Whole Exome Sequencing and In Silico Analysis of Human Sertoli in Patients with Non-Obstructive Azoospermia
Abstract
1. Introduction
2. Results
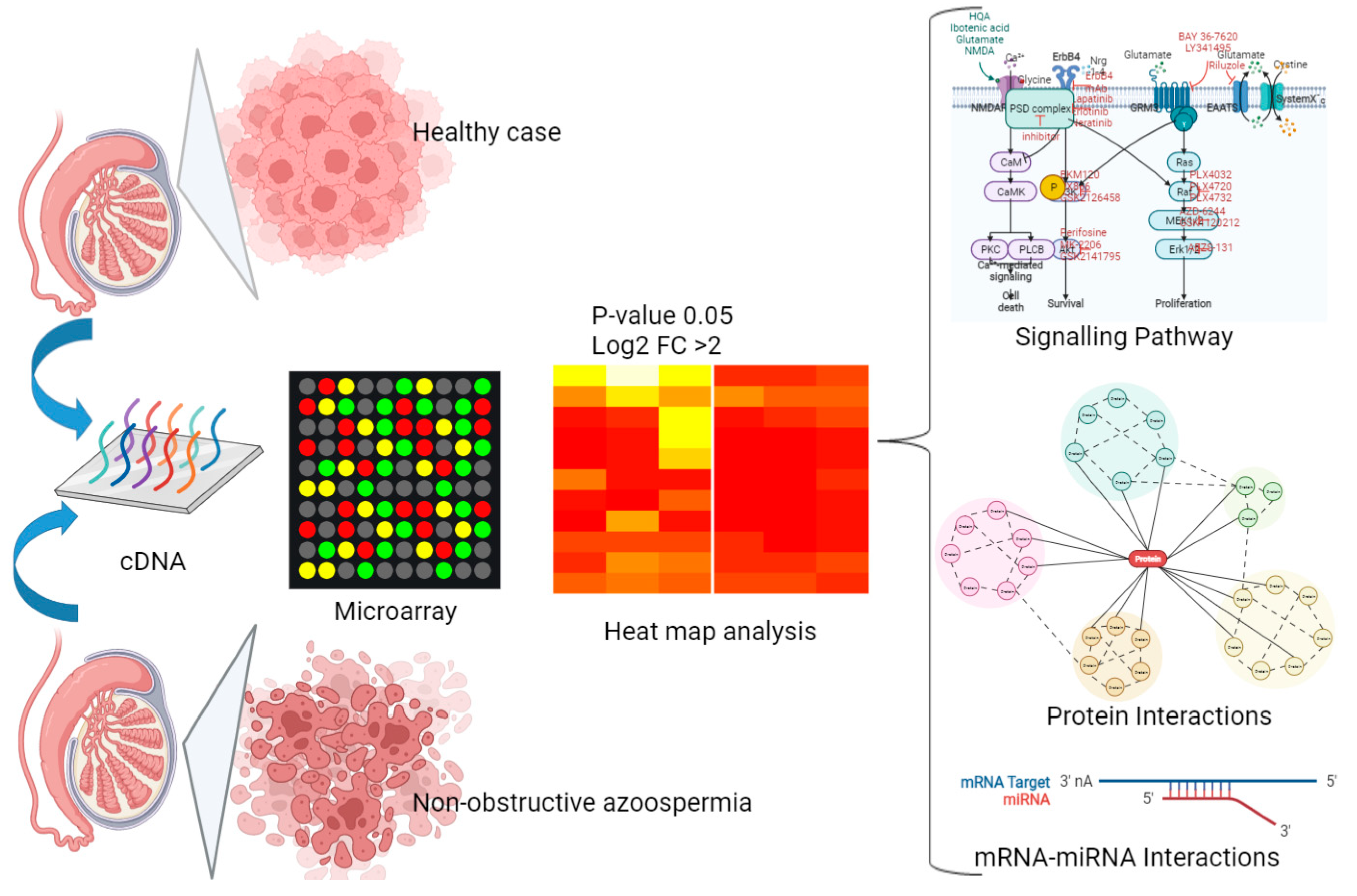
2.1. Isolation of Sertoli Cells
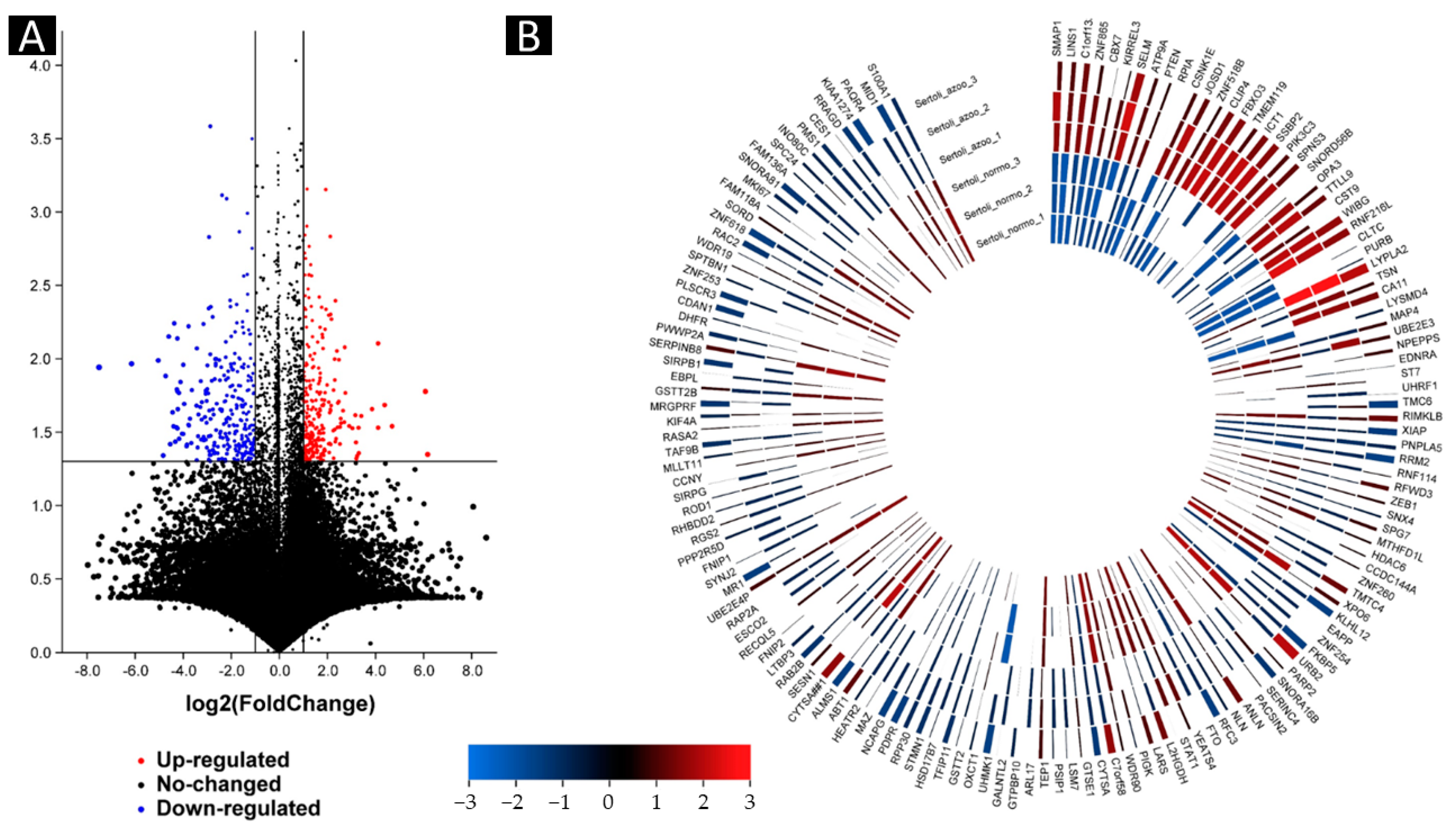
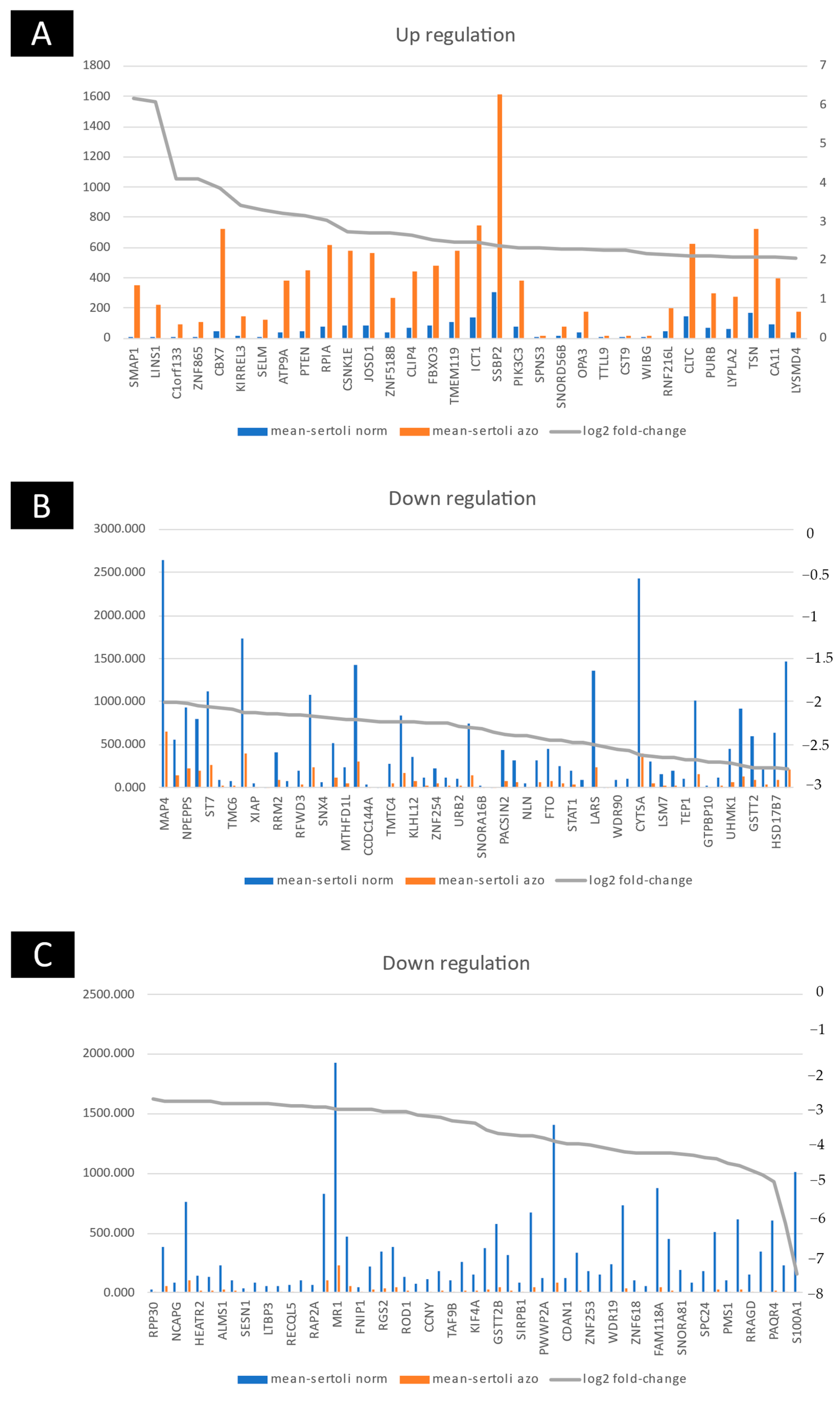
2.2. Up- and Downregulated Gene Expressions between Non-Obstructive Azoospermia and a Normal Case by Microarray
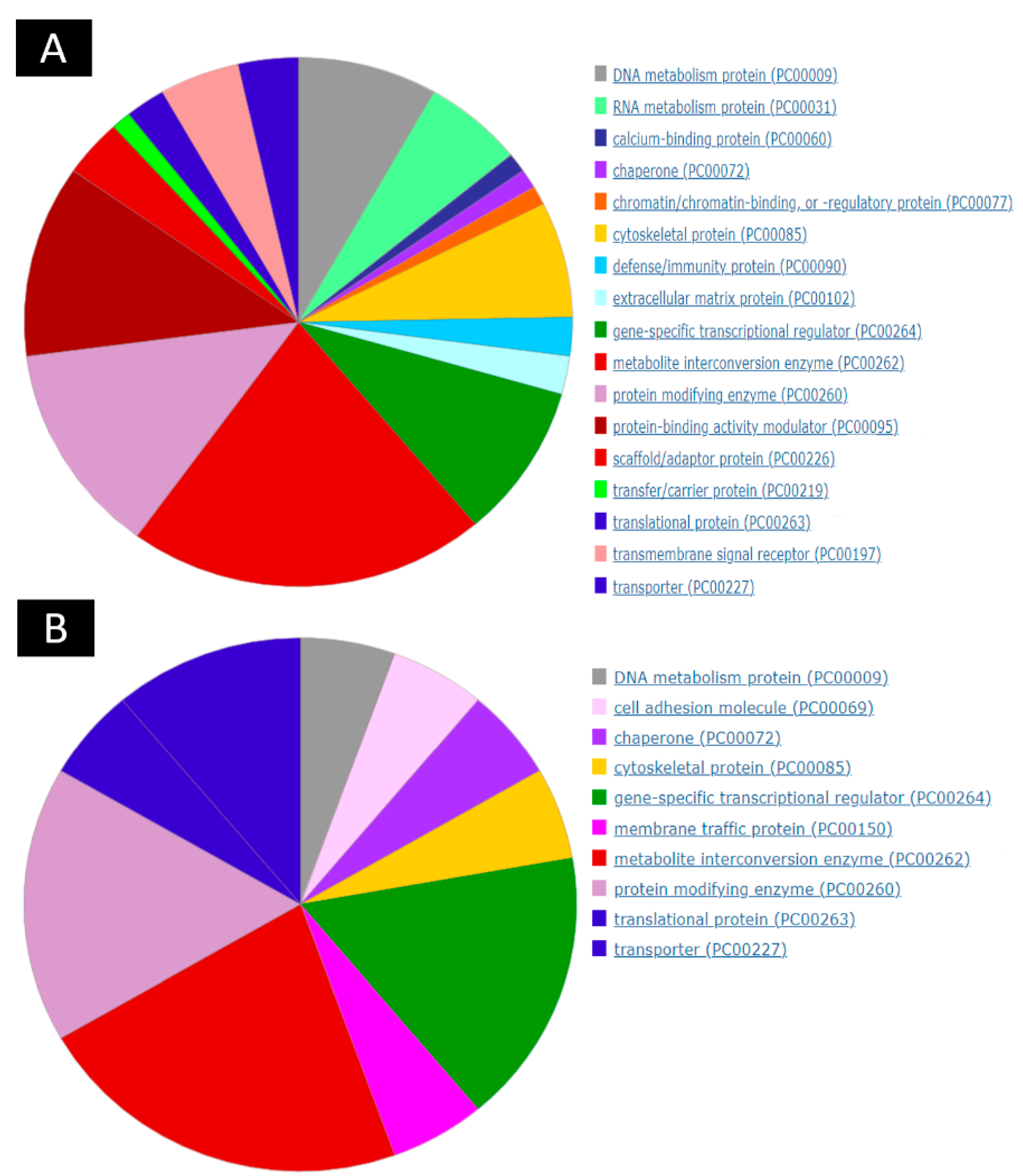
2.3. Group Comparison and Protein Class Sorting
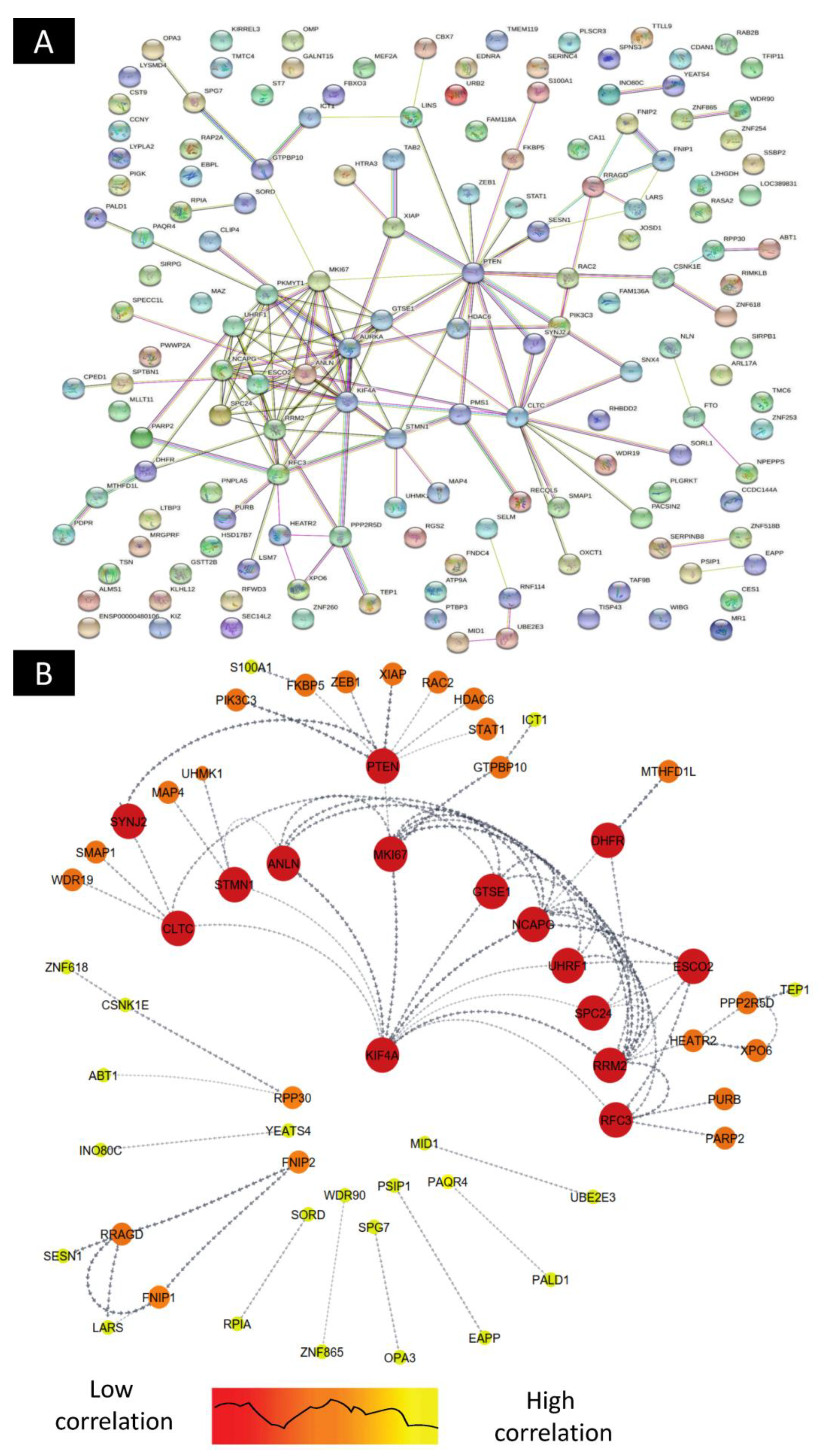
2.4. Construction of Differentially Expressed Gene–Protein–Protein Interaction(PPI) Network
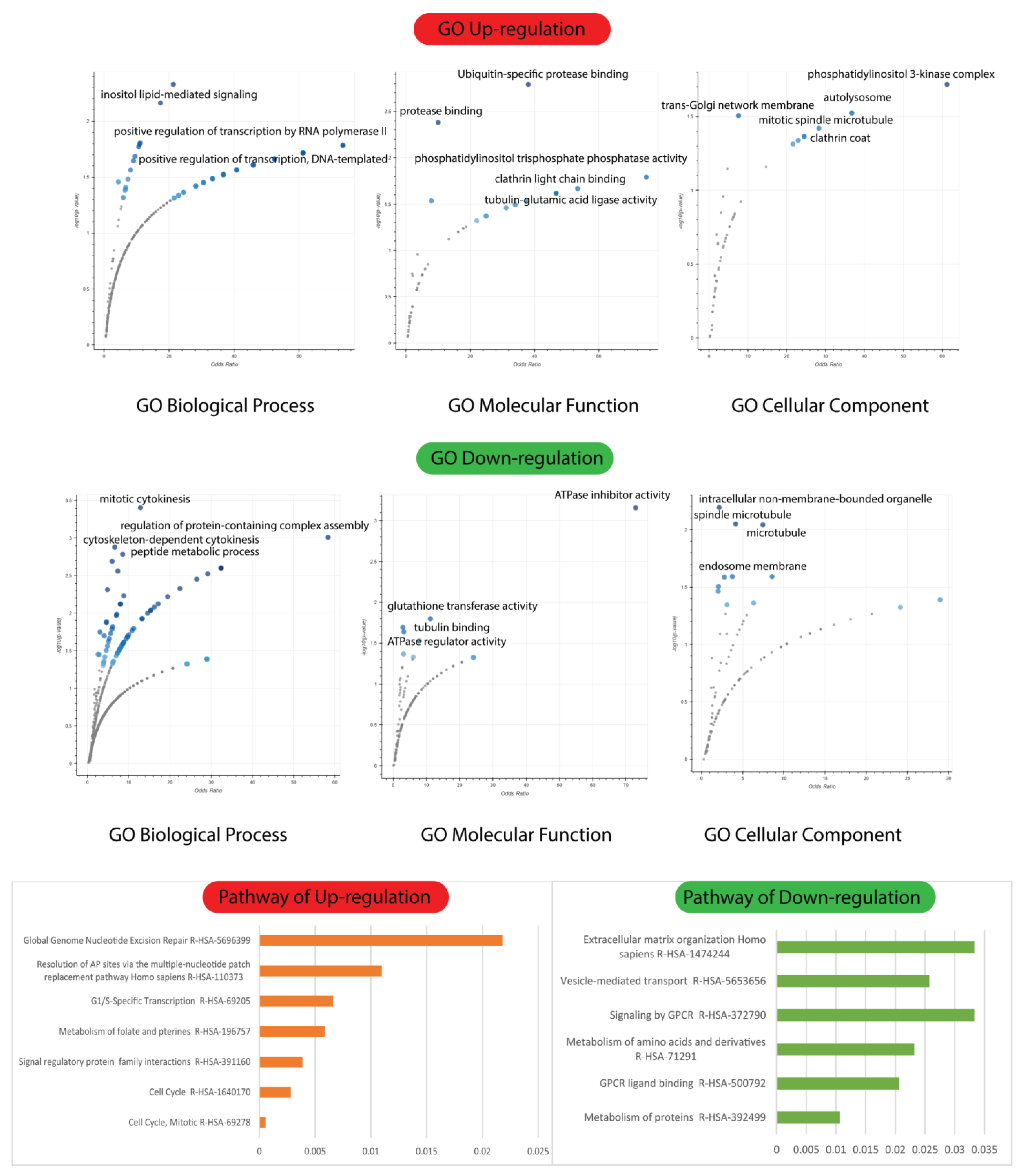
2.5. Biological Process, Molecular Functions, and Cellular Components of Enrichment Analysis
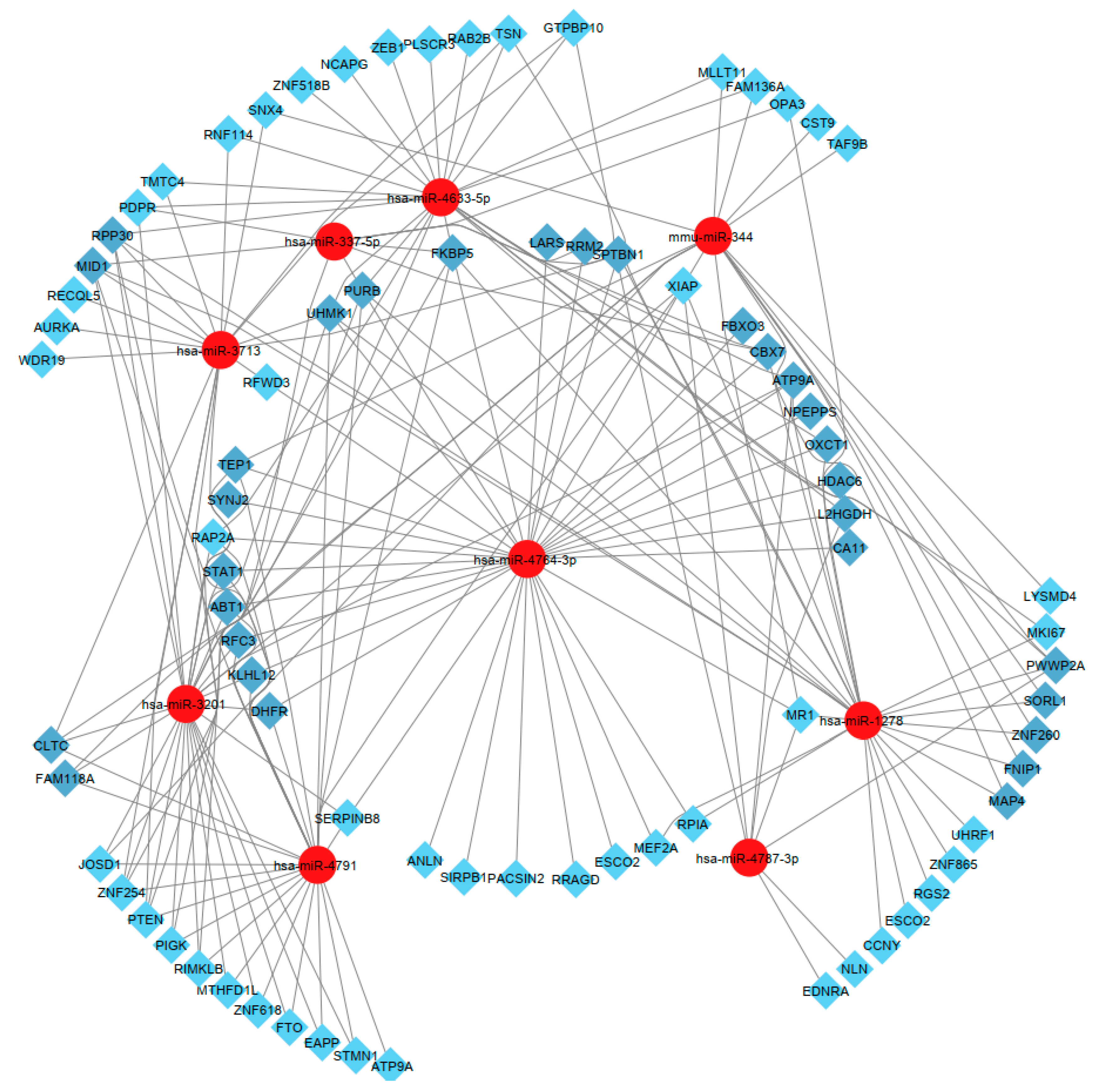
2.6. Isolated and Selected Candidate MicroRNAs
3. Discussion
4. Material and Methods
4.1. Patient and Control Selection
4.2. Surgical and Sample Processing
4.3. Selection of Human Sertoli Cells
4.4. Sertoli Cell Isolation
4.5. Collection of Single Cells from the Population of the Enriched Human Sertoli Cell Population with a Micromanipulation System
RNA Extraction and WES Preparation
4.6. Microarray Analysis
4.7. Microarray Data Normalization and Analyses
4.8. Data Processing and Differential Expression Analysis
4.9. Group Comparison and Protein Class Sorting
4.10. Gene ontology (GO) Investigation
4.11. Target Transcript Protein–Protein Interaction Network Construction
4.12. Target Transcript Prediction and miRNA Regulatory Network Construction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Figueiredo, A.; França, L.; Hess, R.; Costa, G. Sertoli cells are capable of proliferation into adulthood in the transition region between the seminiferous tubules and the rete testis in Wistar rats. Cell Cycle 2016, 15, 2486–2496. [Google Scholar] [CrossRef] [PubMed]
- Chojnacka, K.; Zarzycka, M.; Mruk, D.D. Biology of the Sertoli cell in the fetal, pubertal, and adult mammalian testis. Mol. Mech. Cell Differ. Gonad Dev. 2016, 58, 225–251. [Google Scholar]
- O’Donnell, L.; Smith, L.B.; Rebourcet, D. Sertoli cells as key drivers of testis function. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Stanton, P.G. Regulation of the blood-testis barrier. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 166–173. [Google Scholar]
- Makise, M.; Nakamura, H.; Kuniyasu, A. The role of vimentin in the tumor marker Nup88-dependent multinucleated phenotype. BMC Cancer 2018, 18, 519. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.-D.; Hao, S.-L.; Yang, W.-X. Multiple signaling pathways in Sertoli cells: Recent findings in spermatogenesis. Cell Death Dis. 2019, 10, 541. [Google Scholar] [CrossRef]
- Meroni, S.B.; Galardo, M.N.; Rindone, G.; Gorga, A.; Riera, M.F.; Cigorraga, S.B. Molecular mechanisms and signaling pathways involved in sertoli cell proliferation. Front. Endocrinol. 2019, 10, 224. [Google Scholar] [CrossRef]
- Rashid, M.-u.; Zahedi-Amiri, A.; Glover, K.K.; Gao, A.; Nickol, M.E.; Kindrachuk, J.; Wilkins, J.A.; Coombs, K.M. Zika virus dysregulates human Sertoli cell proteins involved in spermatogenesis with little effect on tight junctions. PLoS Negl. Trop. Dis. 2020, 14, e0008335. [Google Scholar] [CrossRef] [PubMed]
- Kuyucu, Y.; Coşkun, G.; Şaker, D.; Karaoğlan, Ö.; Ürünsak, İ.F.; İzol, V.; Arıdoğan, İ.A.; Erdoğan, Ş.; Özgür, H.; Polat, S. Immunohistochemical examination of androgen receptor and estrogen receptor alpha expressions in obstructive and non-obstructive azoospermia. Syst. Biol. Reprod. Med. 2021, 67, 463–470. [Google Scholar] [CrossRef]
- Babakhanzadeh, E.; Nazari, M.; Ghasemifar, S.; Khodadadian, A. Some of the factors involved in male infertility: A prospective review. Int. J. Gen. Med. 2020, 13, 29. [Google Scholar] [CrossRef]
- Jodar, M.; Soler-Ventura, A.; Oliva, R.; Molecular Biology of Reproduction and Development Research Group. Semen proteomics and male infertility. J. Proteom. 2017, 162, 125–134. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, Y.; Gao, Y.; Lin, Z.; Yang, S.; Wang, T.; Wang, Q.; Xie, N.; Hua, R.; Liu, M. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018, 28, 879–896. [Google Scholar] [CrossRef]
- Herman, S.; Lipiński, P.; Ogórek, M.; Starzyński, R.; Grzmil, P.; Bednarz, A.; Lenartowicz, M. Molecular regulation of copper homeostasis in the male gonad during the process of spermatogenesis. Int. J. Mol. Sci. 2020, 21, 9053. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Tian, R.; Xu, J.; Ji, Z.; Zhang, Y.; Zhao, L.; Yang, C.; Li, P.; Zhi, E.; Bai, H. Novel copy number variations within SYCE1 caused meiotic arrest and non-obstructive azoospermia. BMC Med. Genom. 2022, 15, 137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, P.; Liu, N.; Jing, T.; Ji, Z.; Yang, C.; Zhao, L.; Tian, R.; Chen, H.; Huang, Y. Novel Bi-Allelic Variants of FANCM Cause Sertoli Cell-Only Syndrome and Non-Obstructive Azoospermia. Front. Genet. 2021, 12, 799886. [Google Scholar] [CrossRef] [PubMed]
- Ronen, A.; Glickman, B.W. Human DNA repair genes. Envron. Mol. Mutagen 2001, 37, 241–283. [Google Scholar] [CrossRef]
- Dias, T.R.; Martins, A.D.; Reis, V.P.; Socorro, S.; Silva, B.M.; Alves, M.G.; Oliveira, P.F. Glucose transport and metabolism in sertoli cell: Relevance for male fertility. Curr. Chem. Biol. 2013, 7, 282–293. [Google Scholar] [CrossRef]
- Kaiser, G.R.; Monteiro, S.C.; Gelain, D.P.; Souza, L.F.; Perry, M.L.; Bernard, E.A. Metabolism of amino acids by cultured rat Sertoli cells. Metabolism 2005, 54, 515–521. [Google Scholar] [CrossRef]
- Grootegoed, J.; Oonk, R.; Jansen, R.; Van der Molen, H. Metabolism of radiolabelled energy-yielding substrates by rat Sertoli cells. Reproduction 1986, 77, 109–118. [Google Scholar] [CrossRef]
- Gao, H. Amino acids in reproductive nutrition and health. Amino Acids Nutr. Health 2020, 1265, 111–131. [Google Scholar] [CrossRef]
- Zhou, R.; Wu, J.; Liu, B.; Jiang, Y.; Chen, W.; Li, J.; He, Q.; He, Z. The roles and mechanisms of Leydig cells and myoid cells in regulating spermatogenesis. Cell. Mol. Life Sci. 2019, 76, 2681–2695. [Google Scholar] [CrossRef]
- Houda, A.; Nyaz, S.; Sobhy, B.M.; Bosilah, A.H.; Romeo, M.; Michael, J.P.; Eid, H.M. Seminiferous Tubules and Spermatogenesis. In Male Reproductive Anatomy; IntechOpen: London, UK, 2021. [Google Scholar]
- Saeidikhoo, S.; Ezi, S.; Khatmi, A.; Aghajanpour, F.; Soltani, R.; Abdollahifar, M.A.; Jahanian, A.; Aliaghaei, A. Effect of Sertoli cell transplantation on reducing neuroinflammation-induced necroptosis and improving motor coordination in the rat model of cerebellar ataxia induced by 3-acetylpyridine. J. Mol. Neurosci. 2020, 70, 1153–1163. [Google Scholar] [CrossRef]
- Rudolph, L.; Bentley, G.; Calandra, R.S.; Paredes, A.; Tesone, M.; Wu, T.; Micevych, P. Peripheral and central mechanisms involved in the hormonal control of male and female reproduction. J. Neuroendocrinol. 2016, 28. [Google Scholar] [CrossRef] [PubMed]
- Monsees, T.K.; Görnig, M.; Schill, W.B.; Miska, W. Possible involvement of proteases in the regulation of spermatogenesis. Andrologia 1998, 30, 185–191. [Google Scholar] [CrossRef]
- Yang, C.-X.; Yang, Y.-W.; Mou, Q.; Chen, L.; Wang, C.; Du, Z.-Q. Proteomic changes induced by ascorbic acid treatment on porcine immature Sertoli cells. Theriogenology 2022, 118, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Yu, C.; Zhang, Q. Ubiquitin-Proteasome System–Regulated Protein Degradation in Spermatogenesis. Cells 2022, 11, 1058. [Google Scholar] [CrossRef] [PubMed]
- Bragoszewski, P.; Turek, M.; Chacinska, A. Control of mitochondrial biogenesis and function by the ubiquitin–proteasome system. Open Biol. 2017, 7, 170007. [Google Scholar] [CrossRef] [PubMed]
- Zapata, H.; Morales, P.; Jara, M. Role of Ubiquitin-proteasome system in Espematogenesis. Int. J. Med. Surg. Sci. 2015, 2, 621–634. [Google Scholar] [CrossRef]
- Ma, T.; Hou, J.; Zhou, Y.; Chen, Y.; Qiu, J.; Wu, J.; Ding, J.; Han, X.; Li, D. Dibutyl phthalate promotes juvenile Sertoli cell proliferation by decreasing the levels of the E3 ubiquitin ligase Pellino 2. Environ. Health 2020, 19, 1–12. [Google Scholar] [CrossRef]
- Kishi, K.; Uchida, A.; Takase, H.M.; Suzuki, H.; Kurohmaru, M.; Tsunekawa, N.; Kanai-Azuma, M.; Wood, S.A.; Kanai, Y. Spermatogonial deubiquitinase USP9X is essential for proper spermatogenesis in mice. Reproduction 2017, 154, 135–143. [Google Scholar] [CrossRef]
- Li, X.Y.; Zhang, Y.; Wang, X.X.; Jin, C.; Wang, Y.Q.; Sun, T.C.; Li, J.; Tang, J.X.; Batool, A.; Deng, S.L. Regulation of blood-testis barrier assembly in vivo by germ cells. FASEB J. 2018, 32, 1653–1664. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Y. Action and Interaction between Retinoic Acid Signaling and Blood–Testis Barrier Function in the Spermatogenesis Cycle. Cells 2022, 11, 352. [Google Scholar] [CrossRef]
- Venkatesh, D.; Mruk, D.; Herter, J.M.; Cullere, X.; Chojnacka, K.; Cheng, C.Y.; Mayadas, T.N. AKAP9, a regulator of microtubule dynamics, contributes to blood-testis barrier function. Am. J. Pathol. 2016, 186, 270–284. [Google Scholar] [CrossRef] [PubMed]
- Gunes, S.; Al-Sadaan, M.; Agarwal, A. Spermatogenesis, DNA damage and DNA repair mechanisms in male infertility. Reprod. Biomed. Online 2015, 31, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, A.; Bhandary, B.; Potter, S.J.; Ratner, N.; DeFalco, T. Cdc42 activity in Sertoli cells is essential for maintenance of spermatogenesis. Cell Rep. 2021, 37, 109885. [Google Scholar] [CrossRef]
- Xiong, Z.; Wang, C.; Wang, Z.; Dai, H.; Song, Q.; Zou, Z.; Xiao, B.; Zhao, A.Z.; Bai, X.; Chen, Z. Raptor directs Sertoli cell cytoskeletal organization and polarity in the mouse testis. Biol. Reprod. 2018, 99, 1289–1302. [Google Scholar] [CrossRef]
- Pradhan, R.; Ngo, P.A.; Martínez-Sánchez, L.d.C.; Neurath, M.F.; López-Posadas, R. Rho GTPases as key molecular players within intestinal mucosa and GI diseases. Cells 2021, 10, 66. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Xiao, X.; Lui, W.-y.; Lee, W.M.; Mruk, D.; Cheng, C.Y. Cell polarity proteins and spermatogenesis. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 62–70. [Google Scholar]
- Palmer, N.; Talib, S.Z.A.; Kaldis, P. Diverse roles for CDK-associated activity during spermatogenesis. FEBS Lett. 2019, 593, 2925–2949. [Google Scholar] [CrossRef] [PubMed]
- Lord, T.; Nixon, B. Metabolic changes accompanying spermatogonial stem cell differentiation. Dev. Cell 2020, 52, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Griswold, M.D. 50 years of spermatogenesis: Sertoli cells and their interactions with germ cells. Biol. Reprod. 2018, 99, 87–100. [Google Scholar] [CrossRef]
- Griswold, M.D. Spermatogenesis: The commitment to meiosis. Physiol. Rev. 2016, 96, 1–17. [Google Scholar] [CrossRef]
- Puzuka, A.; Alksere, B.; Gailite, L.; Erenpreiss, J. Idiopathic infertility as a feature of genome instability. Life 2021, 11, 628. [Google Scholar] [CrossRef]
- Morris, B.J.; Willcox, B.J.; Donlon, T.A. Genetic and epigenetic regulation of human aging and longevity. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2019, 1865, 1718–1744. [Google Scholar] [CrossRef] [PubMed]
- Tournaye, H.; Krausz, C.; Oates, R.D. Novel concepts in the aetiology of male reproductive impairment. Lancet Diabetes Endocrinol. 2017, 5, 544–553. [Google Scholar] [CrossRef]
- Krausz, C.; Cioppi, F.; Riera-Escamilla, A. Testing for genetic contributions to infertility: Potential clinical impact. Expert Rev. Mol. Diagn. 2018, 18, 331–346. [Google Scholar] [CrossRef] [PubMed]
- Skakkebaek, N.E.; Rajpert-De Meyts, E.; Buck Louis, G.M.; Toppari, J.; Andersson, A.-M.; Eisenberg, M.L.; Jensen, T.K.; Jørgensen, N.; Swan, S.H.; Sapra, K.J. Male reproductive disorders and fertility trends: Influences of environment and genetic susceptibility. Physiol. Rev. 2016, 96, 55–97. [Google Scholar] [CrossRef] [PubMed]
- Dunleavy, J.E.; O’Bryan, M.K.; Stanton, P.G.; O’Donnell, L. The cytoskeleton in spermatogenesis. Reproduction 2019, 157, R53–R72. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Ren, W.; Huang, D.; Sang, Y.; Cao, L.; Huang, J. Cell structure and physiology. In Cell Movement in Health and Disease; Elsevier: Amsterdam, The Netherlands, 2022; pp. 3–16. [Google Scholar]
- Teves, M.E.; Roldan, E.R.; Krapf, D.; Strauss III, J.F.; Bhagat, V.; Sapao, P. Sperm differentiation: The role of trafficking of proteins. Int. J. Mol. Sci. 2020, 21, 3702. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Q.; Xu, S.; Xiao, Y.; Wang, Y.; Feng, C.; Xue, R.; Zhao, H.; Song, Z.; Li, J. Transcriptome dynamics during turbot spermatogenesis predicting the potential key genes regulating Male germ cell proliferation and maturation. Sci. Rep. 2018, 8, 15825. [Google Scholar] [CrossRef]
- Conrad, S.; Azizi, H.; Hatami, M.; Kubista, M.; Bonin, M.; Hennenlotter, J.; Sievert, K.-D.; Skutella, T. Expression of genes related to germ cell lineage and pluripotency in single cells and colonies of human adult germ stem cells. Stem Cells Int. 2016, 2016, 8582526. [Google Scholar] [CrossRef]
- Niazi Tabar, A.; Azizi, H.; Hashemi Karoii, D.; Skutella, T. Testicular Localization and Potential Function of Vimentin Positive Cells during Spermatogonial Differentiation Stages. Animals 2022, 12, 268. [Google Scholar] [CrossRef]
- Hashemi Karoii, D.; Azizi, H. A review of protein-protein interaction and signaling pathway of Vimentin in cell regulation, morphology and cell differentiation in normal cells. J. Recept. Signal Transduct. 2022, 16, 512–520. [Google Scholar] [CrossRef]
- Karoii, D.H.; Azizi, H.; Amirian, M. Signaling Pathways and Protein–Protein Interaction of Vimentin in Invasive and Migration Cells: A Review. Cell Reprogram. 2022, 24, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Azizi, H.; Hamidabadi, H.G.; Skutella, T. Differential proliferation effects after short-term cultivation of mouse spermatogonial stem cells on different feeder layers. Cell J. 2019, 21, 186. [Google Scholar] [PubMed]
- Azizi, H.; Conrad, S.; Hinz, U.; Asgari, B.; Nanus, D.; Peterziel, H.; Hajizadeh Moghaddam, A.; Baharvand, H.; Skutella, T. Derivation of pluripotent cells from mouse SSCs seems to be age dependent. Stem Cells Int. 2016, 2016, 8216312. [Google Scholar] [CrossRef] [PubMed]
- Hashemi Karoii, D.; Azizi, H.; Skutella, T. Microarray and in silico analysis of DNA repair genes between human testis of patients with nonobstructive azoospermia and normal cells. Cell Biochem. Funct. 2022, 40, 1–15. [Google Scholar] [CrossRef] [PubMed]
| Patient (Sample) | Indication | Histopathological Diagnosis | Sperm Quality |
|---|---|---|---|
| Patient 1 | Non-obstructive Azoospermia | Hypospermatogenesis | Rare, Concentration: 60 × 106 spermatozoa/mL Motility: 70% motile Morphology: 31% normal |
| Patient 2 | Non-obstructive Azoospermia | Hypospermatogenesis | Rare, Concentration: 200 × 106 spermatozoa/mL Motility: 50% motile Morphology: 48% normal |
| Patient 3 | Non-obstructive Azoospermia | Hypospermatogenesis | Rare, Concentration: 80 × 106 spermatozoa/mL Motility: 50% motile Morphology: 26% normal |
| Normal | Normozoospermia | / | Abundant, motile spermatozoa |
| Normal | Normozoospermia | / | Abundant, motile spermatozoa |
| Normal | Normozoospermia | / | Abundant, motile spermatozoa |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azizi, H.; Hashemi Karoii, D.; Skutella, T. Whole Exome Sequencing and In Silico Analysis of Human Sertoli in Patients with Non-Obstructive Azoospermia. Int. J. Mol. Sci. 2022, 23, 12570. https://doi.org/10.3390/ijms232012570
Azizi H, Hashemi Karoii D, Skutella T. Whole Exome Sequencing and In Silico Analysis of Human Sertoli in Patients with Non-Obstructive Azoospermia. International Journal of Molecular Sciences. 2022; 23(20):12570. https://doi.org/10.3390/ijms232012570
Chicago/Turabian StyleAzizi, Hossein, Danial Hashemi Karoii, and Thomas Skutella. 2022. "Whole Exome Sequencing and In Silico Analysis of Human Sertoli in Patients with Non-Obstructive Azoospermia" International Journal of Molecular Sciences 23, no. 20: 12570. https://doi.org/10.3390/ijms232012570
APA StyleAzizi, H., Hashemi Karoii, D., & Skutella, T. (2022). Whole Exome Sequencing and In Silico Analysis of Human Sertoli in Patients with Non-Obstructive Azoospermia. International Journal of Molecular Sciences, 23(20), 12570. https://doi.org/10.3390/ijms232012570







