Response of Ruminal Microbiota–Host Gene Interaction to High-Altitude Environments in Tibetan Sheep
Abstract
1. Introduction
2. Results
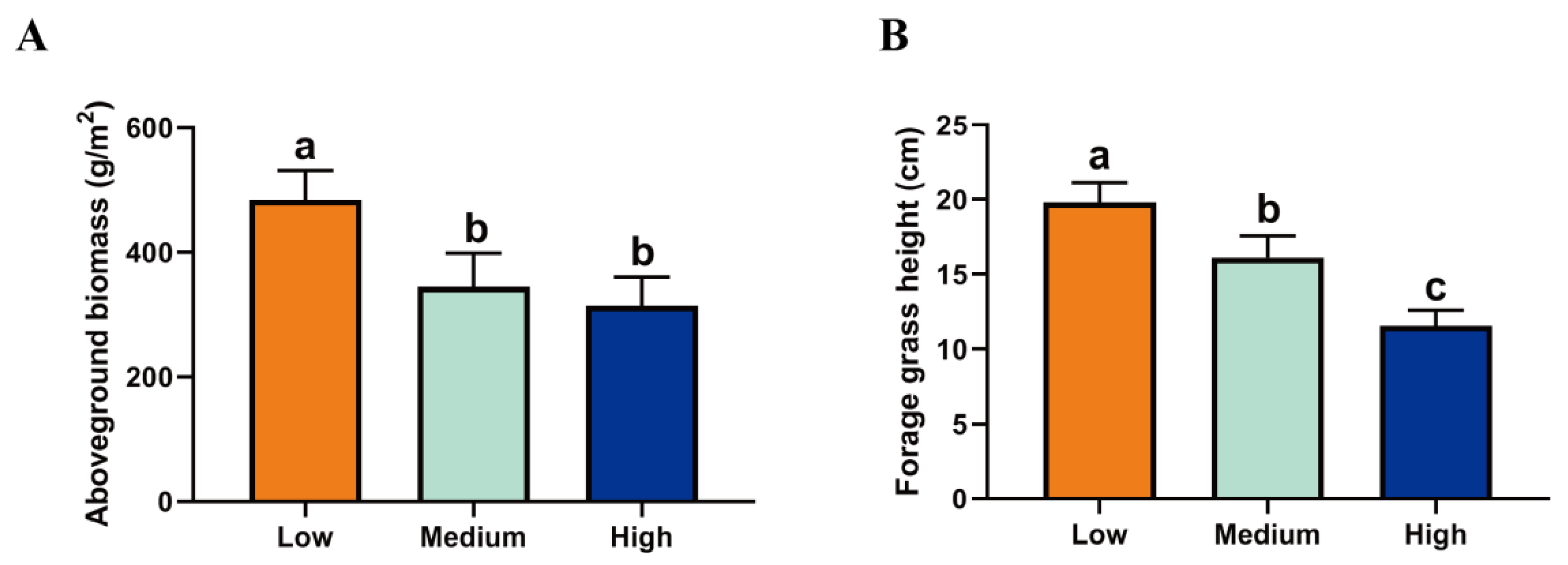
2.1. Forage Species and Nutrient Composition at Different Altitudes
2.2. Serum Immune Levels of Tibetan Sheep at Different Altitudes
2.3. Characteristics of Rumen VFAs and Epithelial Morphology
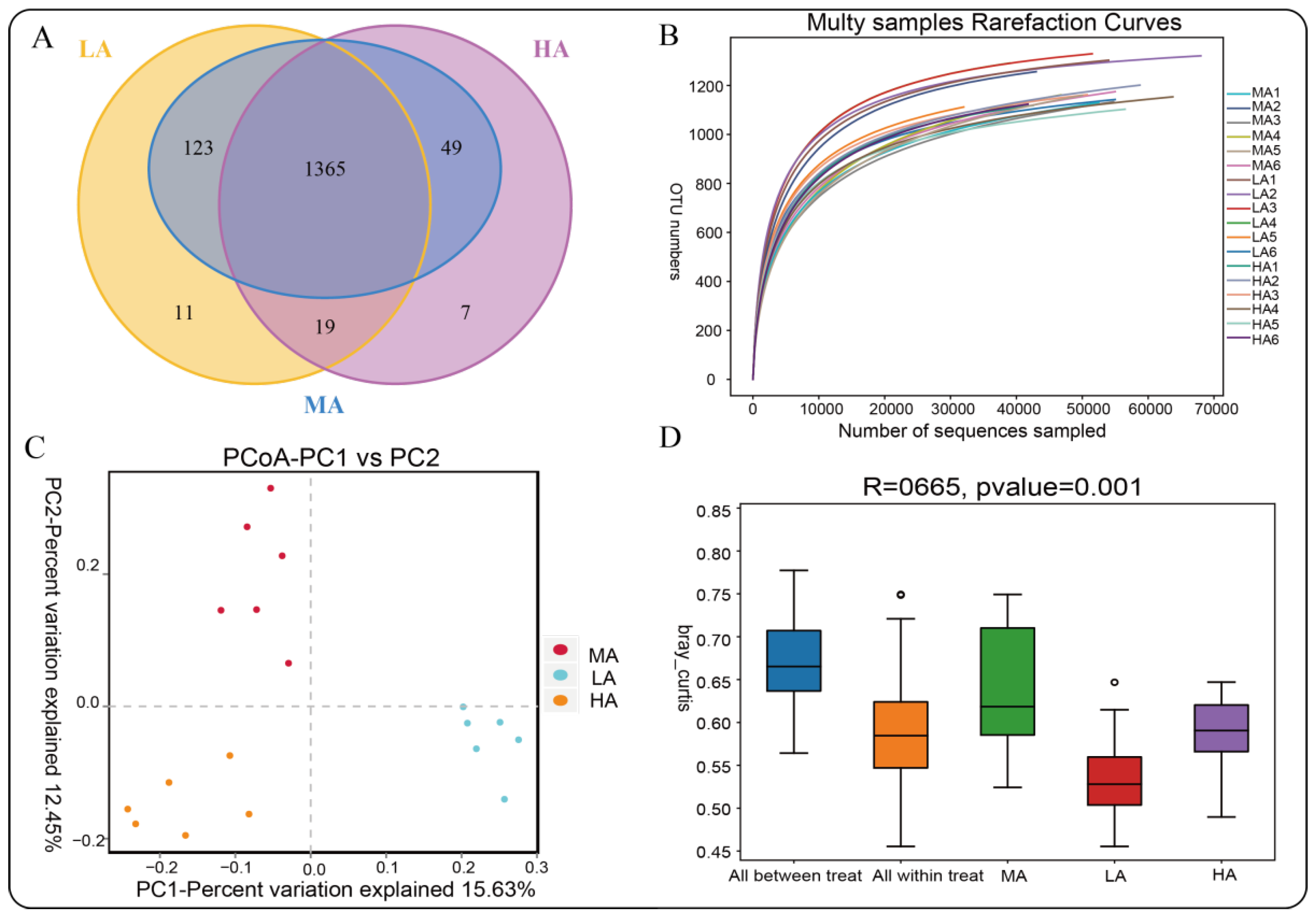
2.4. Microbial Diversity of Tibetan Sheep at Different Altitudes
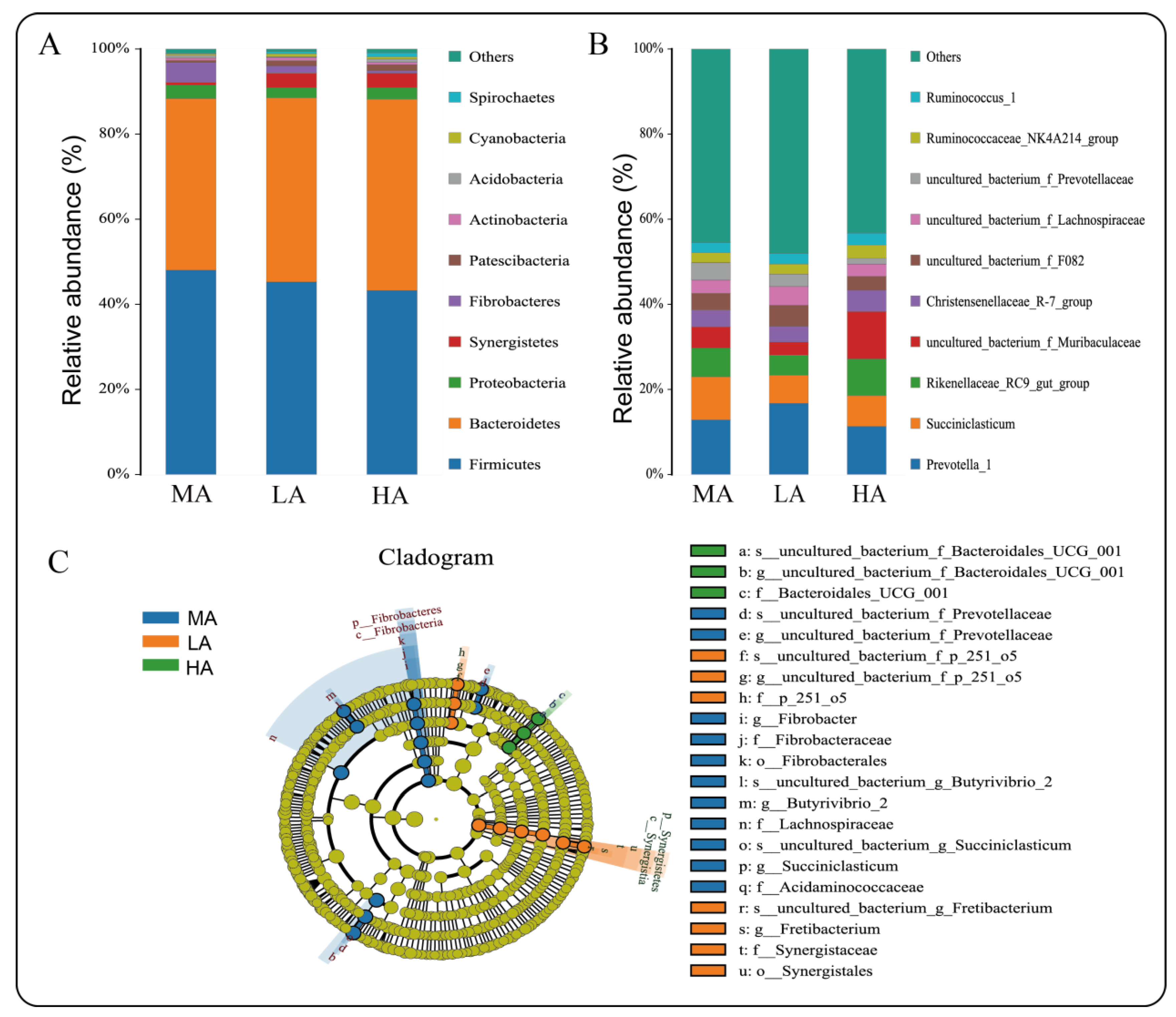
2.5. Microbial Composition and Abundance of Tibetan Sheep at Different Altitudes
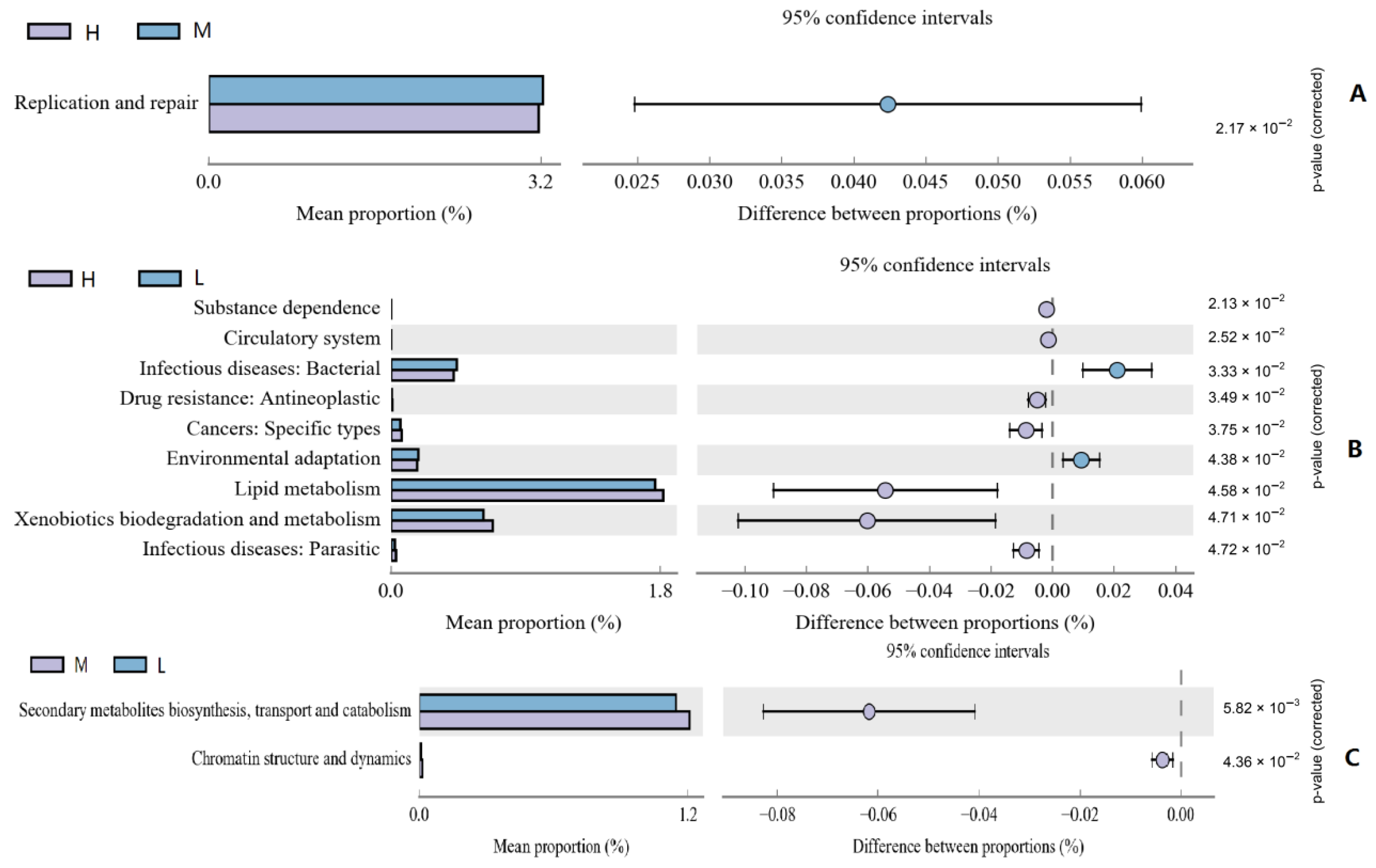
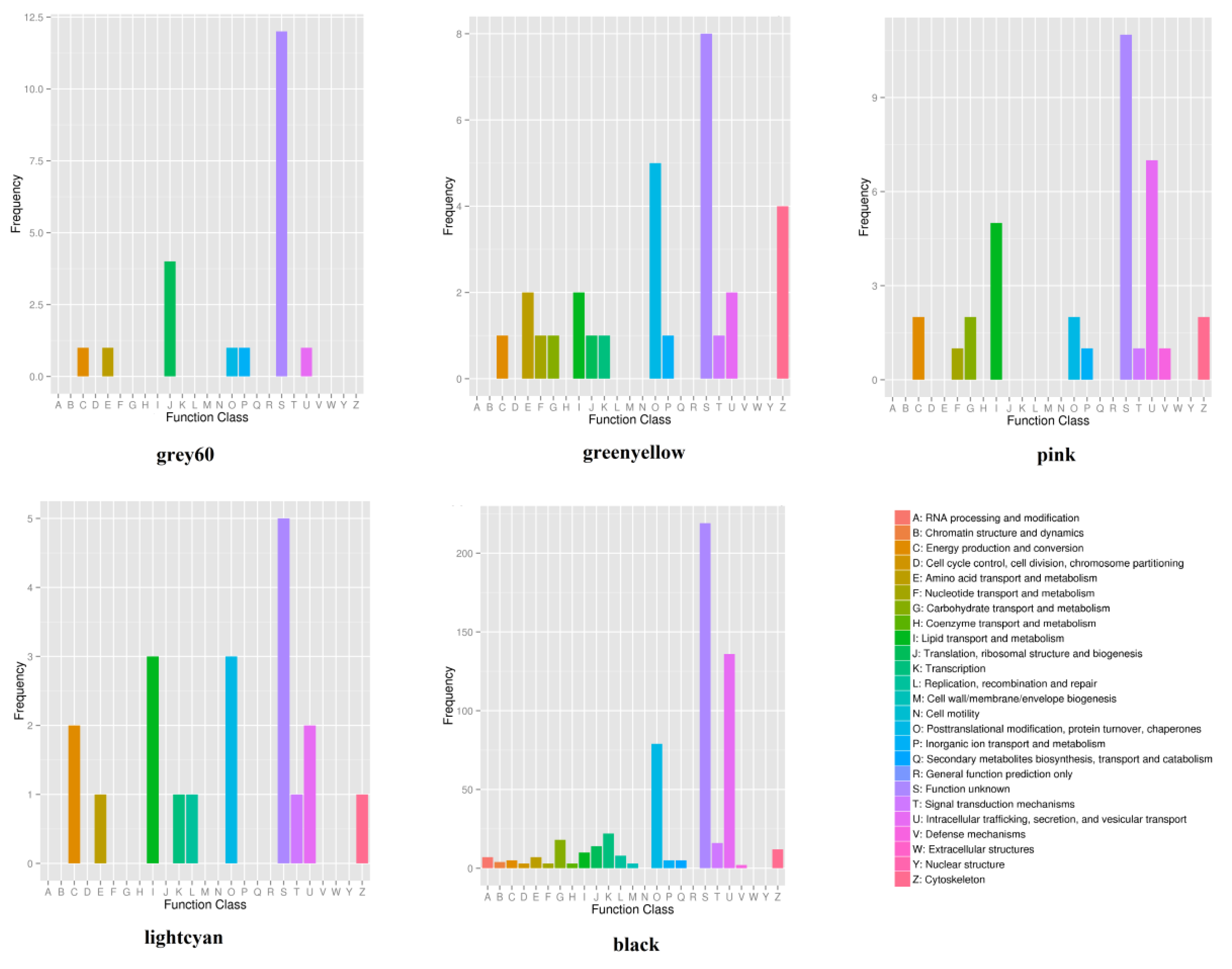
2.6. Microbial Gene Function
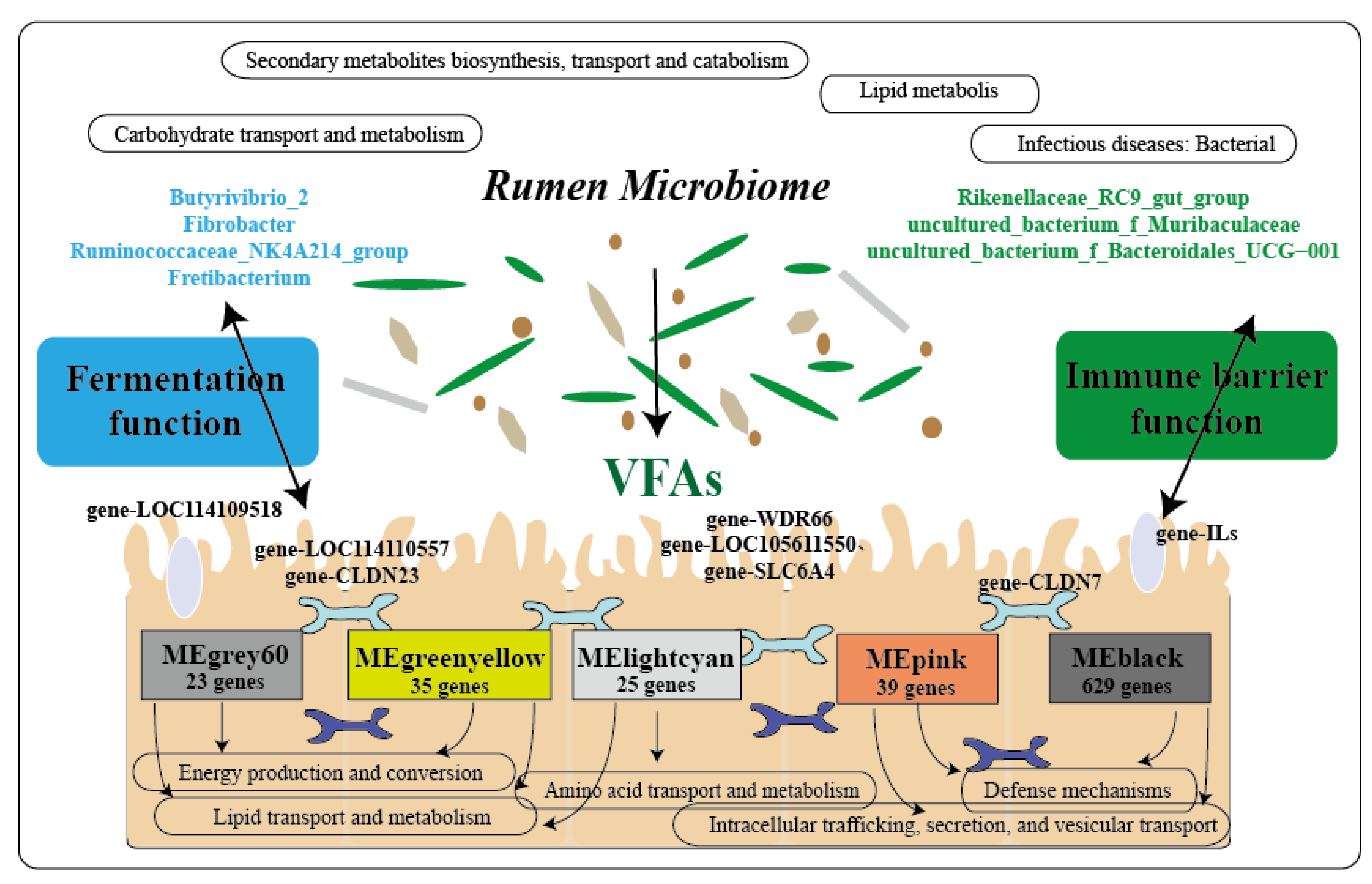
2.7. Microbiota–Host Gene Interactions Influence Rumen Fermentation and the Epithelial Barrier
2.7.1. WGCNA of Host Transcriptome and Phenotypic Traits
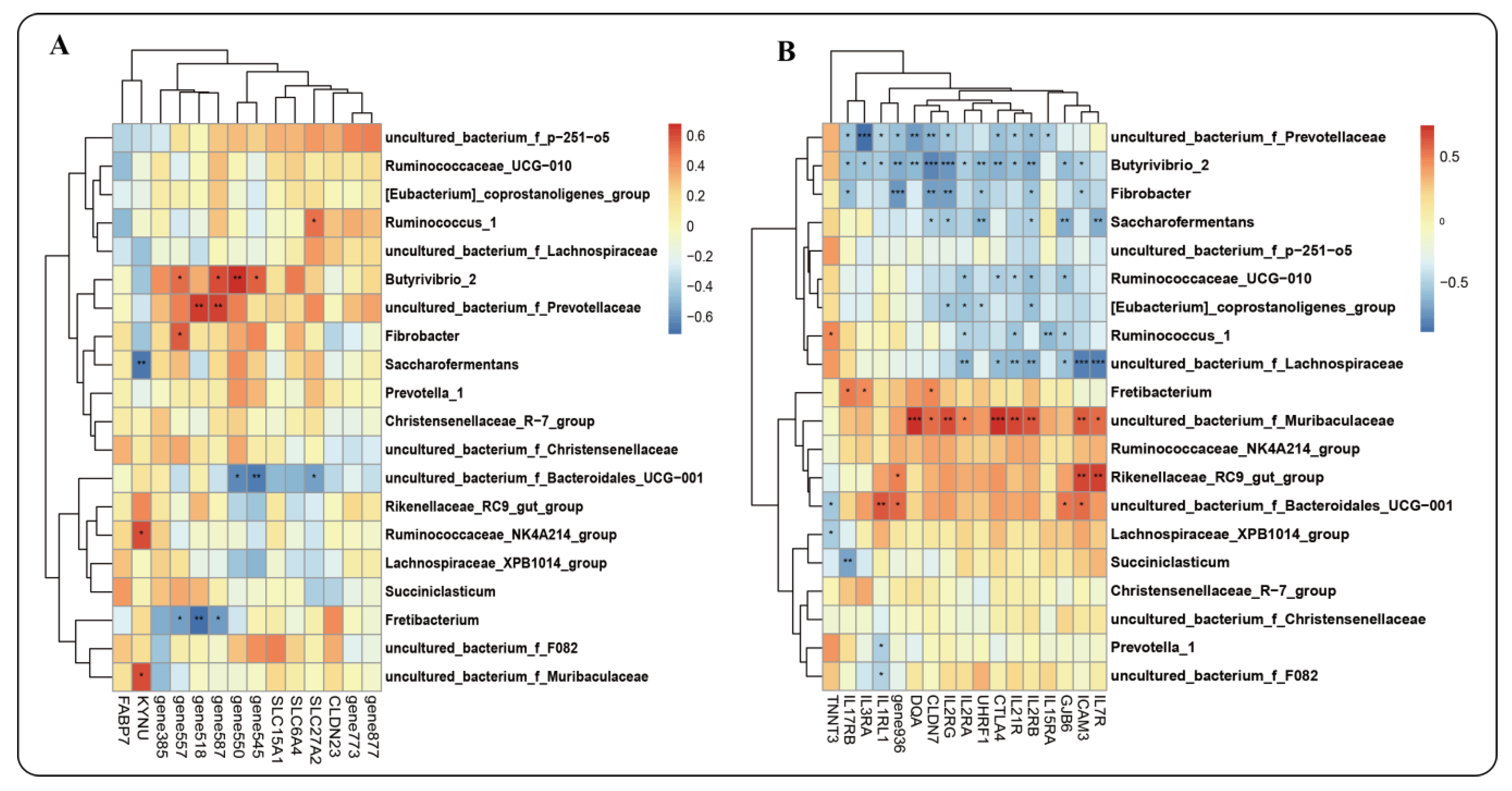
2.7.2. Interaction between Host Genes and Microbiota
3. Discussion
4. Materials and Methods
4.1. Experimental Design and Location
 , n = 6/group, Non-pregnant) were selected as research subjects. All sheep were under traditional local natural grazing management and no supplementary feeding was carried out, and grazed on fixed pastures from 8 am to 7 pm. In August 2020, samples were collected in Zhuoni (LA, 2500 m, Gansu province), Haiyan (MA, 3500 m, Qinghai province), and Yushu (HA, 4500 m, Qinghai province) in the Qinghai–Tibet Plateau region (Figure 12), and the three sampling sites belong to Qinghai Plateau and its adjacent areas.
, n = 6/group, Non-pregnant) were selected as research subjects. All sheep were under traditional local natural grazing management and no supplementary feeding was carried out, and grazed on fixed pastures from 8 am to 7 pm. In August 2020, samples were collected in Zhuoni (LA, 2500 m, Gansu province), Haiyan (MA, 3500 m, Qinghai province), and Yushu (HA, 4500 m, Qinghai province) in the Qinghai–Tibet Plateau region (Figure 12), and the three sampling sites belong to Qinghai Plateau and its adjacent areas.4.2. Sample Collection
4.3. Determination of Forage Nutrient Composition and Rumen VFAs
4.4. Serum Immune Index Determination
4.5. Morphological Analysis
4.6. DNA Extraction and 16S rRNA Sequencing
4.7. RNA Extraction and Transcriptome Sequencing
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, Y.; Xie, M.; Chen, W.; Talbot, R.; Maddox, J.F.; Faraut, T.; Wu, C.; Muzny, D.M.; Li, Y.; Zhang, W.; et al. The sheep genome illuminates biology of the rumen and lipid metabolism. Science 2014, 344, 1168–1173. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wu, H.; Liu, S.; Chai, S.; Meng, Q.; Zhou, Z. Dynamic alterations in yak rumen bacteria community and metabolome characteristics in response to feed type. Front. Microbiol. 2019, 10, 1116. [Google Scholar] [CrossRef] [PubMed]
- Bergman, E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990, 70, 567–590. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Miyamoto, N.; Shibata, K.; Valasek, M.A.; Motoike, T.; Kedzierski, R.M.; Yanagisawa, M. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc. Natl. Acad. Sci. USA 2004, 101, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Kang, S.G.; Park, J.H.; Yanagisawa, M.; Kim, C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013, 145, 396–406. [Google Scholar] [CrossRef]
- Martin, R.; Nauta, A.J.; Ben, A.K.; Knippels, L.M.; Knol, J.; Garssen, J. Early life: Gut microbiota and immune development in infancy. Benef. Microbes 2010, 1, 367–382. [Google Scholar] [CrossRef]
- Tlaskalová-Hogenová, H.; Stěpánková, R.; Kozáková, H.; Hudcovic, T.; Vannucci, L.; Tučková, L.; Rossmann, P.; Hrnčíř, T.; Kverka, M.; Zákostelská, Z.; et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: Contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol. Immunol. 2011, 8, 110–120. [Google Scholar] [CrossRef]
- Shabat, S.K.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Berg, M.M.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The gut microbiota--masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Bevins, C.L.; Salzman, N.H. The potter’s wheel: The host’s role in sculpting its microbiota. Cell Mol. Life Sci. 2011, 68, 3675–3685. [Google Scholar] [CrossRef]
- Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011, 9, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.; Zilber-Rosenberg, I. Do microbiotas warm their hosts? Gut Microbes 2016, 7, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Sha, Y.; Dingkao, R.; Zhang, W.; Lv, W.; Wei, H.; Shi, H.; Hu, J.; Wang, J.; Li, S.; et al. Interactions between rumen microbes, VFAs, and host genes regulate nutrient absorption and epithelial barrier function during cold season nutritional stress in Tibetan sheep. Front. Microbiol. 2020, 11, 593062. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Guan, L.L. Understanding host-microbial interactions in rumen: Searching the best opportunity for microbiota manipulation. J. Anim. Sci. Biotechnol. 2017, 8, 8. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, D.; Wang, L.; Hao, J.; Wang, J.; Zhou, X.; Wang, W.; Qiu, Q.; Huang, X.; Zhou, J.; et al. Convergent evolution of rumen microbiomes in high-altitude mammals. Curr. Biol. 2016, 26, 1873–1879. [Google Scholar] [CrossRef]
- Han, N.; Pan, Z.; Liu, G.; Yang, R.; Yujing, B. Hypoxia: The "Invisible Pusher" of gut microbiota. Front. Microbiol. 2021, 12, 690600. [Google Scholar] [CrossRef]
- Wu, D.; Vinitchaikul, P.; Deng, M.; Zhang, G.; Sun, L.; Wang, H.; Gou, X.; Mao, H.; Yang, S. Exploration of the effects of altitude change on bacteria and fungi in the rumen of yak (Bos grunniens). Arch. Microbiol. 2021, 203, 835–846. [Google Scholar] [CrossRef]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.W.; Soh, E.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic microbiome evolution in social bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef]
- Ross, A.A.; Müller, K.M.; Weese, J.S.; Neufeld, J.D. Comprehensive skin microbiome analysis reveals the uniqueness of human skin and evidence for phylosymbiosis within the class Mammalia. Proc. Natl. Acad. Sci. USA 2018, 115, E5786–E5795. [Google Scholar] [CrossRef]
- Zepeda, M.M.; Xiong, Z.; Escalera-Zamudio, M.; Runge, A.K.; Thézé, J.; Streicker, D.; Frank, H.K.; Loza-Rubio, E.; Liu, S.; Ryder, O.A.; et al. Hologenomic adaptations underlying the evolution of sanguivory in the common vampire bat. Nat. Ecol. Evol. 2018, 2, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Zhang, S.; Xu, H.; Kong, F.; Yu, X.; Wang, P.; Yang, M.; Li, D.; Zhang, M.; Ni, Q.; et al. Gut microbiota of Tibetans and Tibetan pigs varies between high and low altitude environments. Microbiol. Res. 2020, 235, 126447. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Wanapat, M.; Yan, T.; Hou, F. Altitude influences microbial diversity and herbage fermentation in the rumen of yaks. BMC Microbiol. 2020, 20, 370. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Y.; Sun, Y.; Wang, R.; Zhang, J.; Jia, Z. Plateau hypoxia attenuates the metabolic activity of intestinal flora to enhance the bioavailability of nifedipine. Drug Deliv. 2018, 25, 1175–1181. [Google Scholar] [CrossRef]
- Ma, Y.; Ma, S.; Chang, L.; Wang, H.; Ga, Q.; Ma, L.; Bai, Z.; Shen, Y.; Ge, R.L. Gut microbiota adaptation to high altitude in indigenous animals. Biochem. Biophys. Res. Commun. 2019, 516, 120–126. [Google Scholar] [CrossRef]
- Suzuki, T.A.; Martins, F.M.; Nachman, M.W. Altitudinal variation of the gut microbiota in wild house mice. Mol. Ecol. 2019, 28, 2378–2390. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Xie, F.; Sun, D.; Liu, J.; Zhu, W.; Mao, S. Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome 2019, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.A.; Phifer-Rixey, M.; Mack, K.L.; Sheehan, M.J.; Lin, D.; Bi, K.; Nachman, M.W. Host genetic determinants of the gut microbiota of wild mice. Mol. Ecol. 2019, 28, 3197–3207. [Google Scholar] [CrossRef]
- Amato, K.R.; Leigh, S.R.; Kent, A.; Mackie, R.I.; Yeoman, C.J.; Stumpf, R.M.; Wilson, B.A.; Nelson, K.E.; White, B.A.; Garber, P.A. The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra). Microb. Ecol. 2015, 69, 434–443. [Google Scholar] [CrossRef]
- Zhu, L.; Wu, Q.; Deng, C.; Zhang, M.; Zhang, C.; Chen, H.; Lu, G.; Wei, F. Adaptive evolution to a high purine and fat diet of carnivorans revealed by gut microbiomes and host genomes. Environ. Microbiol. 2018, 20, 1711–1722. [Google Scholar] [CrossRef]
- An, D.; Dong, X.; Dong, Z. Prokaryote diversity in the rumen of yak (Bos grunniens) and Jinnan cattle (Bos taurus) estimated by 16S rDNA homology analyses. Anaerobe 2005, 11, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Cao, G.; Wang, Q.; Jing, Z.; Ding, L.; Long, R. Changes in plant biomass and species composition of alpine Kobresia meadows along altitudinal gradient on the Qinghai-Tibetan Plateau. Sci. China C Life Sci. 2008, 51, 86–94. [Google Scholar] [CrossRef]
- Russell, J.B.; Rychlik, J.L. Factors that alter rumen microbial ecology. Science 2001, 292, 1119–1122. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, T.; Xu, S.; Ma, L.; Han, X.; Wang, X.; Zhang, X.; Hu, L.; Zhao, N.; Chen, Y.; et al. Effect of dietary concentrate to forage ratio on growth performance, rumen fermentation and bacterial diversity of Tibetan sheep under barn feeding on the Qinghai-Tibetan plateau. Peerj 2019, 7, e7462. [Google Scholar] [CrossRef]
- Polyorach, S.; Wanapat, M.; Cherdthong, A. Influence of yeast fermented cassava chip protein (yefecap) and roughage to concentrate ratio on ruminal fermentation and microorganisms using in vitro gas production technique. Asian-Australas J. Anim. Sci. 2014, 27, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Wathes, D.C.; Cheng, Z.; Chowdhury, W.; Fenwick, M.A.; Fitzpatrick, R.; Morris, D.G.; Patton, J.; Murphy, J.J. Negative energy balance alters global gene expression and immune responses in the uterus of postpartum dairy cows. Physiol. Genom. 2009, 39, 1–13. [Google Scholar] [CrossRef]
- Graham, C.; Simmons, N.L. Functional organization of the bovine rumen epithelium. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 288, R173–R181. [Google Scholar] [CrossRef]
- Rickard, M.D.; Ternouth, J.H. The effect of the increased dietary volatile fatty acids on the morphological and physiological development of lambs with particular reference to the rumen. J. Agric. Sci. 1965, 65, 371–377. [Google Scholar] [CrossRef]
- Baldwin, R.T.; Jesse, B.W. Technical note: Isolation and characterization of sheep ruminal epithelial cells. J. Anim. Sci. 1991, 69, 3603–3609. [Google Scholar] [CrossRef]
- Bell, A.W. Regulation of organic nutrient metabolism during transition from late pregnancy to early lactation. J. Anim. Sci. 1995, 73, 2804–2819. [Google Scholar] [CrossRef]
- Fan, Q.; Cui, X.; Wang, Z.; Chang, S.; Wanapat, M.; Yan, T.; Hou, F. Rumen microbiota of tibetan sheep (ovis aries) adaptation to extremely cold season on the qinghai-tibetan plateau. Front. Vet Sci. 2021, 8, 673822. [Google Scholar] [CrossRef]
- Jia, P.; Cui, K.; Ma, T.; Wan, F.; Wang, W.; Yang, D.; Wang, Y.; Guo, B.; Zhao, L.; Diao, Q. Influence of dietary supplementation with Bacillus licheniformis and Saccharomyces cerevisiae as alternatives to monensin on growth performance, antioxidant, immunity, ruminal fermentation and microbial diversity of fattening lambs. Sci. Rep. 2018, 8, 16712. [Google Scholar] [CrossRef]
- Liu, H.; Hu, L.; Han, X.; Zhao, N.; Xu, T.; Ma, L.; Wang, X.; Zhang, X.; Kang, S.; Zhao, X.; et al. Tibetan sheep adapt to plant phenology in alpine meadows by changing rumen microbial community structure and function. Front. Microbiol. 2020, 11, 587558. [Google Scholar] [CrossRef]
- Nuriel-Ohayon, M.; Neuman, H.; Koren, O. Microbial changes during pregnancy, birth, and infancy. Front. Microbiol. 2016, 7, 1031. [Google Scholar] [CrossRef]
- Ransom-Jones, E.; Jones, D.L.; McCarthy, A.J.; McDonald, J.E. The Fibrobacteres: An important phylum of cellulose-degrading bacteria. Microb. Ecol. 2012, 63, 267–281. [Google Scholar] [CrossRef]
- Seshadri, R.; Leahy, S.C.; Attwood, G.T.; Teh, K.H.; Lambie, S.C.; Cookson, A.L.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Hadjithomas, M.; Varghese, N.J.; et al. Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat. Biotechnol. 2018, 36, 359–367. [Google Scholar] [CrossRef]
- Zhang, T.; Sun, P.; Geng, Q.; Fan, H.; Gong, Y.; Hu, Y.; Shan, L.; Sun, Y.; Shen, W.; Zhou, Y. Disrupted spermatogenesis in a metabolic syndrome model: The role of vitamin A metabolism in the gut-testis axis. Gut 2021, 71, 78–87. [Google Scholar] [CrossRef]
- Xue, D.; Chen, H.; Chen, F.; He, Y.; Zhao, C.; Zhu, D.; Zeng, L.; Li, W. Analysis of the rumen bacteria and methanogenic archaea of yak (Bos grunniens) steers grazing on the Qinghai-Tibetan Plateau. Livest. Sci. 2016, 188, 61–71. [Google Scholar] [CrossRef]
- Pan, X.; Cai, Y.; Li, Z.; Chen, X.; Heller, R.; Wang, N.; Wang, Y.; Zhao, C.; Wang, Y.; Xu, H.; et al. Modes of genetic adaptations underlying functional innovations in the rumen. Sci. China Life Sci. 2021, 64, 1–21. [Google Scholar] [CrossRef]
- Madsen, D.H.; Szabo, R.; Molinolo, A.A.; Bugge, T.H. TMPRSS13 deficiency impairs stratum corneum formation and epidermal barrier acquisition. Biochem. J. 2014, 461, 487–495. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, C.; Kemmner, W. Wdr66 is a novel marker for risk stratification and involved in epithelial-mesenchymal transition of esophageal squamous cell carcinoma. BMC Cancer 2013, 13, 137. [Google Scholar] [CrossRef]
- Lane, M.A.; Baldwin, R.T.; Jesse, B.W. Developmental changes in ketogenic enzyme gene expression during sheep rumen development. J. Anim. Sci. 2002, 80, 1538–1544. [Google Scholar] [CrossRef]
- Ren, Z.; Yao, R.; Liu, Q.; Deng, Y.; Shen, L.; Deng, H.; Zuo, Z.; Wang, Y.; Deng, J.; Cui, H.; et al. Effects of antibacterial peptides on rumen fermentation function and rumen microorganisms in goats. PLoS ONE 2019, 14, e221815. [Google Scholar] [CrossRef]
- Gaffen, S.L. An overview of IL-17 function and signaling. Cytokine 2008, 43, 402–407. [Google Scholar] [CrossRef]
- Wang, J.; Chai, J. Molecular actions of NLR immune receptors in plants and animals. Sci. China Life Sci. 2020, 63, 1303–1316. [Google Scholar] [CrossRef]
- Sommer, F.; Nookaew, I.; Sommer, N.; Fogelstrand, P.; Bäckhed, F. Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol. 2015, 16, 62. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Liang, G.; Guan, L.L. Regulation of rumen development in neonatal ruminants through microbial metagenomes and host transcriptomes. Genome Biol. 2019, 20, 172. [Google Scholar] [CrossRef]
- Yang, L.Y.; Chen, J.; Cheng, X.L.; Xi, D.M.; Yang, S.L.; Deng, W.D.; Mao, H.M. Phylogenetic analysis of 16S rRNA gene sequences reveals rumen bacterial diversity in Yaks (Bos grunniens). Mol. Biol. Rep. 2010, 37, 553–562. [Google Scholar] [CrossRef]
- Liu, R.Z.; Mita, R.; Beaulieu, M.; Gao, Z.; Godbout, R. Fatty acid binding proteins in brain development and disease. Int. J. Dev. Biol. 2010, 54, 1229–1239. [Google Scholar] [CrossRef]
- Xing, T.; Benderman, L.J.; Sabu, S.; Parker, J.; Yang, J.; Lu, Q.; Ding, L.; Chen, Y.H. Tight junction protein claudin-7 is essential for intestinal epithelial stem cell self-renewal and differentiation. Cell Mol. Gastroenterol. Hepatol. 2020, 9, 641–659. [Google Scholar] [CrossRef] [PubMed]
- Spolski, R.; Gromer, D.; Leonard, W.J. The γ (c) family of cytokines: Fine-tuning signals from IL-2 and IL-21 in the regulation of the immune response. F1000Research 2017, 6, 1872. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.S.; Sansom, D.M. The emerging role of CTLA4 as a cell-extrinsic regulator of T cell responses. Nat. Rev. Immunol. 2011, 11, 852–863. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Liu, D.; Wang, Y.; Liu, L.; Li, J.; Yuan, J.; Jiang, Z.; Jiang, Z.; Hsiao, W.W.; Liu, H.; et al. Ginseng polysaccharides alter the gut microbiota and kynurenine/tryptophan ratio, potentiating the antitumour effect of antiprogrammed cell death 1/programmed cell death ligand 1 (anti-PD-1/PD-L1) immunotherapy. Gut 2021, 71, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Xu, S.; Liu, H.; Xu, T.; Hu, L.; Zhao, N.; Han, X.; Zhang, X. Yak rumen microbial diversity at different forage growth stages of an alpine meadow on the Qinghai-Tibet Plateau. Peerj 2019, 7, e7645. [Google Scholar] [CrossRef] [PubMed]
- Midkiff, V.C. The history of feed analysis, as chronicled in the development of AOAC official methods, 1884–1984. J. Assoc. Off. Anal. Chem. 1984, 67, 851–860. [Google Scholar] [CrossRef]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, H.F.; He, Y.; Wu, J.Y.; Jiang, Y.X.; Tam, N.F.; Zhou, H.W. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl. Environ. Microbiol. 2012, 78, 8264–8271. [Google Scholar] [CrossRef]
- Dubois, P.C.; Trynka, G.; Franke, L.; Hunt, K.A.; Romanos, J.; Curtotti, A.; Zhernakova, A.; Heap, G.A.; Adány, R.; Aromaa, A.; et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010, 42, 295–302. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, W.; Zeng, P.; Wang, J.; Geng, B.; Yang, J.; Cui, Q. LncTar: A tool for predicting the RNA targets of long noncoding RNAs. Brief. Bioinform. 2015, 16, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]





| LA | MA | HA | SEM | p-Value | |
|---|---|---|---|---|---|
| CP | 11.25 b | 10.06 c | 17.81 a | 0.91 | <0.01 |
| EE | 4.29 a | 3.77 b | 4.18 b | 0.09 | 0.03 |
| Ash | 7.34 b | 4.55 c | 9.18 a | 2.18 | <0.01 |
| NDF | 58.08 b | 70.11 a | 50.40 c | 0.91 | <0.01 |
| ADF | 33.98 b | 36.17 a | 28.14 c | 1.39 | <0.01 |
| HCEL | 24.10 b | 33.94 a | 22.26 c | 0.14 | <0.01 |
| NFC | 19.02 a | 11.50 b | 18.43 a | 0.95 | <0.00 |
| Ca | 0.89 b | 0.65 b | 1.80 a | 0.10 | <0.01 |
| P | 1.21 a | 0.70 b | 1.52 a | 0.91 | <0.01 |
| LA | MA | HA | SEM | p-Value | |
|---|---|---|---|---|---|
| Acetic Acid (mmol/L) | 53.59b b | 71.06 a | 36.21 c | 3.76 | <0.00 |
| Propanoic Acid (mmol/L) | 7.64 b | 11.75 a | 7.05 b | 0.68 | 0.00 |
| Isobutyric acid (mmol/L) | 1.47 a | 1.53 a | 0.73 b | 0.09 | <0.00 |
| Butyric acid (mmol/L) | 7.92 a | 5.89 b | 3.37 c | 0.53 | <0.00 |
| Isovaleric acid (mmol/L) | 1.94 a | 1.81 a | 0.84 b | 0.12 | <0.00 |
| Valeric Acid (mmol/L) | 0.25 | 0.28 | 0.52 | 0.08 | 0.35 |
| Total-VFAs (mmol/L) | 72.79 b | 92.32 a | 48.72 c | 4.79 | <0.00 |
| A/P | 7.03 a | 6.22 b c | 5.37 c | 0.28 | 0.04 |
| Index | LA | MA | HA | SEM | p-Value |
|---|---|---|---|---|---|
| Shannon | 8.33 a | 7.62 b | 7.62 b | 0.10 | 0.00 |
| Simpson | 0.99 | 0.99 | 0.98 | 0.00 | 0.25 |
| ACE | 1325.88 a | 1317.05 a | 1241.72 b | 15.58 | 0.04 |
| Chao1 | 1346.56 | 1355.22 | 1281.25 | 15.02 | 0.08 |
| PD_whole_tree | 69.27 | 67.39 | 67.04 | 0.63 | 0.31 |
| Coverage | 0.99695 | 0.995867 | 0.996917 | 0.00 | 0.10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sha, Y.; Ren, Y.; Zhao, S.; He, Y.; Guo, X.; Pu, X.; Li, W.; Liu, X.; Wang, J.; Li, S. Response of Ruminal Microbiota–Host Gene Interaction to High-Altitude Environments in Tibetan Sheep. Int. J. Mol. Sci. 2022, 23, 12430. https://doi.org/10.3390/ijms232012430
Sha Y, Ren Y, Zhao S, He Y, Guo X, Pu X, Li W, Liu X, Wang J, Li S. Response of Ruminal Microbiota–Host Gene Interaction to High-Altitude Environments in Tibetan Sheep. International Journal of Molecular Sciences. 2022; 23(20):12430. https://doi.org/10.3390/ijms232012430
Chicago/Turabian StyleSha, Yuzhu, Yue Ren, Shengguo Zhao, Yanyu He, Xinyu Guo, Xiaoning Pu, Wenhao Li, Xiu Liu, Jiqing Wang, and Shaobin Li. 2022. "Response of Ruminal Microbiota–Host Gene Interaction to High-Altitude Environments in Tibetan Sheep" International Journal of Molecular Sciences 23, no. 20: 12430. https://doi.org/10.3390/ijms232012430
APA StyleSha, Y., Ren, Y., Zhao, S., He, Y., Guo, X., Pu, X., Li, W., Liu, X., Wang, J., & Li, S. (2022). Response of Ruminal Microbiota–Host Gene Interaction to High-Altitude Environments in Tibetan Sheep. International Journal of Molecular Sciences, 23(20), 12430. https://doi.org/10.3390/ijms232012430






