelin, A.; Kriz, P.; Elashnikov, R.; Vokata, B.; Ulbrich, P.; Pershina, A.; Svorcik, V.; Guselnikova, O.; Lyutakov, O. SERS and advanced chemometrics—Utilization of Siamese neural network for picomolar identification of beta-lactam antibiotics resistance gene fragment. Anal. Chim. Acta 2022, 1192, 339373. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, X.-H.; Su, L.; Wang, H.-Q.; Lin, T.-F.; Fang, Y.-P.; Zhao, H.-M.; Lu, W.-J.; Liu, M.-J.; Liu, W.-B.; et al. Rapid, Label-free prediction of antibiotic resistance in Salmonella typhimurium by surface-enhanced Raman spectroscopy. Int. J. Mol. Sci. 2022, 23, 1356. [Google Scholar] [CrossRef]
- Lin, S.-J.; Chao, P.-H.; Cheng, H.-W.; Wang, J.-K.; Wang, Y.-L.; Han, Y.-Y.; Huang, N.-T. An antibiotic concentration gradient microfluidic device integrating surface-enhanced Raman spectroscopy for multiplex antimicrobial susceptibility testing. Lab A Chip 2022, 22, 1805–1814. [Google Scholar] [CrossRef] [PubMed]
- Bashir, S.; Nawaz, H.; Irfan Majeed, M.; Mohsin, M.; Nawaz, A.; Rashid, N.; Batool, F.; Akbar, S.; Abubakar, M.; Ahmad, S.; et al. Surface-enhanced Raman spectroscopy for the identification of tigecycline-resistant E. coli strains. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 258, 119831. [Google Scholar] [CrossRef] [PubMed]
- Samek, O.; Bernatová, S.; Dohnal, F. The potential of SERS as an AST methodology in clinical settings. Nanophotonics 2021, 10, 2537–2561. [Google Scholar] [CrossRef]
- Szaniawska, A.; Mazur, K.; Kwarta, D.; Pyrak, E.; Kudelski, A. How Surface-Enhanced Raman Spectroscopy Could Contribute to Medical Diagnoses. Chemosensors 2022, 10, 190. [Google Scholar] [CrossRef]
- Izuchukwu, U.D.; Asogwa, F.C.; Louis, H.; Uchenna, E.F.; Gber, T.E.; Chinasa, U.M.; Chinedum, N.J.; Eze, B.O.; Adeyinka, A.S.; Chris, O.U. Synthesis, vibrational analysis, molecular property investigation, and molecular docking of new benzenesulphonamide-based carboxamide derivatives against Plasmodium falciparum. J. Mol. Struct. 2022, 1269, 133796. [Google Scholar] [CrossRef]
- Asogwa, F.; Agwamba, E.C.; Louis, H.; Muozie, M.; Benjamin, I.; Gber, T.; Mathias, G.; Adeyinka, A.; Ikeuba, A. Structural Benchmarking, Density Functional Theory simulation, spectroscopic investigation and molecular docking of N-(1H-pyrrol-2-yl) methylene)-4-methylaniline as castration-resistant prostate cancer chemotherapeutic agent. Chem. Phys. Impact 2022, 5, 100091. [Google Scholar] [CrossRef]
- Benjamin, I.; Gber, T.; Louis, H.; Ntui, T.; Oyo-Ita, E.; Unimuke, T.; Edim, M.; Adeyinka, A. Modelling of aminothiophene-carbonitrile derivatives as potential drug candidates for hepatitis B and C. Iran. J. Sci. Technol. Trans. A Sci. 2022, 46, 1–14. [Google Scholar] [CrossRef]
- Politzer, P.; Murray, J.S. The fundamental nature and role of the electrostatic potential in atoms and molecules. Theor. Chem. Acc. 2002, 108, 134–142. [Google Scholar] [CrossRef]
- Koopmans, T. Über die Zuordnung von Wellenfunktionen und Eigenwerten zu den Einzelnen Elektronen Eines Atoms. Physica 1934, 1, 104–113. [Google Scholar] [CrossRef]
- Parr, R.G.; Pearson, R.G. Absolute hardness: Companion parameter to absolute electronegativity. J. Am. Chem. Soc. 1983, 105, 7512–7516. [Google Scholar] [CrossRef]
- Parr, R.G.; Donnelly, R.A.; Levy, M.; Palke, W.E. Electronegativity: The density functional viewpoint. J. Chem. Phys. 1978, 68, 3801–3807. [Google Scholar] [CrossRef]
- Maynard, A.T.; Huang, M.; Rice, W.G.; Covell, D.G. Reactivity of the HIV-1 nucleocapsid protein p7 zinc finger domains from the perspective of Density Functional Theory. Proc. Natl. Acad. Sci. USA 1998, 95, 11578–11583. [Google Scholar] [CrossRef] [PubMed]
- Parr, R.G.; Szentpály, L.v.; Liu, S. Electrophilicity Index. J. Am. Chem. Soc. 1999, 121, 1922–1924. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Stăncioiu, L.; Gherman, A.M.R.; Brezeștean, I.; Dina, N.E. Vibrational spectral analysis of Sorafenib and its molecular docking study compared to other TKIs. J. Mol. Struct. 2022, 1248, 131507. [Google Scholar] [CrossRef]
- Breilh, D.; Texier-Maugein, J.; Allaouchiche, B.; Saux, M.C.; Boselli, E. Carbapenems. J. Chemother. 2013, 25, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, G.A. An Introduction to Hydrogen Bonding; Oxford University Press: Oxford, UK, 1997. [Google Scholar]
- Evans, J.; Honnoodee, M.; Micah, W. Amoxacillin clavulanate; Stat Pearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Cole, M. Biochemistry and action of clavulanic acid. Scott. Med. J. 1982, 27 (Suppl. S1), S10–S16. [Google Scholar] [CrossRef]
- Kesado, T.; Hashizume, T.; Asahi, Y. Antibacterial activities of a new stabilized thienamycin, N-formimidoyl thienamycin, in comparison with other antibiotics. Antimicrob. Agents Chemother. 1980, 17, 912–917. [Google Scholar] [CrossRef]
- Dina, N.E.; Zhou, H.; Colniţă, A.; Leopold, N.; Szoke-Nagy, T.; Coman, C.; Haisch, C. Rapid single-cell detection and identification of pathogens by using surface-enhanced Raman spectroscopy. Analyst 2017, 142, 1782–1789. [Google Scholar] [CrossRef]
- Dina, N.E.; Tahir, M.A.; Bajwa, S.Z.; Amin, I.; Valev, V.K.; Zhang, L. SERS-based antibiotic susceptibility testing: Towards point-of-care clinical diagnosis. Biosens. Bioelectron. 2022, accepted. [Google Scholar]
- Cozar, I.B.; Colniţă, A.; Szöke-Nagy, T.; Gherman, A.M.R.; Dina, N.E. Label-free detection of bacteria using surface-enhanced Raman scattering and principal component analysis. Anal. Lett. 2019, 52, 177–189. [Google Scholar] [CrossRef]
- Gherman, A.M.R.; Dina, N.E.; Chiș, V.; Wieser, A.; Haisch, C. Yeast cell wall—Silver nanoparticles interaction: A synergistic approach between surface-enhanced Raman scattering and computational spectroscopy tools. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 222, 117223. [Google Scholar] [CrossRef] [PubMed]
- Stăncioiu, L.; Gherman, A.M.R.; Dina, N.E. Finding specific spectral features for surface-enhanced Raman response of E. faecalis assisted by multivariate analysis when using common silver sols. Rom. Rep. Phys. 2021, 73, 604. [Google Scholar]
- Frisch, M.J.; Trucks, H.B.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.; et al. Gaussian 09, Revision B.01; Gaussian, Inc.: Wallingford, CT, USA, 2010. [Google Scholar]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Vosko, S.H.; Wilk, L.; Nusair, M. Accurate spin-dependent electron liquid correlation energies for local spin density calculations: A critical analysis. Can. J. Phys. 1980, 58, 1200–1211. [Google Scholar] [CrossRef]
- Stephens, P.J.; Devlin, F.J.; Chabalowski, C.F.; Frisch, M.J. Ab initio calculation of vibrational absorption and circular dichroism spectra using Density Functional force fields. J. Phys. Chem. 1994, 98, 11623–11627. [Google Scholar] [CrossRef]
- Petersson, G.A.; Bennett, A.; Tensfeldt, T.G.; Al-Laham, M.A.; Shirley, W.A.; Mantzaris, J. Complete basis set model chemistry. I. The total energies of closed-shell atoms and hydrides of the first-row elements. J. Chem. Phys. 1988, 89, 2193–2218. [Google Scholar] [CrossRef]
- Petersson, G.A.; Al-Laham, M.A. A complete basis set model chemistry. II. Open-shell systems and the total energies of the first-row atoms. J. Chem. Phys. 1991, 94, 6081–6090. [Google Scholar] [CrossRef]
- Andersson, M.P.; Uvdal, P. New scale factors for harmonic vibrational frequencies using the B3LYP Density Functional method with the Triple-ζ Basis Set 6-311+G(d,p). J. Phys. Chem. A 2005, 109, 2937–2941. [Google Scholar] [CrossRef]
- Garau, G.; Dideberg, O. Crystal Structure of the Zinc Carbapenemase CphA in Complex with the Antibiotic Biapenem, 2004-08-18 ed.; Protein Data Bank (PDB): San Diego, CA, USA, 2004. [Google Scholar] [CrossRef]
- Kato, Y.; Shimizu-Ibuka, A. Crystal Structure of Metallo-Beta-Lactamase IMP-27 from Morganella morganii, 2019-10-15 ed.; Protein Data Base (PDB): San Diego, CA, USA, 2020. [Google Scholar] [CrossRef]
- Kim, Y.-C.; Maltseva, N.; Endres, M.; Joachimiak, A.; Center for Structural Genomics of Infectious Diseases (CSGID). Crystal Structure of a Class a Beta-Lactamase from Bacillus cereus in the Complex with Beta-Lactamase Inhibitor Clavulanate, 2020-03-08 ed.; Protein Data Base (PDB): San Diego, CA, USA, 2020. [Google Scholar] [CrossRef]
- D’Andréa, É.D.; Moon, T.M.; Peti, W.; Page, R. Crystal Structure of Pencillin Binding Protein 4 (PBP4) from Enterococcus faecalis in the Imipenem-Bound Form, 2018-09-25 ed.; Protein Data Base (PDB): San Diego, CA, USA, 2018. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
 Chemical structure of 6-APA and the position of the side chain R in penicillin’s structure; β-lactam ring is marked in grey. | |||
| COMPOUND FAMILY (GENERATION) | CHEMICAL STRUCTURE | SENSITIVITY | RESISTANCE |
| BPN β-lactamase sensitive (1st generation) |  C16H18N2O4S | narrow spectrum Gram-positive (GP) bacteria Neisseria gonorrhoeae Leptospira weilii | Escherichia coli Salmonella typhi |
| OXN β-lactamase resistant (2nd generation) | 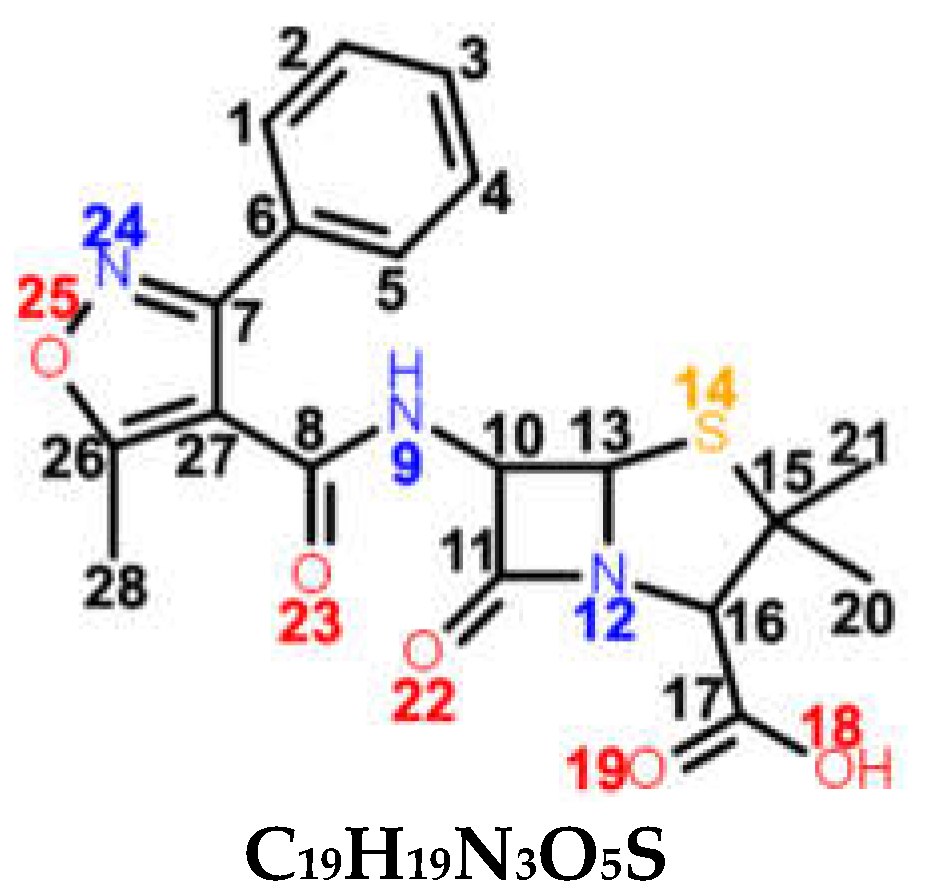 C19H19N3O5S | narrow spectrum Gram-positive bacteria PRSA | ORSA, MRSA |
| APN aminopenicillin (3rd generation) |  C16H19N3O4S | broad spectrum Streptococcus spp. Enterococcus spp. Neisseria meningitidis Haemophilus influenzae | MRSA, PRSA Enterobacteriaceae spp. Pseudomonas spp. |
| CBC carboxypenicillin (4th generation) | 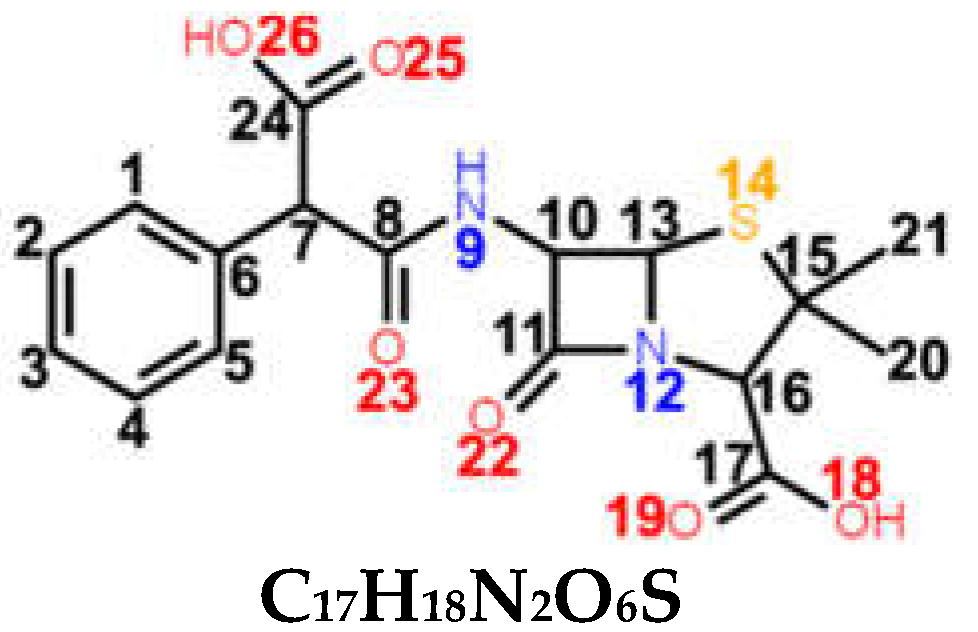 C17H18N2O6S | broad spectrum Escherichia coli Proteus mirabilis Pseudomonas aeruginosa | |
| AZL ureidopenicillin (5th generation) |  C20H23N5O6S | broad spectrum Escherichia coli Haemophilus spp. Pseudomonas aeruginosa | |
| BPN | OXN | APN | CBC | AZL | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 benzene ring | 621 839 983 1002 1029 1582; 1600 | 629 845 982 1002 1026 1578; 1606 | 615 830 993 1003 1027 1585; 1602 | 617 834 957 1004 1032 1582; 1600 | 617 - 988 1003 1030 1585; 1602 | common bands | |||||
| CH3 methyl group | 231 274 292 | 960 1436 1452; 1468 | 250 279 296 | - 1433 1459 | 240 271 291 | 951; 961 1435 1456 | 246 280 - | 948 1436 1457 | 241 - 294 | 948; 958 1437 1458 | |
| COOH carboxyl group | 360 - 807 - | 873 + 895 919 1245 | 368 522 - - | 894; 910 920 - | 360 522 802 847 | 873; 910 - 1249 | 361 525 803 847 | 876;891 920 - | 360 - - 850 | 870;899 - - | |
| NH amide group | 661 1178 | 656 - | 670 1178 | 666 - | 661 1175 | ||||||
| C=O carbonyl group | 402 1638 | 406 1649 | 409 1693 | 405 1666 | 409 1660 | ||||||
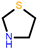 thiazolidine ring | 571 602 919 1292 | 579 616 920 - | - 601 926 - | 577 602 920 1297 | 575 - 913 - | ||||||
 β-lactam ring | 945 - 1775 | 944 - 1758 | - 1156 1764 | - 1156 1763 | - 1158 1775 | ||||||
| CH2 methylene group | 468 1419 | x | x | x | x | specific bands | |||||
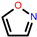 isoxazole ring | x | 250 336 492 648 734 793 | 908 1308 1444 1471 1516 1556 | x | x | x | |||||
| NH2 amino group | x | x | 465 830 1119 | 1186 1512 1638 | x | x | |||||
| COOH carboxyl group | x | x | x | 666 746 1126 | 1180 1372 1666 1763 | x | |||||
 imidazolidine ring | x | x | x | x | 465 714 958 | 1125 1239 1397 | |||||
| NH amide group | x | x | x | x | 641 1532 | ||||||
| BPN | OXN | APN | CBC | AZL | |
|---|---|---|---|---|---|
| EHOMO | −7.06380 | −7.11904 | −7.00911 | −6.98598 | −6.83060 |
| ELUMO | −1.34288 | −1.56275 | −1.39540 | −1.37880 | −1.18804 |
| I | 7.06380 | 7.11904 | 7.00911 | 6.98598 | 6.83060 |
| A | 1.34288 | 1.56275 | 1.39540 | 1.37880 | 1.18804 |
| HLG | 5.72092 | 5.55629 | 5.61371 | 5.60718 | 5.64256 |
| η | 2.86046 | 2.77815 | 2.80686 | 2.80359 | 2.82128 |
| σ | 0.34959 | 0.35995 | 0.35627 | 0.35669 | 0.35445 |
| χ | 4.20334 | 4.34090 | 4.20226 | 4.18239 | 4.00932 |
| μ | −4.20334 | −4.34090 | −4.20226 | −4.18239 | −4.00932 |
| ω | 3.08833 | 3.39136 | 3.14568 | 3.11964 | 2.84882 |
| Class | Pathogen (PDB id) | Ligand | Binding Energy (kcal/mol) | Number of Conformers | HB (D…A) | Type | HB (Å) |
|---|---|---|---|---|---|---|---|
| Gram-negative | A. hydrophila (1x8i) | BIAPENEM (co-crystalized ligand) | −9.4 (MIN) | - | HIS196…O3 | NE2…HE2 | 2.34 |
| LYS224…O4 | NZ…HZ | 2.38 | |||||
| LYS224…O5 | NZ…HZ | 2.39 | |||||
| ASN233…O4 | N…HN | 1.88 | |||||
| BPN | −8.0 (MIN) −7.83 ± 0.21 (AVE) 2.68% (sd) | 29 | HIS118…O19 | ND1…HD | 1.94 | ||
| HIS196…O22 | NE2…HE2 | 2.03–2.16 | |||||
| LYS224…O19 | NZ…HZ | 2.07–2.16 | |||||
| LYS224…O18 | NZ…HZ | 2.28–2.40 | |||||
| ASN233…O18 | N…HN | 1.84–1.94 | |||||
| O18…ASN233 | OH…O | 2.53–2.66 | |||||
| O18…THR119 | OH…OG1 | 2.11 | |||||
| OXN | −8.8 (MIN) −8.70 ± 0.02 (AVE) 0.23% (sd) | 4 | HIS196…N24 | NE2…HE2 | 2.21–2.23 | ||
| LYS224…O25 | NZ…HN | 2.12–2.18 | |||||
| ASN233…O25 | N…HN | 2.00–2.03 | |||||
| O18…THR157 | OH…O | 2.48 | |||||
| O18…PHE156 | OH…O | 2.25 | |||||
| APN | −7.7 (MIN) −7.51 ± 0.08 (AVE) 1.07% (sd) | 6 | HIS118…O19 | ND1…HD | 1.89–1.93 | ||
| LYS224…O22 | NZ…HZ | 2.22 | |||||
| ASN233…O22 | N…HN | 2.13 | |||||
| N24…PHE156 | NH…O | 2.40–2.51 | |||||
| O18…THR119 | OH…OG1 | 2.06 | |||||
| CBC | −8.3 (MIN) −8.20 ±0.10 (AVE) 1.22% (sd) | 38 | HIS118…O26 | ND1…HD | 2.21–2.29 | ||
| HIS196…O23 | NE2…HE2 | 2.22–2.29 | |||||
| LYS224…O22 | NZ…HZ | 2.34–2.39 | |||||
| ASN233…O22 | N…HN | 2.20–2.30 | |||||
| O26…ASP120 | OH…OD1 | 2.04–2.28 | |||||
| AZL | −8.3 (MIN) −7.95 ± 0.19 (AVE) 2.39 (sd) | 4 | GLY160…O18 | N…HN | 2.57–2.62 | ||
| HIS196…O32 | NE2…HE2 | 2.05–2.13 | |||||
| HIS263…O26 | NE2…HE2 | 2.35–2.47 | |||||
| O18…THR157 | OH…O | 2.33–2.46 | |||||
| O18…PHE156 | OH…O | 2.07 | |||||
| M. morganii (6l3s) | BPN | −7.4 (MIN) −7.13 ± 0.13 (AVE) 1.82% (sd) | 2 | SER98…O19 | N…HN | 2.15–2.17 | |
| ASN185…O23 | N…HN | 2.28–2.32 | |||||
| O18…SER98 | OH…OG | 2.33 | |||||
| OXN | −8.0 (MIN) −7.81 ± 0.06 (AVE) 0.77% (sd) | 4 | SER98…O19 | N…HN | 2.15–2.21 | ||
| ASP98…O18 | N…HN | 2.34–2.35 | |||||
| O18…SER98 | OH…OG | 2.08 | |||||
| APN | −7.3 (MIN) −7.16 ± 0.07 (AVE) 0.98% (sd) | 10 | SER98…O19 | N…HN | 2.08–2.23 | ||
| ASP99…O18 | N…HN | 2.23–2.27 | |||||
| ASN185…O23 | N…HN | 2.33–2.42 | |||||
| O18…SER98 | OH…OG | 2.38–2.59 | |||||
| CBC | −7.6 (MIN) −7.40 ± 0.16 (AVE) 2.16% (sd) | 34 | SER98…O19 | N…HN | 1.90–1.96 | ||
| ASP99…O18 | N…HN | 2.33–2.38 | |||||
| LYS179…O26 | NZ…HZ | 2.13–2.26 | |||||
| ASN185…O25 | N…HN | 2.06–2.26 | |||||
| O18…SER98 | OH…OG | 2.30–2.40 | |||||
| AZL | −8.0 (MIN) −7.86 ± 0.08 (AVE) 1.02% (sd) | 10 | SER98…O19 | N…HN | 2.11–2.21 | ||
| ASP99…O18 | N…HN | 2.17–2.26 | |||||
| LYS179…O26 | NZ…HZ | 1.89–1.95 | |||||
| LYS179…O32 | NZ…HZ | 2.11–2.18 | |||||
| ASN185…O23 | N…HN | 2.19–2.32 | |||||
| N30…GLY182 | NH…O | 2.36–2.41 | |||||
| Gram-positive | B. cereus (6w33) | CLAVULANATE (co-crystalized ligand) | −4.5 (MIN) | - | N2…SER258 N2…SER258 O1…SER258 | NH…O NH…OG O1H…H | 2.41 2.13 2.19 |
| BPN | −7.6 (MIN) −7.26 ± 0.14 (AVE) 1.93% (sd) | 2 | SER153…N12 | OG…HG | 2.37–2.38 | ||
| SER153…O18 | OG…HG | 2.16–2.20 | |||||
| ASN155…O23 | ND2…HD | 2.16–2.22 | |||||
| ALA260…O22 | N…HN | 2.17–2.36 | |||||
| ARG266…O19 | NH1…HH | 2.22–2.38 | |||||
| O18…SER258 | OH…OG | 2.13 | |||||
| OXN | −8.5 (MIN) −8.06 ± 0.13 (AVE) 1.61% (sd) | 5 | ASN155…N24 | ND2…HD | 2.09–2.24 | ||
| ASN193…O25 | ND2…HD | 2.17–2.22 | |||||
| SER258…O18 | OG…HG | 2.20–2.27 | |||||
| ARG266…O22 | NH1…HH | 1.99–2.08 | |||||
| ARG266…O18 | NH1…HH | 2.38–2.44 | |||||
| O18…THR239 | OH…O | 2.36 | |||||
| APN | −7.7 (MIN) −7.51 ± 0.10 (AVE) 1.33% (sd) | 1 | SER258…O18 | OG…HG | 2.03 | ||
| ALA260…O23 | N…HN | 2.11 | |||||
| ARG266…O22 | NH1…HH | 1.93 | |||||
| CBC | −8.2 (MIN) −8.09 ± 0.06 (AVE) 0.74% (sd) | 3 | LYS98…O23 | NZ…HZ | 2.71–2.72 | ||
| ASN155…O25 | ND2…HD | 2.15–2.36 | |||||
| SER258…O18 | OG…HG | 2.00 | |||||
| ARG266…O22 | NH1…HH | 2.00–2.01 | |||||
| ARG266…O18 | NH1…HH | 2.31 | |||||
| ARG266…O18 | NH2…HH | 2.05 | |||||
| O26…ASN193 | OH…OD1 | 2.53 | |||||
| O26…ASN155 | OH…DO1 | 2.41 | |||||
| O18…SER258 | OH…OG | 1.99 | |||||
| O26…GLU189 | OH…OE1 | 2.23 | |||||
| AZL | −8.6 (MIN) −8.32 ± 0.12 1.44% (sd) | 5 | ASN155…O26 | ND2…HD | 2.36–2.39 | ||
| ASN155…O32 | ND2…HD | 2.22–2.26 | |||||
| ASN193…O32 | ND2…HD | 2.53–2.57 | |||||
| SER258…O22 | OG…HG | 2.34–2.37 | |||||
| ARG266…O22 | NH1…HH | 2.34–2.39 | |||||
| ARG266…O23 | NH1…HH | 2.54 | |||||
| ARG266…O18 | NH2…HH | 2.52–2.56 | |||||
| ARG266…O22 | NH2…HH | 1.82 | |||||
| N30…ASN193 | NH…OD1 | 1.94–1.96 | |||||
| O18…THR239 | OH…O | 1.87 | |||||
| E. faecalis (6mkh) | IMIPENEM (co-crystalized ligand) | −5.5 (MIN) | - | SER482…O1 ASN484…O3 N3…THR620 O3…SER424 | OG…HG ND2…HD NH…OG1 OH…OG | 2.15 2.11 2.36 1.94 | |
| BPN | −7.9 (MIN) −7.61 ± 0.20 (AVE) 1.93% (sd) | 4 | SER424…O22 | OG…HG | 1.92–1.98 | ||
| THR465…O18 | OG1…HG | 2.45–2.50 | |||||
| ASN484…O19 | ND2…HD | 2.12–2.21 | |||||
| THR620…O23 | OG1…HG | 2.18–2.25 | |||||
| OXN | −7.8 (MIN) −7.70 ± 0.03 (AVE) 0.39% (sd) | 6 | SER658…O22 | OG…HG | 1.87–1.90 | ||
| O18…SER658 | OH…OG | 2.20 | |||||
| O18…GLU635 | OH…OE1 | 1.93 | |||||
| APN | −7.5 (MIN) −7.27 ± 0.09 (AVE) 1.24% (sd) | 1 | ASN484…O22 | ND2…HD | 2.28 | ||
| N24…SER482 | NH…OG | 2.32 | |||||
| O18…THR465 | OH…O | 2.21 | |||||
| CBC | −7.7 (MIN) −7.50 ± 0.10 (AVE) 1.33% (sd) | 5 | SER424…O22 | OG…HG | 1.81–2.23 | ||
| ASN484…N12 | OG…HG | 1.99–2.06 | |||||
| THR620…O25 | ND2…HD | 2.40–2.46 | |||||
| THR622…O18 | N…HN | 2.39–2.59 | |||||
| AZL | −8.6 (MIN) −8.44 ± 0.18 (AVE) 2.13% (sd) | 46 | SER424…O22 | OG…HG | 1.76–2.05 | ||
| THR465…O32 | N…HN | 1.91–2.00 | |||||
| SER482…N12 | OG…HG | 2.06–2.22 | |||||
| ASN484…O23 | ND2…HD | 1.92–2.04 | |||||
| ASN484…O32 | ND2…HD | 1.83–1.92 | |||||
| GLN542…O26 | NE2…HE | 2.17–2.36 | |||||
| THR620…O18 | OG1…HG | 2.29–2.53 | |||||
| THR622…O22 | N…HN | 2.39–2.56 | |||||
| N30…PHE463 | NH…O | 2.17–2.34 | |||||
| N24…THR465 | NH…OG1 | 2.37–2.53 | |||||
| O18…THR620 | OH…OG1 | 2.08–2.27 |
| BPN | OXN | APN | CBC | AZL | TRC | ||
|---|---|---|---|---|---|---|---|
| GN | A. hydrophila PAI-45 | R | R | R | R | 14.3 | 19.9 |
| A. hydrophila PI-88 | R | R | 14.2 | 20.7 | 14.5 | 12.3 | |
| M. morganii PI-81 | R | R | R | R | R | R | |
| GP | B. cereus ESN-09 | R | R | R | R | R | - |
| E. lactis CE-13 | 14.7 | R | 22.7 | 14.2 | 18.7 | - | |
| E. durans CI-28 | 16.3 | R | 24.0 | 16.1 | 20.1 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gherman, A.M.R.; Dina, N.E.; Chiș, V. Cheminformatics Study on Structural and Bactericidal Activity of Latest Generation β-Lactams on Widespread Pathogens. Int. J. Mol. Sci. 2022, 23, 12685. https://doi.org/10.3390/ijms232012685
Gherman AMR, Dina NE, Chiș V. Cheminformatics Study on Structural and Bactericidal Activity of Latest Generation β-Lactams on Widespread Pathogens. International Journal of Molecular Sciences. 2022; 23(20):12685. https://doi.org/10.3390/ijms232012685
Chicago/Turabian StyleGherman, Ana Maria Raluca, Nicoleta Elena Dina, and Vasile Chiș. 2022. "Cheminformatics Study on Structural and Bactericidal Activity of Latest Generation β-Lactams on Widespread Pathogens" International Journal of Molecular Sciences 23, no. 20: 12685. https://doi.org/10.3390/ijms232012685
APA StyleGherman, A. M. R., Dina, N. E., & Chiș, V. (2022). Cheminformatics Study on Structural and Bactericidal Activity of Latest Generation β-Lactams on Widespread Pathogens. International Journal of Molecular Sciences, 23(20), 12685. https://doi.org/10.3390/ijms232012685








