Exogenous Nitric Oxide Alleviates the Damage Caused by Tomato Yellow Leaf Curl Virus in Tomato through Regulation of Peptidase Inhibitor Genes
Abstract
1. Introduction
2. Results
2.1. Virus Detection
2.2. Effect of Exogenous NO on Phenotypic and Growth Indicators of Virus-Inoculated Tomatoes
2.3. Effect of Exogenous NO on the Control of TYLCV
2.4. Transcriptome Sequencing Data and Analysis
2.4.1. Analysis of Differentially Expressed Genes (DEGs)
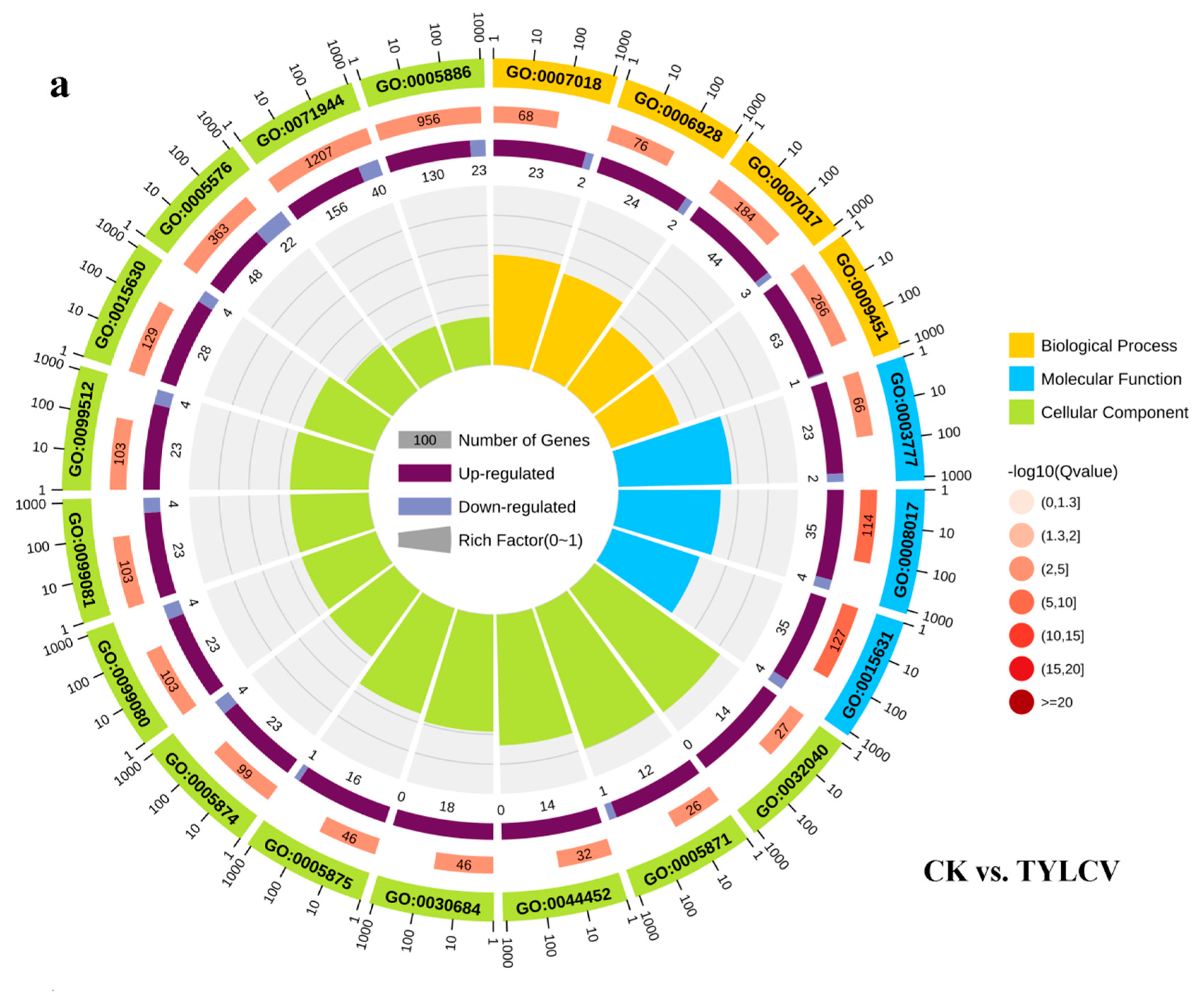
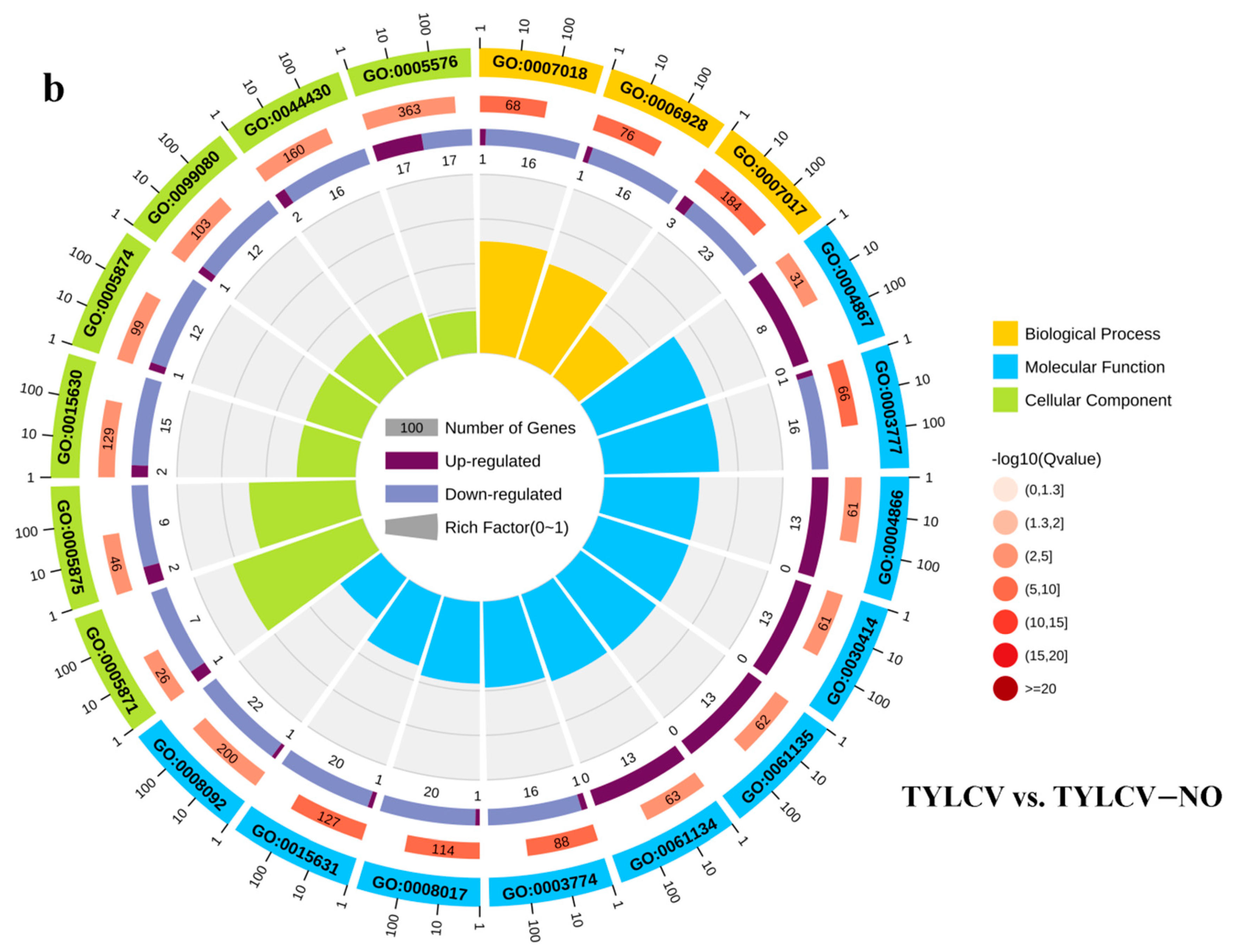
2.4.2. Gene Ontology Enrichment Analysis
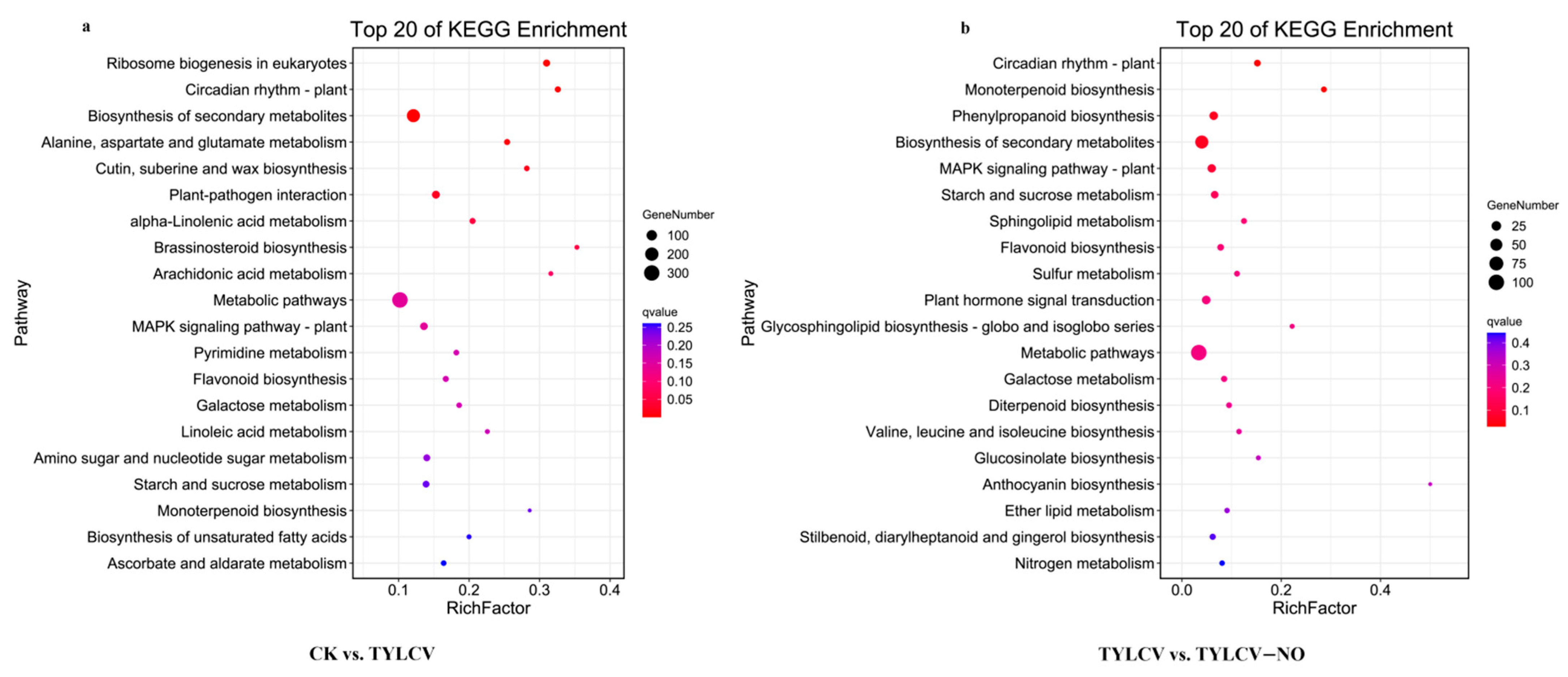
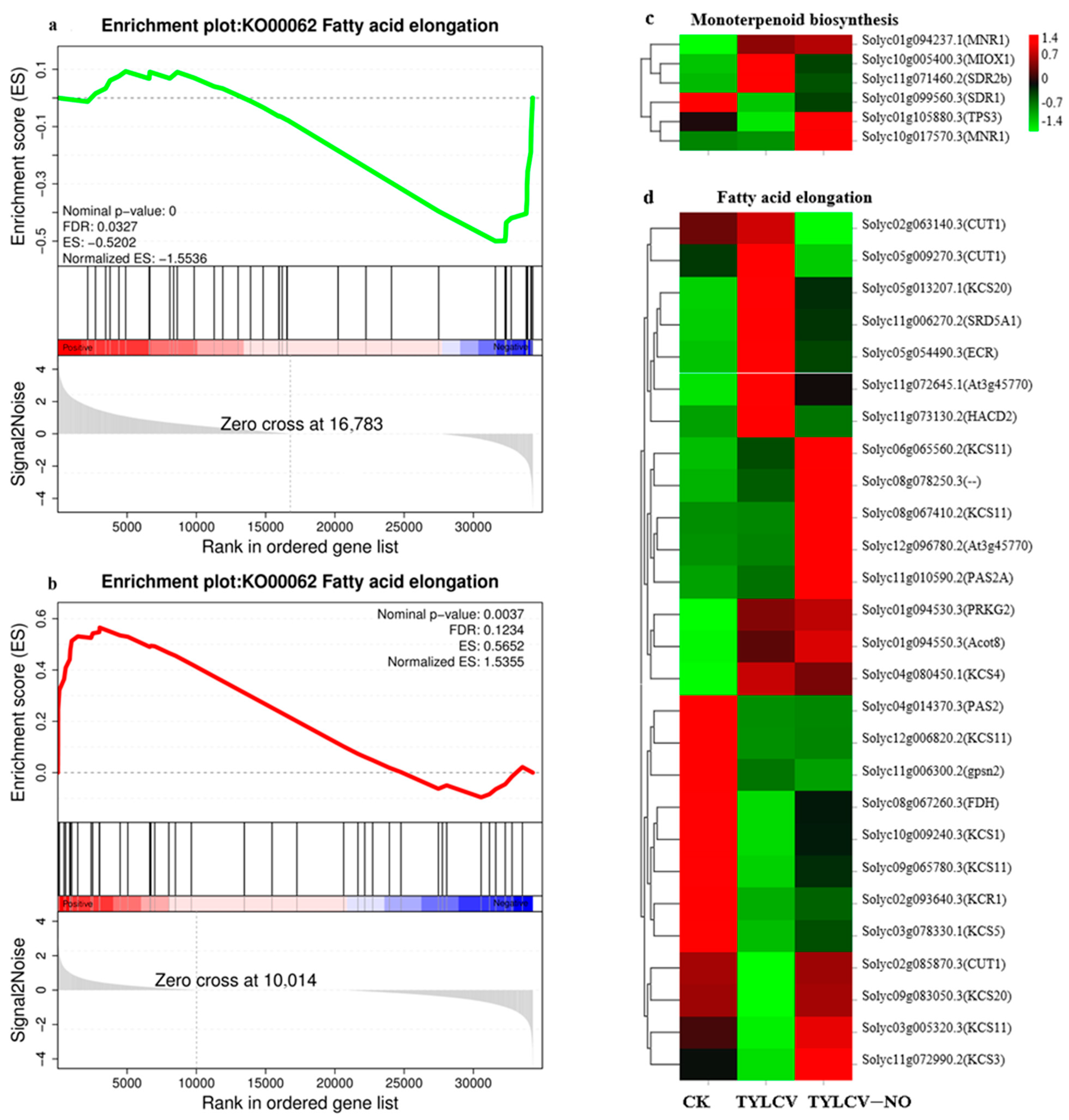
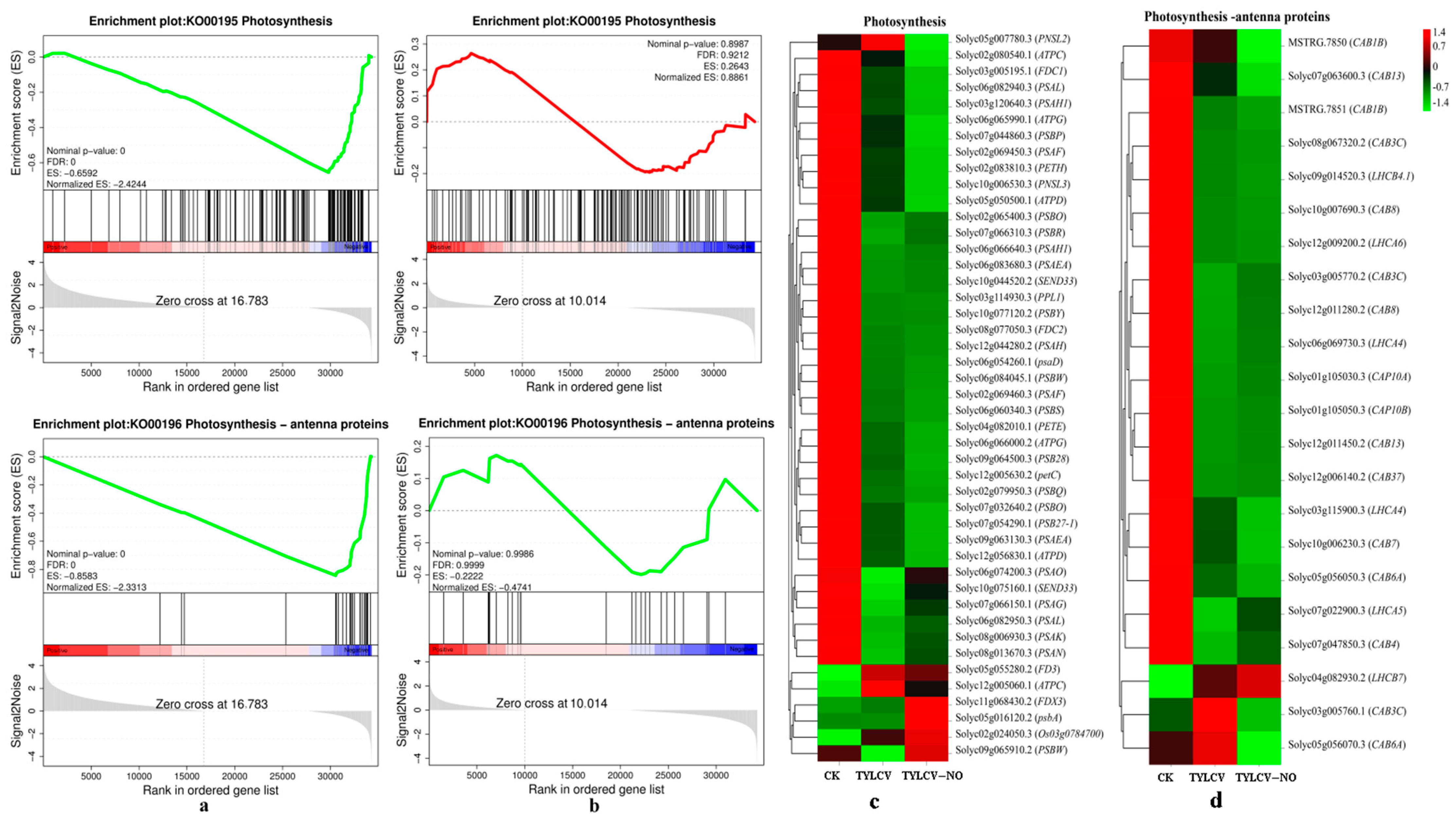
2.4.3. KEGG Enrichment Analysis
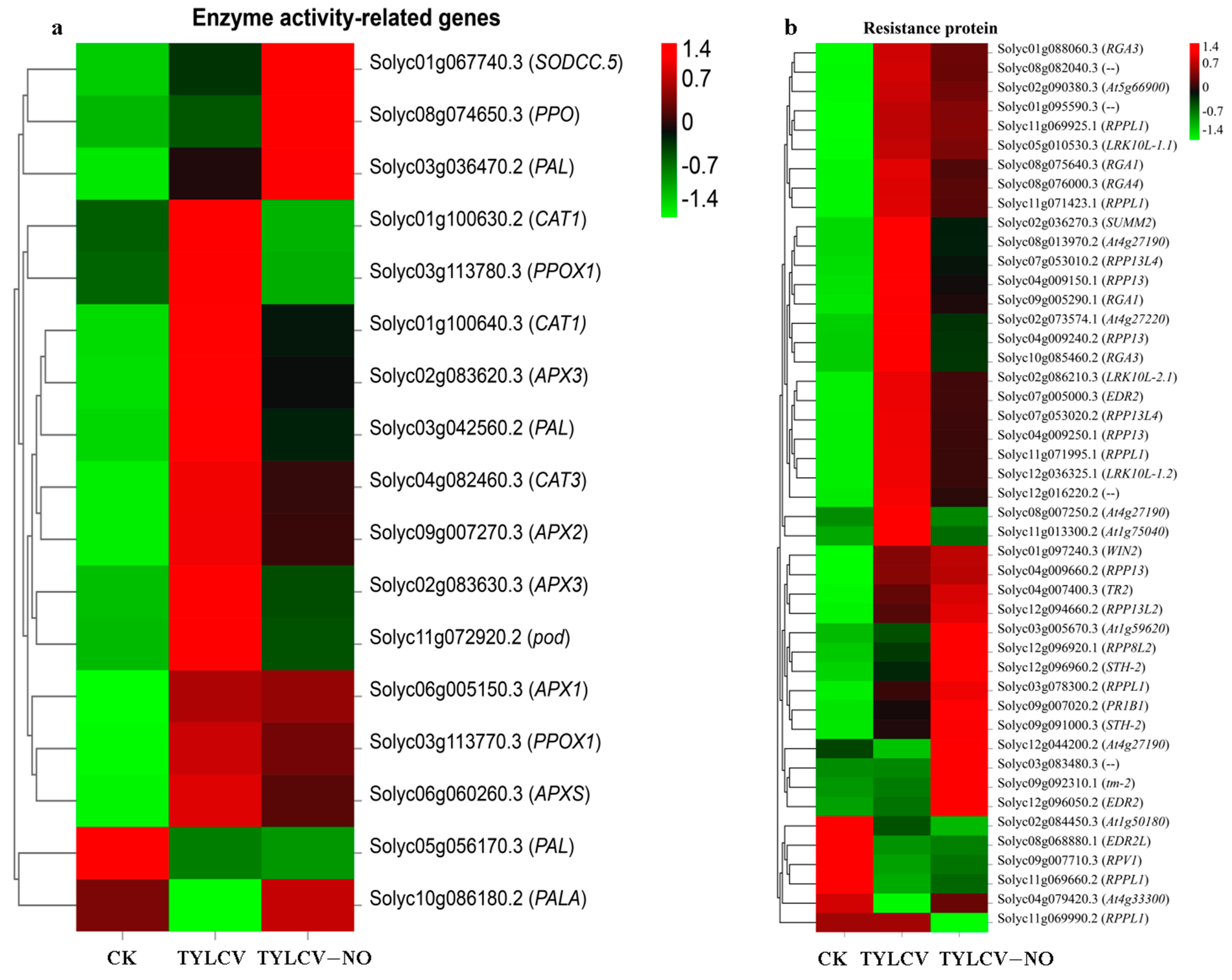
2.4.4. Regulation of Enzyme Activity-Related Genes and Resistance Proteins by Exogenous NO
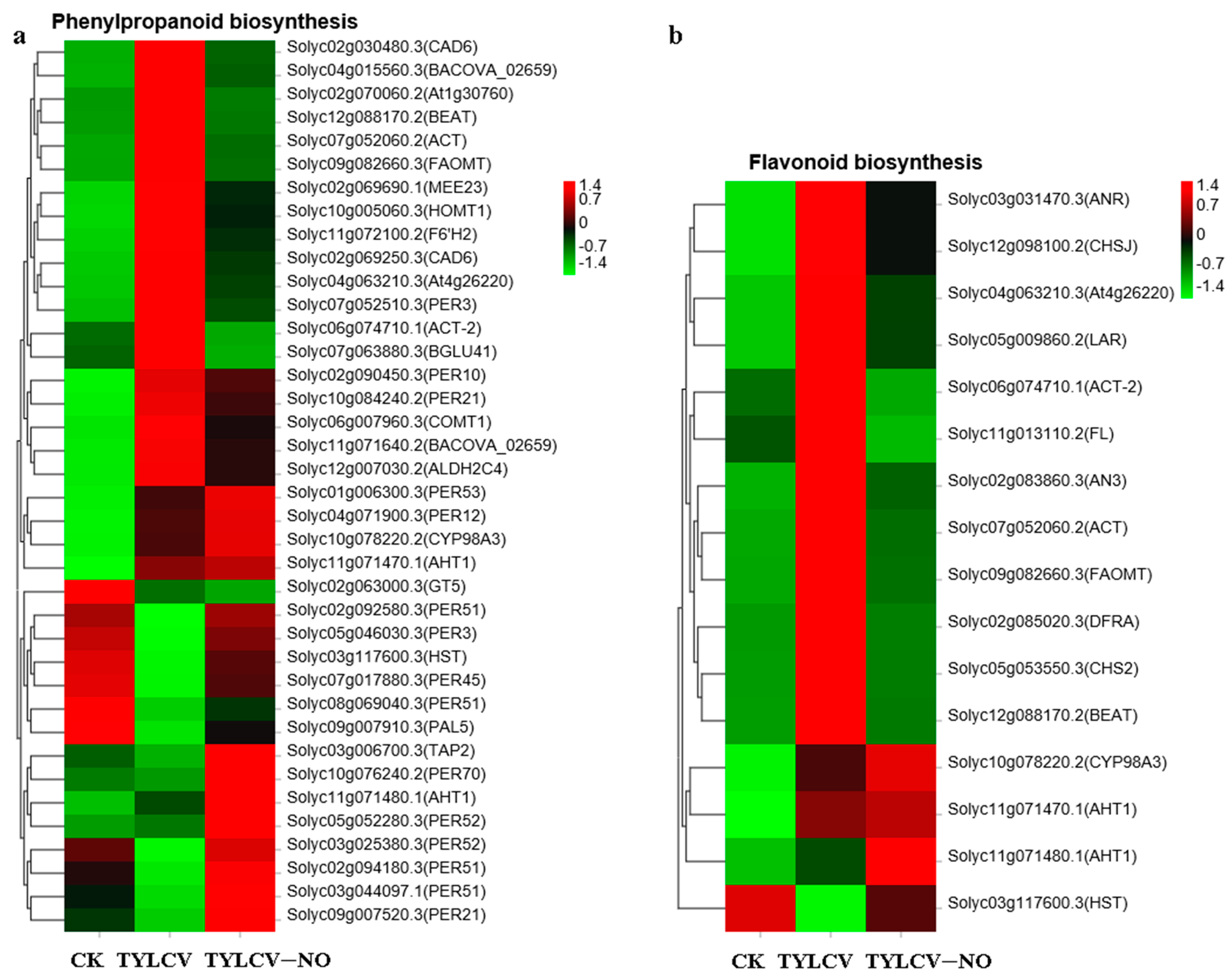
2.4.5. Exogenous NO-Mediated Key Secondary Metabolite Genes
2.4.6. Exogenous NO-Mediated Key Metabolite Genes
2.4.7. Effect of Exogenous NO on Photosynthesis
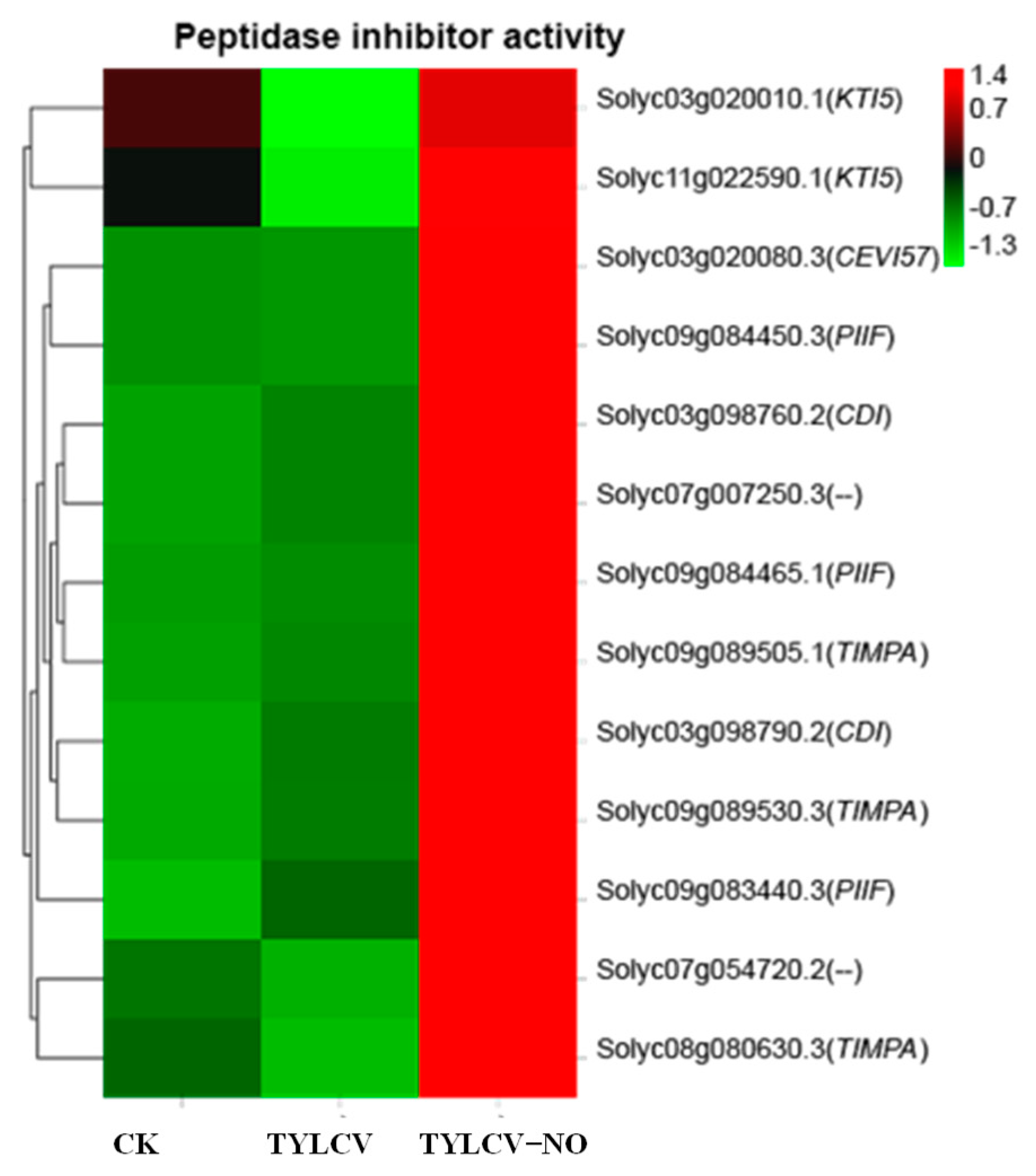
2.4.8. Exogenous NO-Mediated Peptidase Inhibitors Improve Disease Resistance in Tomato
2.4.9. qRT-PCR Validation of RNA-Seq with Viral Gene Copy Number Detection
2.5. Validation of the Mechanism of Exogenous NO Alleviation of Tomato Diseases by Physiological Parameters
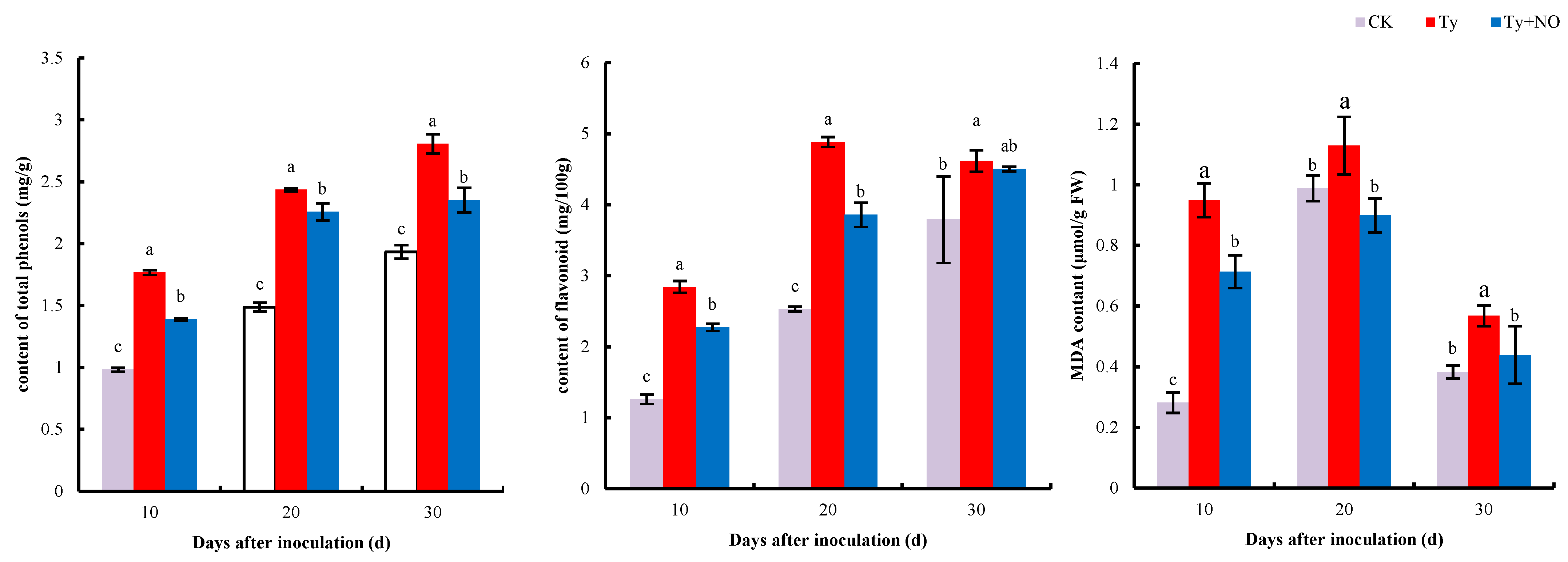
2.5.1. Changes in the Extent of Membrane Lipid Peroxidation and Secondary Metabolites
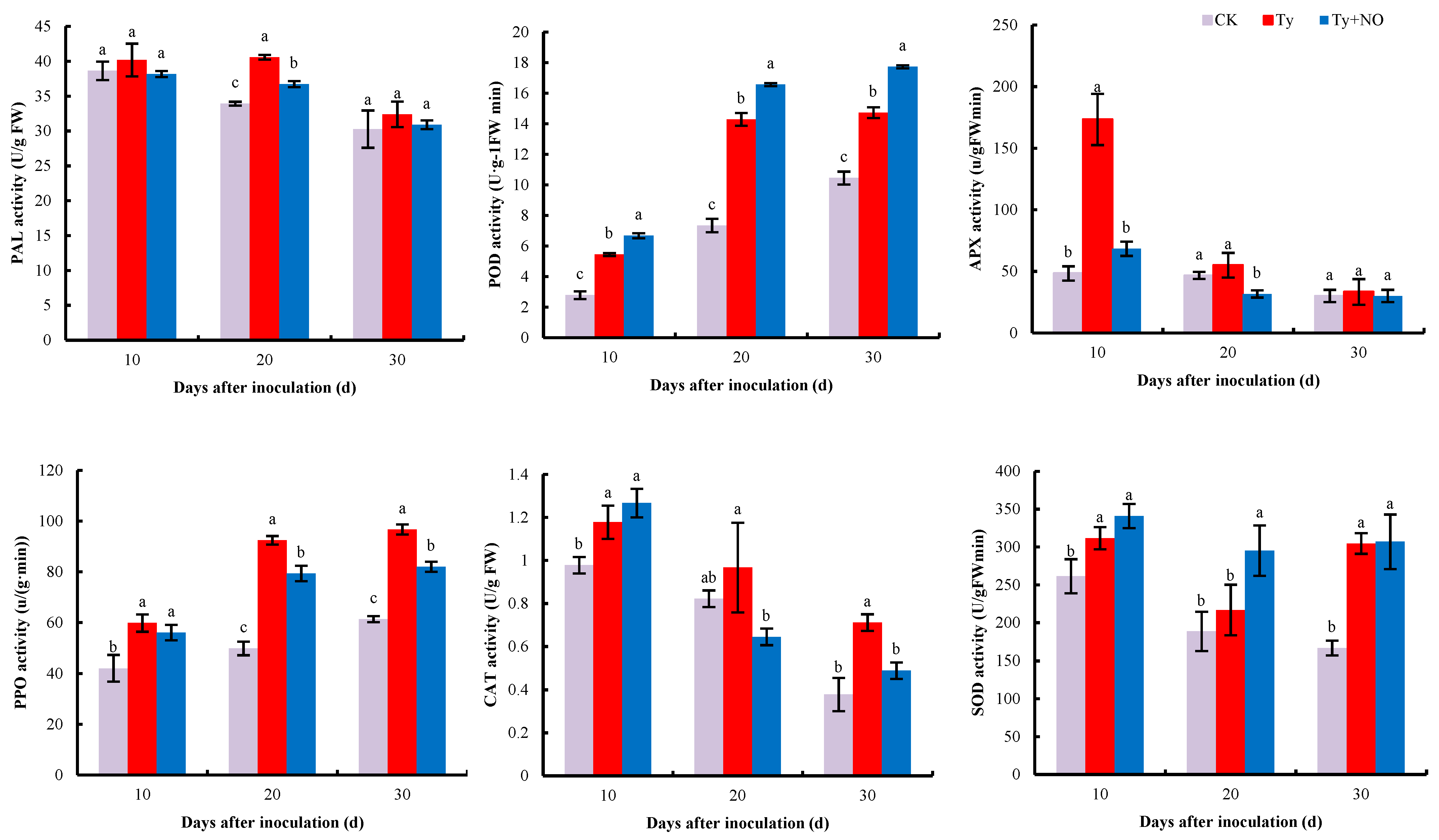
2.5.2. Changes in the Activity of Antioxidant and Defense Enzymes
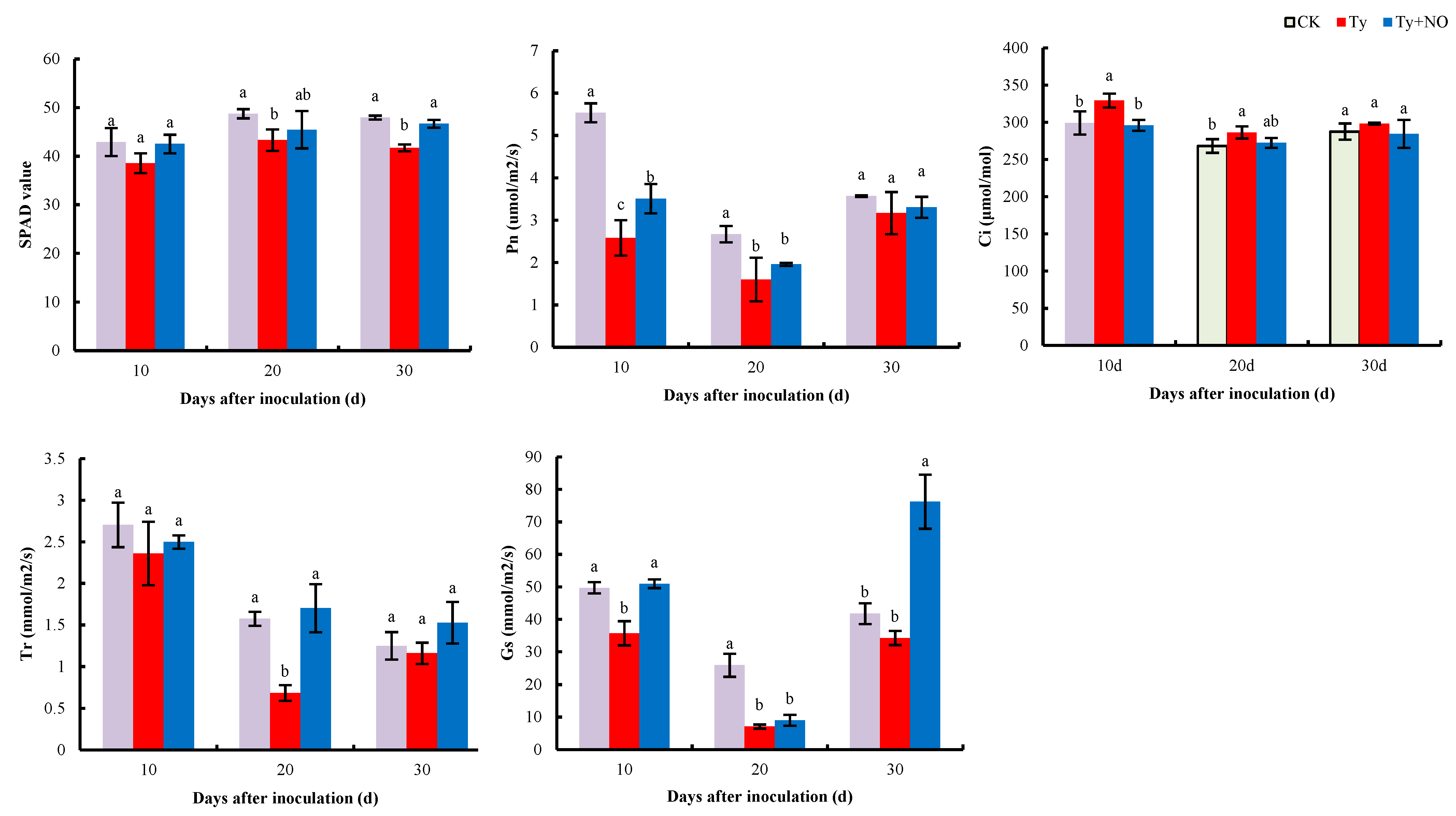
2.5.3. Changes in Photosynthetic Parameters
2.6. Effect of Exogenous NO on the Amplification of TYLCV in Tomato
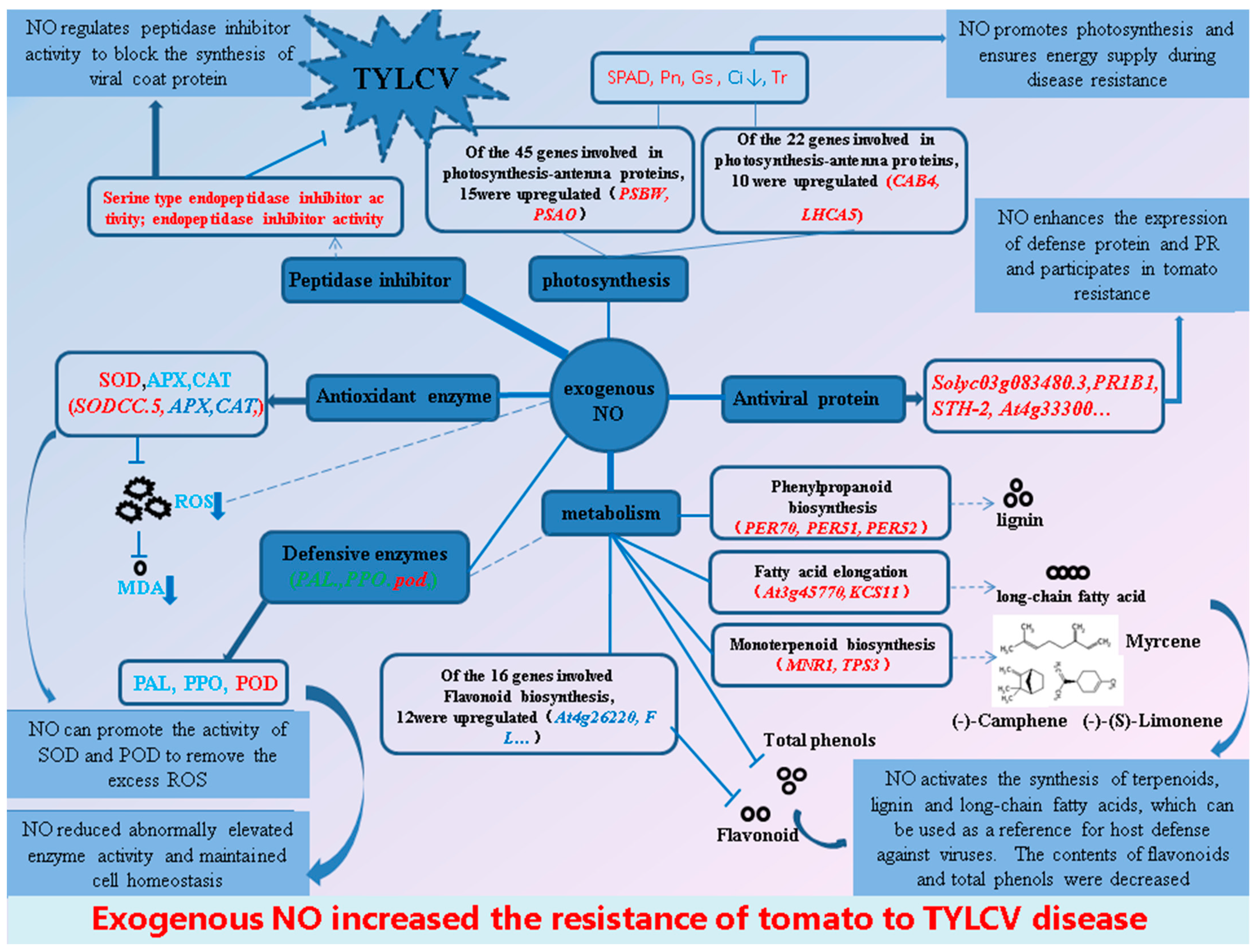
3. Discussion
4. Materials and Methods
4.1. Experimental Materials and Virus Inoculation
4.2. Experimental Treatments
4.3. Virus PCR Detection
4.4. Measurement of Phenotypic Indicators and Analysis of Disease Severity
4.5. Detection of Physiological Indices
4.6. RNA Extraction, Library Construction, and Sequencing
4.7. RNA-Seq Data Analysis
4.8. GO and KEGG Enrichment Analysis
4.9. qRT-PCR Analysis
4.10. Statistical Analysis of Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmad, P.; Ahanger, M.; Alyemeni, M.; Wijaya, L.; Alam, P. Exogenous application of nitric oxide modulates osmolyte metabolism, antioxidants, enzymes of ascorbate-glutathione cycle and promotes growth under cadmium stress in tomato. Protoplasma 2018, 255, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Dhaliwal, M.S.; Jindal, S.K.; Sharma, A.; Prasanna, H.C. Tomato yellow leaf curl virus disease of tomato and its management through resistance breeding: A review. J. Hortic. Sci. Biotechnol. 2020, 95, 425–444. [Google Scholar] [CrossRef]
- Scholthof, K.B.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef] [PubMed]
- Riley, D.; Srinivasan, R. Integrated Management of Tomato Yellow Leaf Curl Virus and its Whitefly Vector in Tomato. J. Econ. Entomol. 2019, 112, 1526–1540. [Google Scholar] [CrossRef] [PubMed]
- Sade, D.; Sade, N.; Brotman, Y.; Czosnek, H. Tomato yellow leaf curl virus (TYLCV)-resistant tomatoes share molecular mechanisms sustaining resistance with their wild progenitor Solanum habrochaites but not with TYLCV-susceptible tomatoes. Plant Sci. 2020, 295, 110439. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, A.; Sinha, S.K.; Praveen, S. Structure of replication initiator protein unites diverse viruses causing tomato leaf curl disease (ToLCD). Plant Sci. 2004, 166, 1063–1067. [Google Scholar] [CrossRef]
- Hormuzdi, S.G.; Bisaro, D.M. Genetic analysis of beet curly top virus: Examination of the roles of L2 and L3 genes in viral pathogenesis. Virology 1995, 206, 1044–1054. [Google Scholar] [CrossRef][Green Version]
- Kong, L.J. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 2000, 19, 3485–3495. [Google Scholar] [CrossRef]
- Mati, S.; Pegoraro, M.; Noris, E. The C2 protein of tomato yellow leaf curl Sardinia virus acts as a pathogenicity determinant and a 16-amino acid domain is responsible for inducing a hypersensitive response in plants. Virus Res. 2016, 215, 12–19. [Google Scholar] [CrossRef]
- Selth, L.A.; Dogra, S.C.; Rash Eed, M.S.; Randles, J.W.; Rezaian, M.A. Identification and Characterization of a Host Reversibly Glycosylated Peptide that Interacts with the Tomato leaf curl virus V1 Protein. Plant Mol. Biol. 2006, 61, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; He, S.; Geng, Q.; Duan, Y.; Guo, M.; Li, J.; Cao, Y. Synthesis and biological activity evaluation of novel amino acid derivatives as potential elicitors against Tomato yellow leaf curl virus. Amino Acids 2015, 47, 2495–2503. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, M.; Friedmann, M.; Lachman, O.; Yehezkel, A.; Nahon, S.; Cohen, S.; Pilowsky, M. Comparison of Resistance Level to Tomato Yellow Leaf Curl Virus Among Commercial Cultivars and Breeding Lines. Plant Dis. 1997, 81, 1425–1428. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; He, J.; Liu, H.; Liu, G.; Ren, X. Nitric oxide alleviates deterioration and preserves antioxidant properties in ‘Tainong’ mango fruit during ripening. Hortic. Environ. Biotechnol. 2017, 58, 27–37. [Google Scholar] [CrossRef]
- Del, C.F.; Nejamkin, A.; Cassia, R.; Correa-Aragunde, N.; Fernández, B.; Foresi, N.; Lombardo, C.; Ramirez, L.; Lamattina, L. The era of nitric oxide in plant biology: Twenty years tying up loose ends. Nitric Oxide Biol. Chem. 2019, 85, 17–27. [Google Scholar]
- Hong, J.K.; Yun, B.W.; Kang, J.G.; Raja, M.U.; Kwon, E.; Sorhagen, K.; Chu, C.; Wang, Y.; Loake, G.J. Nitric oxide function and signalling in plant disease resistance. J. Exp. Bot. 2008, 59, 147–154. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, S.; Zeng, K. Exogenous nitric oxide-induced postharvest disease resistance in citrus fruit to Colletotrichum gloeosporioides. J. Sci. Food Agr. 2016, 96, 505–512. [Google Scholar] [CrossRef]
- Lu, R.; Liu, Z.; Shao, Y.; Su, J.; Li, X.; Sun, F.; Zhang, Y.; Li, S.; Zhang, Y.; Cui, J.; et al. Nitric Oxide Enhances Rice Resistance to Rice Black-Streaked Dwarf Virus Infection. Rice 2020, 13, 24–36. [Google Scholar] [CrossRef]
- Liu, L.; Yu, M.; Zheng, Y.; Sheng, J.; Shen, L. Effect of Postharvest nitric oxide treatment on jasmonates biosynthesis and disease incidence to botrytis cinerea pers. in tomato fruits. Food Sci. 2010, 31, 457–461. [Google Scholar]
- Kobeasy, M.I.; El-Beltagi, H.S.; El-Shazly, M.A.; Khattab, E.A.H. Induction of resistance in Arachis hypogaea L. against Peanut mottle virus by nitric oxide and salicylic acid. Physiol. Mol. Plant Pathol. 2011, 76, 112–118. [Google Scholar] [CrossRef]
- Hermes, V.S.; Asta, P.D.; Amaral, F.P.; Anacleto, K.B. The regulation of transcription of genes related to oxidative stress and glutathione synthesis in Zea mays leaves by nitric oxide. Biol. Plantarum. 2013, 57, 620–626. [Google Scholar] [CrossRef]
- Kelm, M.; Dahmann, R.; Wink, D.; Feelisch, M. The nitric oxide/superoxide assay. Insights into the biological chemistry of the NO/O-2. interaction. J. Biol. Chem. 1997, 272, 9922–9932. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.Z. Study on Identification Method of Tomato Chlorosis Virus Disease and Preliminary Mapping of Resistance Genes. Master’s Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2021. [Google Scholar]
- Jing, C.; Zhang, H.; Feng, M.; Zuo, D.; Jiang, T. Transcriptome analysis of woodland strawberry (Fragaria vesca) response to the infection by Strawberry vein banding virus (SVBV). Virol. J. 2016, 13, 128–134. [Google Scholar]
- Chen, T.; Lv, Y.; Zhao, T.; Li, N.; Yang, Y.; Yu, W.; He, X.; Liu, T.; Zhang, B. Comparative transcriptome profiling of a resistant vs. susceptible tomato (Solanum lycopersicum) cultivar in response to infection by tomato yellow leaf curl virus. PLoS ONE 2013, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Coppola, V.; Coppola, M.; Rocco, M.; Digilio, M.; D’Ambrosio, C.; Renzone, G.; Martinelli, R.; Scaloni, A.; Pennacchio, F.; Rao, R.; et al. Transcriptomic and proteomic analysis of a compatible tomato-aphid interaction reveals a predominant salicylic acid-dependent plant response. BMC Genom. 2013, 14, 515–532. [Google Scholar] [CrossRef] [PubMed]
- Rabie, M.; Ratti, C.; Abdel Aleem, E.; Fattouh, F. Detection and molecular characterization of tomato yellow leaf curl virus naturally infecting Lycopersicon esculentum in Egypt. Acta Virol. 2017, 61, 252–263. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Agurla, S.; Gayatri, G.; Raghavendra, A.S. Nitric oxide as a secondary messenger during stomatal closure as a part of plant immunity response against pathogens. Nitric Oxide 2014, 43, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Bellin, D.; Asai, S.; De Lledonne, M.; Yoshioka, H. Nitric Oxide as a Mediator for Defense Responses. Mol. Plant Microbe. Interact. 2013, 26, 271–277. [Google Scholar] [CrossRef]
- Wendehenne, D.; Durner, J.; Klessig, D.F. Nitric oxide: A new player in plant signalling and defence responses. Curr. Opin. Plant Biol. 2004, 7, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Kim, M.; Kwak, H.; Choi, H.; Nam, M.; Choe, J.; Choi, B.; Han, S.; Kang, J.; Jung, C. Molecular dissection of distinct symptoms induced by tomato chlorosis virus and tomato yellow leaf curl virus based on comparative transcriptome analysis. Virology 2018, 516, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, N.; Singh, M.; Dubey, R.; Venkataramanappa, V.; Datta, D. Phytochemicals and antioxidative enzymes defence mechanism on occurrence of yellow vein mosaic disease of pumpkin (Cucurbita moschata). 3 Biotechnol. 2013, 3, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, Y.; Huang, Y.; Liu, J.; Xing, G.; Sun, S.; Li, S.; Xu, Z.; Xiong, A. A novel plant protein-disulfide isomerase participates in resistance response against the TYLCV in tomato. Planta 2020, 252, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Maddox, C.E.; Laur, L.M.; Tian, L. Antibacterial activity of phenolic compounds against the phytopathogen Xylella fastidiosa. Curr. Microbiol. 2010, 60, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Ganai, B.; Kamili, A.; Bhat, A.; Mir, Z.; Bhat, J.; Tyagi, A.; Islam, S.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212–213, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Gao, L.; Zhang, W.H.; Liu, J.K.; Liu, D.Q. Characteristic expression of wheat PR5 gene in response to infection by the leaf rust pathogen, Puccinia triticina. J. Plant Interact. 2015, 10, 132–141. [Google Scholar] [CrossRef]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Khanal, B.P.; Knoche, M. Mechanical properties of cuticles and their primary determinants. J. Exp. Bot. 2017, 68, 5351–5367. [Google Scholar] [CrossRef]
- Xing, J.; Chin, C.K. Modification of fatty acids in eggplant affects its resistance to Verticilliumdahliae. Physiol. Mol. Plant Pathol. 2000, 56, 217–225. [Google Scholar] [CrossRef]
- Duccio, R.L.; Caccioni, A.; Monica Guizzardi, A.; Daniela, M.; Biondi, B.; Agatino Renda, B.; Giuseppe Ruberto, B. Relationship between volatile components of citrus fruit essential oils and antimicrobial action on Penicillium digitatum and Penicillium italicum. Int. J. Food Microbiol. 1998, 43, 73–79. [Google Scholar]
- Wei, Y. Variation of Defensive Substance and Transcriptome Analysis of Different Resistant Pinus Massoniana Inoculated by pine Wood Nematode. Ph.D. Thesis, China Academy of Forestry Science, Beijing, China, 2016. [Google Scholar]
- Funayama, S.; Sonoike, K.; Terashima, I. Photosynthetic properties of leaves of Eupatorium makinoi infected by a geminivirus. Photosynth. Res. 1997, 53, 253–261. [Google Scholar] [CrossRef]
- Robert, G.N. The Biochemistry and Physiology of Plant Disease. BioScience 1986, 18, 418–419. [Google Scholar]
- Yu, L.; Guo, S.; Zhu, W.; Yan, J.; Hei, Y. Effects of tomato yellow leaf curl virus on photosynthetic characteristics and chloroplast ultra-structure of the tomato leaves. Acta Bot. Boreali-Occident. Sin. 2011, 31, 1355–1359. [Google Scholar]
- Skalamera, D.; Heath, M.C. Changes in the cytoskeleton accompanying infection-induced nuclear movements and the hypersensitive response in plant cells invaded by rust fungi. Plant J. 1998, 16, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Clemente, M.; Corigliano, M.; Pariani, S.; Sánchez-López, E.; Sander, V.; Ramos-Duarte, V. Plant Serine Protease Inhibitors: Biotechnology Application in Agriculture and Molecular Farming. Int. J. Mol. Sci. 2019, 20, 1345. [Google Scholar] [CrossRef] [PubMed]
- Navot, N.; Pichersky, E.; Zeidan, M.; Zamir, D.; Czosnek, H. Tomato yellow leaf curl virus: A whitefly-transmitted geminivirus with a single genomic component. Virology 1991, 185, 151–161. [Google Scholar] [CrossRef]
- Hohlfeld, K.; Tomassi, C.; Wegner, J.R.K.; Kesteleyn, B.; Linclau, B. Disubstituted BIS-THF moieties as new P2 ligands in nonpeptidal HIV-1 protease inhibitors. ACS Med. Chem. Lett. 2015, 58, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Huang, Y.; Xu, Z.-S.; Wang, F.; Xiong, A.-S. Salicylic acid-induced differential resistance to the Tomato yellow leaf curl virus among resistant and susceptible tomato cultivars. BMC Plant Biol. 2019, 19, 173. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, X.Y.; Qian, Y.J.; Zhou, X. Molecular characterization and infectivity of Papaya leaf curl China virus infecting tomato in China. J. Zhejiang Univ. Sci. B 2010, 11, 109–114. [Google Scholar] [CrossRef][Green Version]
- Li, Z.H.; Zhou, X.P.; Zhang, X.; Xie, Y. Molecular characterization of tomato-infecting begomoviruses in Yunnan, China. Arch. Virol. 2004, 149, 1721–1732. [Google Scholar] [CrossRef]
- Lapidot, M.; Friedmann, M.; Pilowsky, M.; Ben-Joseph, R.; Cohen, S. Effect of Host Plant Resistance to Tomato yellow leaf curl virus (TYLCV) on Virus Acquisition and Transmission by Its Whitefly Vector. Phytopathology 2001, 91, 1209–1213. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Huang, Z.; Liu, S.; Fu, J. Growth Response and Gene Expression in Antioxidant-related Enzymes in Two Bermudagrass Genotypes Differing in Salt Tolerance. J. Am. Soc. Hortic. Sci. 2012, 137, 134–143. [Google Scholar] [CrossRef]
- Chahid, K.; Laglaoui, A.; Zantar, S.; Ennabili, A. Antioxidant-enzyme reaction to the oxidative stress due to alpha-cypermethrin, chlorpyriphos, and pirimicarb in tomato (Lycopersicon esculentum Mill.). Environ. Sci. Pollut. Res. Int. 2015, 22, 18115–18126. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Q.; Wang, G.X.; Wang, X.D.; Guo, J.Y. Germination, osmotic adjustment, and antioxidant enzyme activities of gibberellin-pretreated Picea asperata seeds under water stress. New For. 2010, 39, 231–243. [Google Scholar] [CrossRef]
- Yoshiyuki, N.; Kozi, A. Purification of Ascorbate Peroxidase in Spinach Chloroplasts; Its Inactivation in Ascorbate-Depleted Medium and Reactivation by Monodehydroascorbate Radical. Plant Cell Physiol. 1987, 28, 131–140. [Google Scholar]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Gayoso, C.; Pomar, F.; Novo-Uzal, E.; Merino, F.; de Ilárduya, O.M. The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 2010, 10, 232–250. [Google Scholar] [CrossRef]
- Spagna, G.; Barbagallo, R.; Chisari, M.; Branca, F. Characterization of a tomato polyphenol oxidase and its role in browning and lycopene content. J. Agric. Food Chem. 2005, 53, 2032–2038. [Google Scholar] [CrossRef]
- Pastrana-Bonilla, E.; Akoh, C.C.; Sellappan, S.; Krewer, G. Phenolic content and antioxidant capacity of muscadine grapes. J. Agric. Food Chem. 2003, 51, 5497–5503. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Tang, M.; Wu, J. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999, 64, 555–559. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550–570. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Jing, J.; Yin-lei, W.; Li-ping, Z.; Rong, Z.; Ya-ru, L.; Tong-min, Z.; Wen-gui, Y. Selection of tomato reference genes for qRT-PCR. Jiangsu J. Agric. Sci. 2017, 33, 389–396. [Google Scholar]
- Livak, K.J.A.; Schmittgen, T.D.B. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2 ΔΔ C T Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
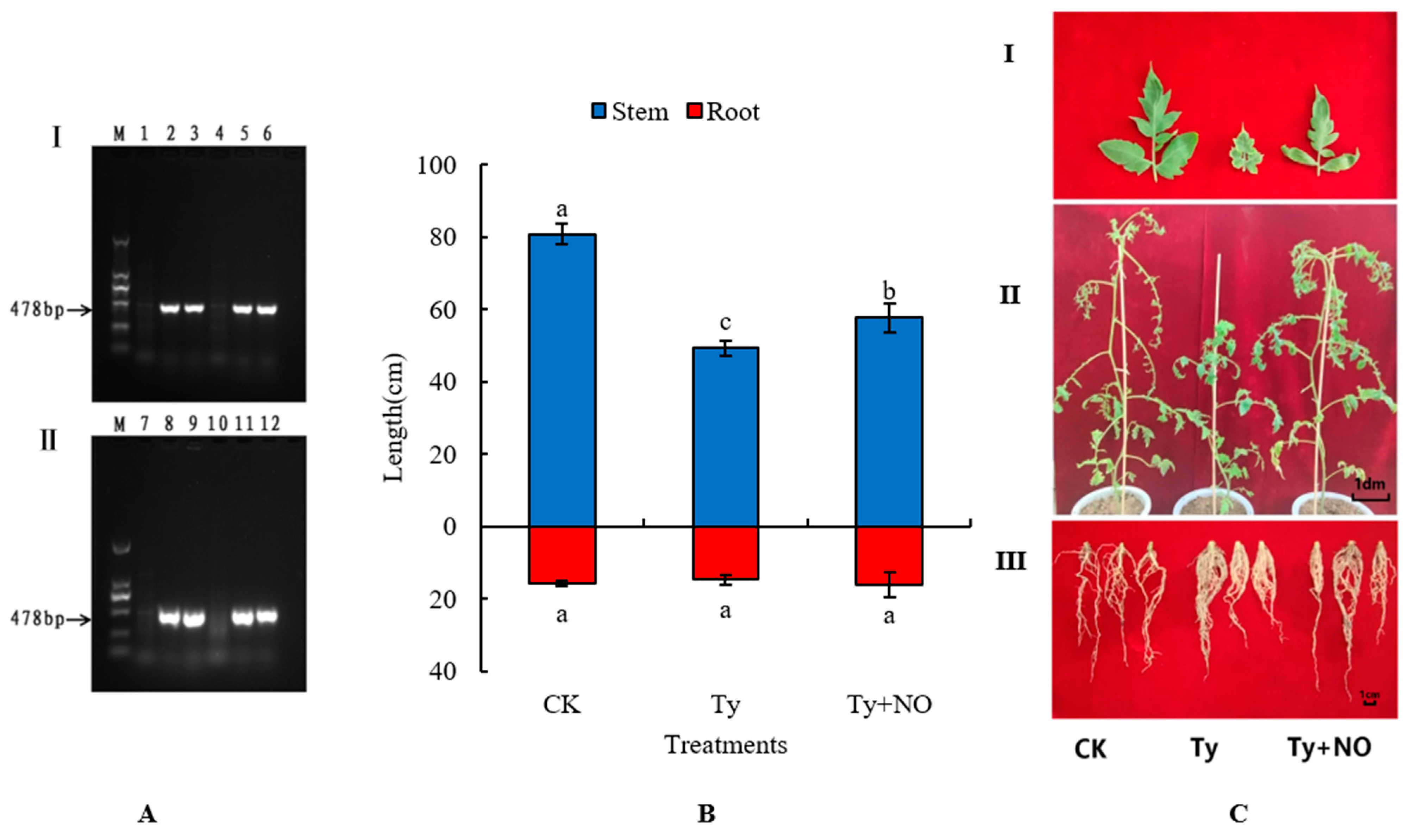

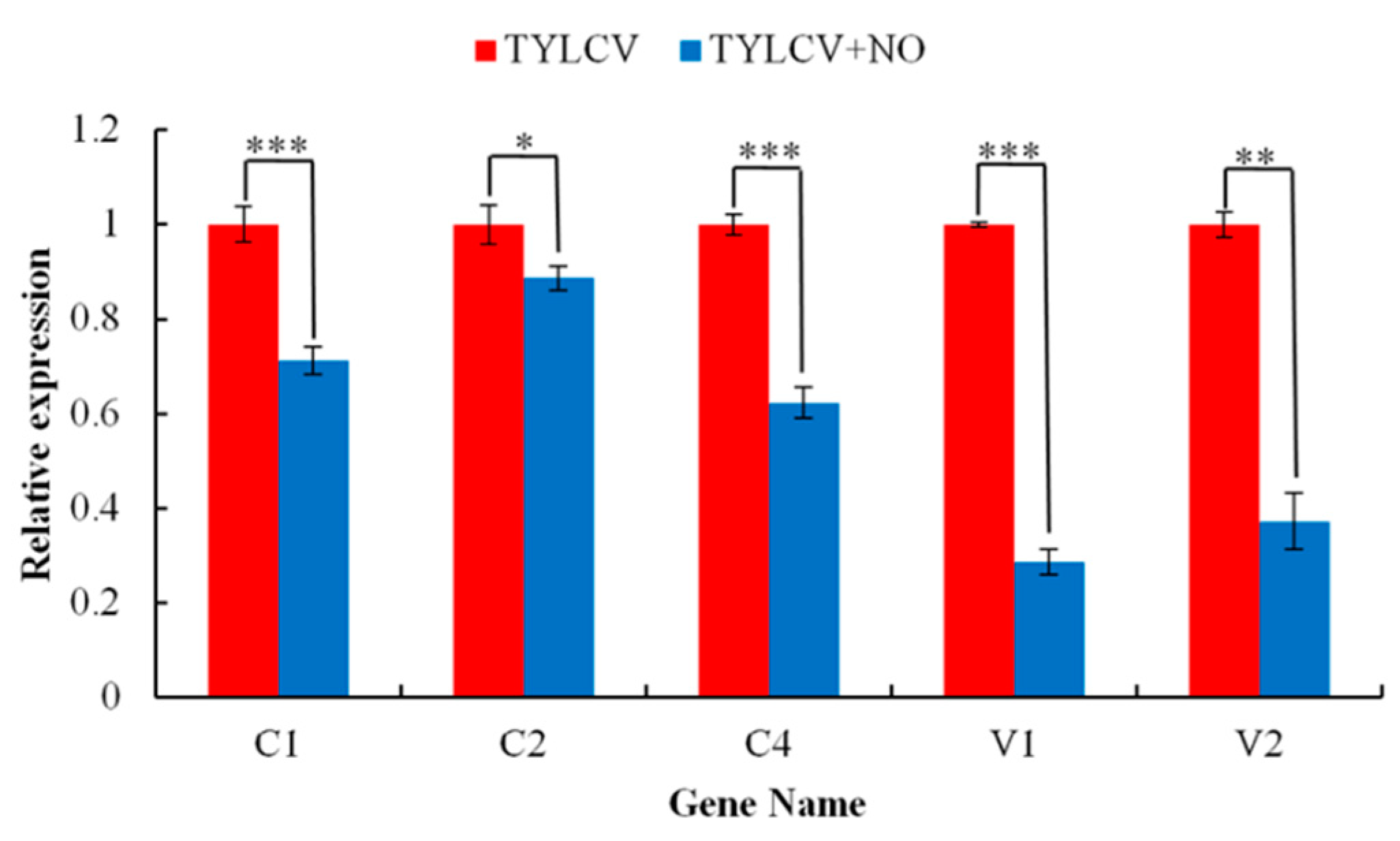
| Treatments | Dry Weight (g) | Fresh Weight (g) | Stem Diameter (mm) | Root Diameter (mm) |
|---|---|---|---|---|
| CK | 2.60 ± 0.44 a | 34.00 ± 1.31 b | 4.71 ± 0.30 b | 2.12 ± 0.58 b |
| TYLCV | 2.17 ± 0.45 a | 23.97 ± 1.67 c | 5.23 ± 0.16 a | 3.56 ± 0.51 a |
| TYLCV + NO | 2.93 ± 0.65 a | 37.13 ± 1.60 a | 5.25 ± 0.23 a | 3.20 ± 0.55 ab |
| Treatments | Total Number of Plants | Number of Plants Infected by TYLCV | Infection Rate (%) | Disease Index (%) | Control Rate (%) |
|---|---|---|---|---|---|
| CK | 20 | 0 | 0% | — | — |
| TYLCV | 20 | 20 | 100% | 62.5% | — |
| TYLCV + NO | 20 | 20 | 100% | 35.0% | 44.0% |
| Sample Name | Concentration (ng/μL) | Volume (μL) | Total (μg) | RIN Value |
|---|---|---|---|---|
| CK-1 | 612 | 40 | 24.48 | 7.7 |
| CK-2 | 486 | 39 | 18.95 | 7.8 |
| CK-3 | 585 | 38 | 22.23 | 7.4 |
| TYLCV-1 | 495 | 38 | 18.81 | 7.6 |
| TYLCV-2 | 621 | 39 | 24.22 | 7.5 |
| TYLCV-3 | 486 | 38 | 18.47 | 7.5 |
| TYLCV-NO-1 | 591 | 39 | 23.05 | 7.6 |
| TYLCV-NO-2 | 573 | 38 | 21.77 | 7.5 |
| TYLCV-NO-3 | 462 | 38 | 17.56 | 7.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Wang, B.; Zhu, X.; Zhao, Y.; Jin, B.; Wei, X. Exogenous Nitric Oxide Alleviates the Damage Caused by Tomato Yellow Leaf Curl Virus in Tomato through Regulation of Peptidase Inhibitor Genes. Int. J. Mol. Sci. 2022, 23, 12542. https://doi.org/10.3390/ijms232012542
Wang X, Wang B, Zhu X, Zhao Y, Jin B, Wei X. Exogenous Nitric Oxide Alleviates the Damage Caused by Tomato Yellow Leaf Curl Virus in Tomato through Regulation of Peptidase Inhibitor Genes. International Journal of Molecular Sciences. 2022; 23(20):12542. https://doi.org/10.3390/ijms232012542
Chicago/Turabian StyleWang, Xian, Baoqiang Wang, Xiaolin Zhu, Ying Zhao, Baoxia Jin, and Xiaohong Wei. 2022. "Exogenous Nitric Oxide Alleviates the Damage Caused by Tomato Yellow Leaf Curl Virus in Tomato through Regulation of Peptidase Inhibitor Genes" International Journal of Molecular Sciences 23, no. 20: 12542. https://doi.org/10.3390/ijms232012542
APA StyleWang, X., Wang, B., Zhu, X., Zhao, Y., Jin, B., & Wei, X. (2022). Exogenous Nitric Oxide Alleviates the Damage Caused by Tomato Yellow Leaf Curl Virus in Tomato through Regulation of Peptidase Inhibitor Genes. International Journal of Molecular Sciences, 23(20), 12542. https://doi.org/10.3390/ijms232012542




