Hop Extract Anti-Inflammatory Effect on Human Chondrocytes Is Potentiated When Encapsulated in Rapeseed Lecithin Nanoliposomes
Abstract
1. Introduction
2. Results
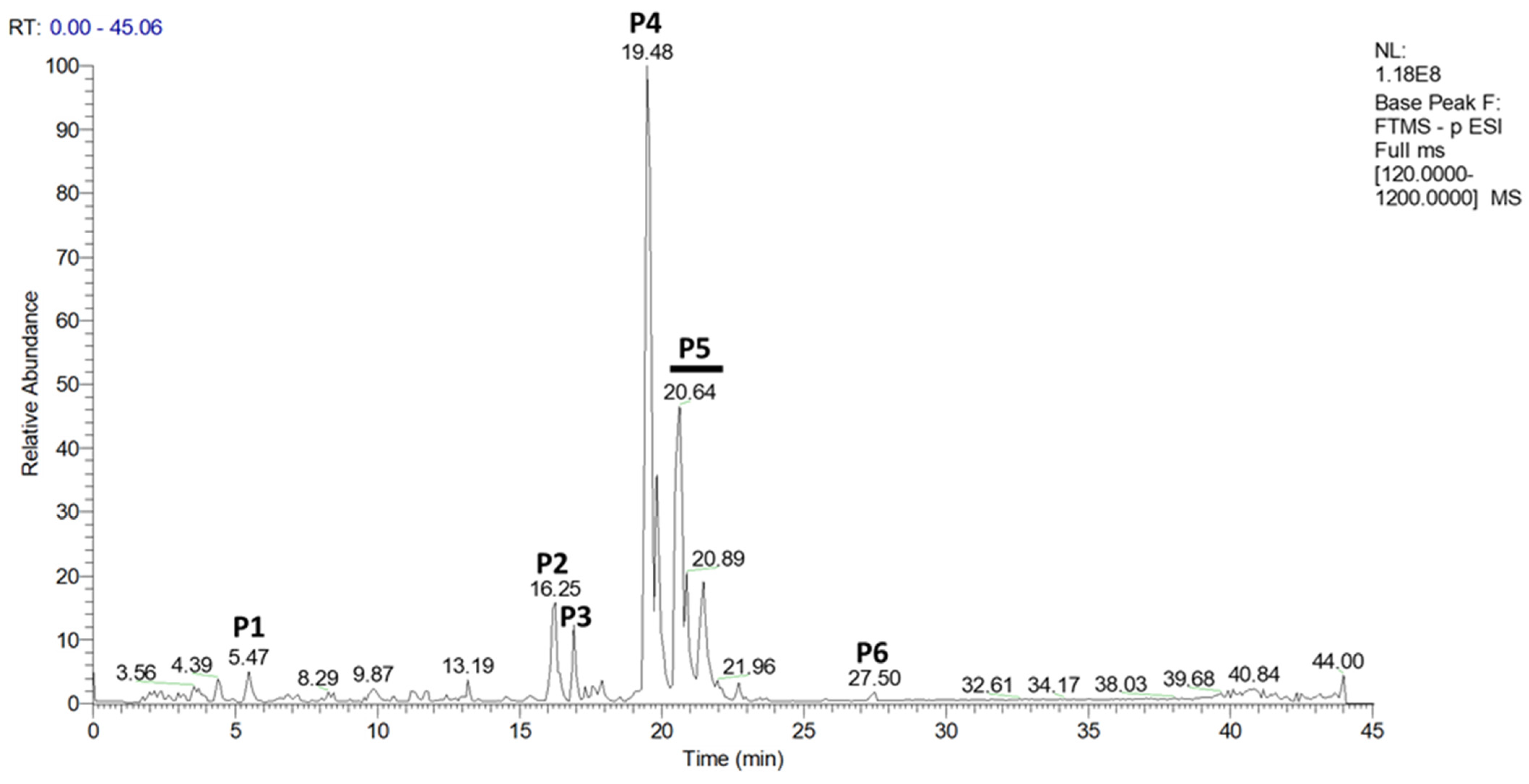
2.1. Characterization of Methanol-Soluble Molecules from Hop Extract
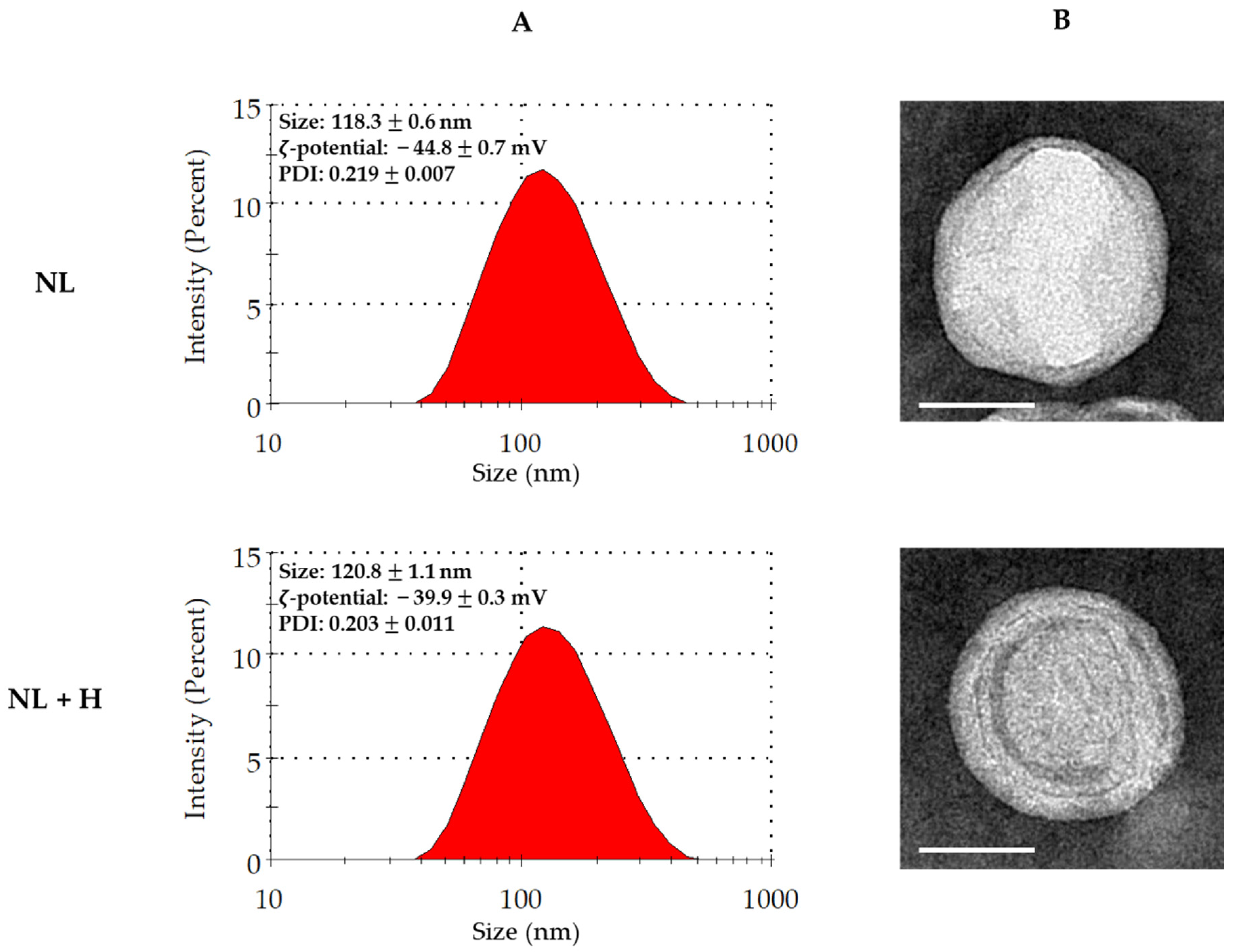
2.2. Physicochemical Characterization of Empty and Hop-Loaded Rapeseed lecithin Liposomes
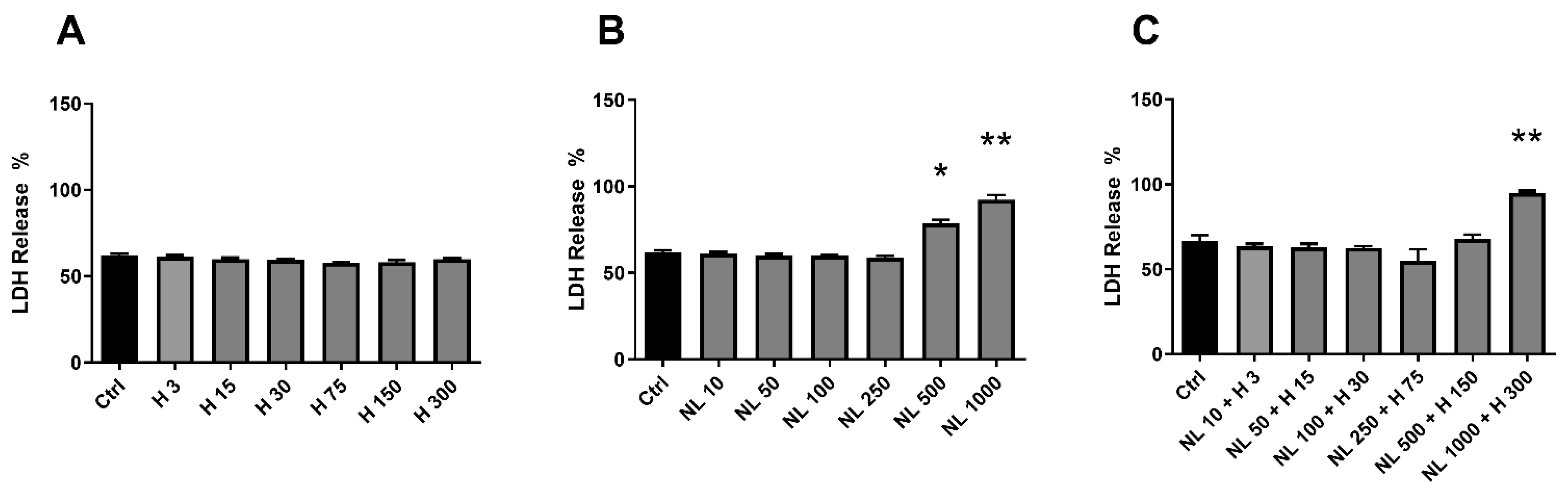
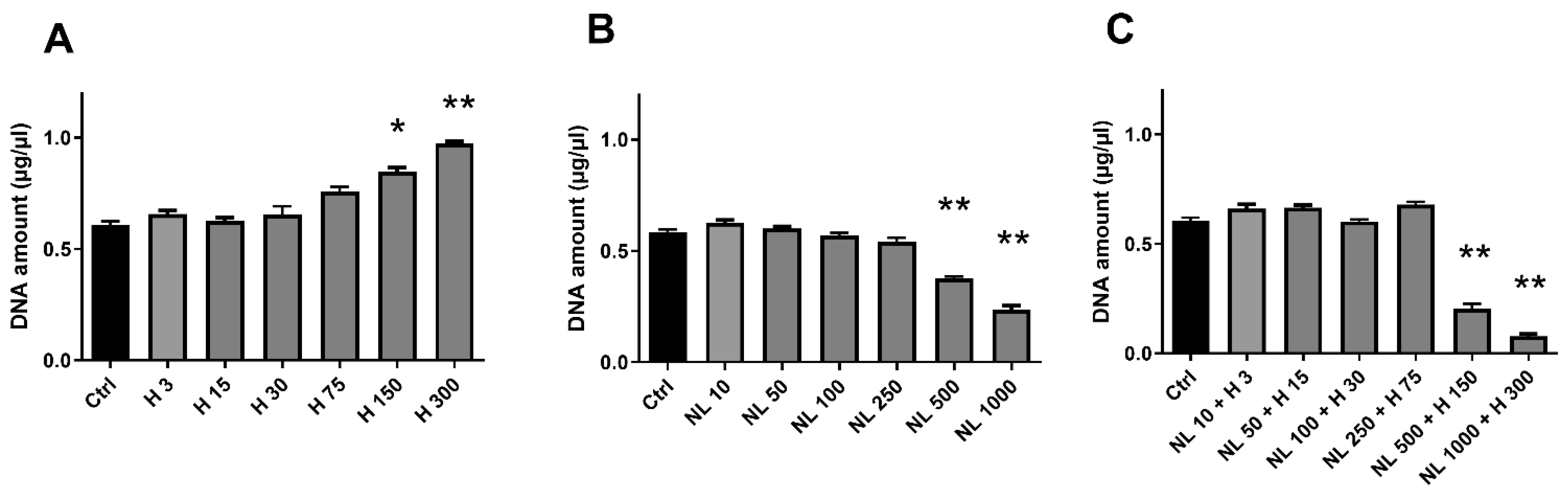
2.3. Biocompatibility of Hop Extract and Empty and Hop-Loaded Rapeseed Lecithin Nanoliposomes
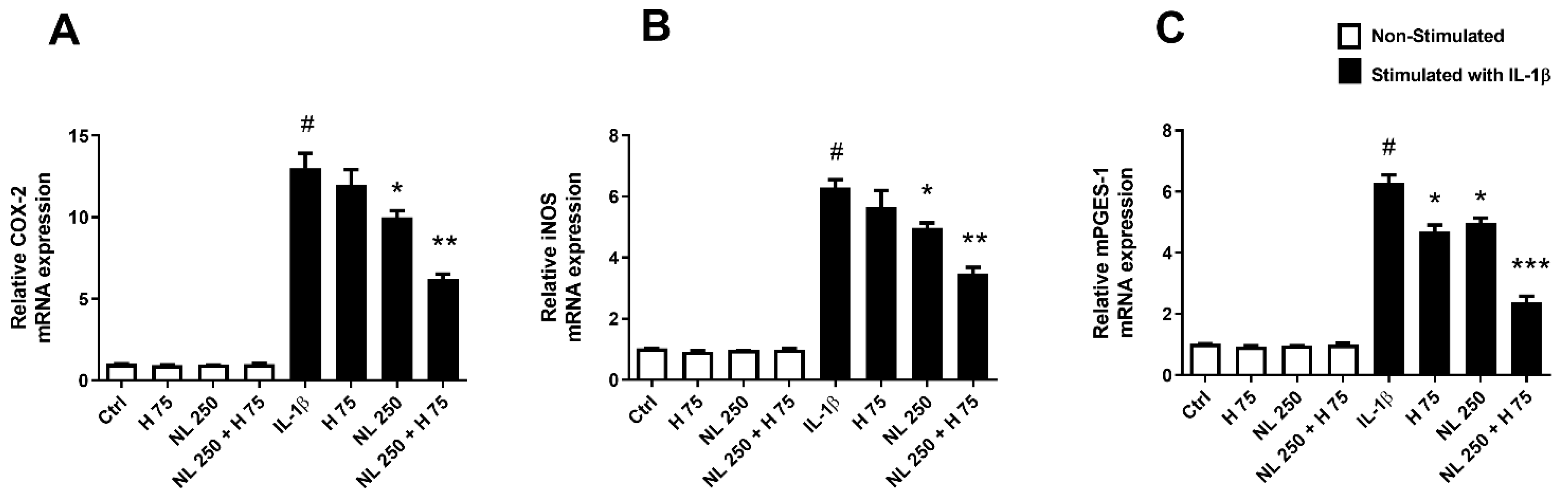
2.4. Effect of Hop Extract and Empty and Hop-Loaded Rapeseed Lecithin Nanoliposomes on Inflammation
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Extraction of Specialized Metabolites from Hop
4.3. Characterization of Specialized Metabolites from Dry Extract
4.4. UHPLC-ESI-MS Analysis
4.5. Molecular Identification
4.6. Nanoliposome Preparation
4.7. Physicochemical Characterization
4.7.1. Dynamic Light Scattering
4.7.2. Transmission Electron Microscopy
4.8. Human Cartilage Specimens and Chondrocyte Culture
4.9. Biocompatibility Assays
4.9.1. Cytotoxicity Assay
4.9.2. Cell Metabolic Activity
4.9.3. Cell Proliferation
4.10. RNA Isolation, Reverse Transcription, and Real-Time Polymerase Chain Reaction (RT-PCR)
4.11. PGE2 Assay
4.12. Nitrite Assay
4.13. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goldring, M.B. Osteoarthritis and Cartilage: The Role of Cytokines. Curr. Rheumatol. Rep. 2000, 2, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Oliveira Silva, M.; Gregory, J.L.; Ansari, N.; Stok, K.S. Molecular Signaling Interactions and Transport at the Osteochondral Interface: A Review. Front. Cell Dev. Biol. 2020, 8, 750. [Google Scholar] [CrossRef]
- Goldring, M.B. Update on the Biology of the Chondrocyte and New Approaches to Treating Cartilage Diseases. Best Pract. Res. Clin. Rheumatol. 2006, 20, 1003–1025. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Hussain, S.; Hu, Y.; Yan, J.; Min, Z.; Lan, X.; Guo, Y.; Zhao, Y.; Huang, H.; Feng, M.; et al. Maintenance of SOX9 Stability and ECM Homeostasis by Selenium-Sensitive PRMT5 in Cartilage. Osteoarthr. Cartil. 2019, 27, 932–944. [Google Scholar] [CrossRef]
- Hunter, D.J.; Bierma-Zeinstra, S. Osteoarthritis. Lancet 2019, 393, 1745–1759. [Google Scholar] [CrossRef]
- Cui, A.; Li, H.; Wang, D.; Zhong, J.; Chen, Y.; Lu, H. Global, Regional Prevalence, Incidence and Risk Factors of Knee Osteoarthritis in Population-Based Studies. EClinicalMedicine 2020, 29, 100587. [Google Scholar] [CrossRef] [PubMed]
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A Disease of the Joint as an Organ. Arthritis Rheum. 2012, 64, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-H.; Jain, V.; Gibson, J.; Attarian, D.E.; Haraden, C.A.; Yohn, C.B.; Laberge, R.-M.; Gregory, S.; Kraus, V.B. Synovial Cell Cross-Talk with Cartilage Plays a Major Role in the Pathogenesis of Osteoarthritis. Sci. Rep. 2020, 10, 10868. [Google Scholar] [CrossRef]
- Terkawi, M.A.; Ebata, T.; Yokota, S.; Takahashi, D.; Endo, T.; Matsumae, G.; Shimizu, T.; Kadoya, K.; Iwasaki, N. Low-Grade Inflammation in the Pathogenesis of Osteoarthritis: Cellular and Molecular Mechanisms and Strategies for Future Therapeutic Intervention. Biomedicines 2022, 10, 1109. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Lefebvre, V.; de Crombrugghe, B. Potent Inhibition of the Master Chondrogenic FactorSox9 Gene by Interleukin-1 and Tumor Necrosis Factor-α. J. Biol. Chem. 2000, 275, 3687–3692. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.P.; DiBattista, J.A.; Roughley, P.; McCollum, R.; Martel-Pelletier, J. Cytokines and Inflammation in Cartilage Degradation. Rheum. Dis. Clin. North Am. 1993, 19, 545–568. [Google Scholar] [CrossRef]
- Kim, H.-A.; Yeo, Y.; Jung, H.A.; Jung, Y.O.; Park, S.J.; Kim, S.J. Phase 2 Enzyme Inducer Sulphoraphane Blocks Prostaglandin and Nitric Oxide Synthesis in Human Articular Chondrocytes and Inhibits Cartilage Matrix Degradation. Rheumatology 2012, 51, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-Y.; Yao, Y.-D.; Luo, J.-F.; Liu, Z.-Q.; Huang, Y.-M.; Wu, F.-C.; Sun, Q.-H.; Liu, J.-X.; Zhou, H. Microsomal Prostaglandin E2 Synthase-1 and Its Inhibitors: Molecular Mechanisms and Therapeutic Significance. Pharmacol. Res. 2022, 175, 105977. [Google Scholar] [CrossRef]
- Xian Bo, S.; Yan Jie, W.; De Chao, C.; Sai, M.; Zhe, W.; Ya Kun, Z.; Hui Hui, G.; Chen, W.; Xiao, M.; Zhong Yao, H.; et al. An Inducible Nitric Oxide Synthase Dimerization Inhibitor Prevents the Progression of Osteoarthritis. Front. Pharmacol. 2022, 13, 861183. [Google Scholar] [CrossRef] [PubMed]
- Goldring, M.B.; Culley, K.L.; Otero, M. Pathogenesis of Osteoarthritis in General. In Cartilage; Grässel, S., Aszódi, A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–25. ISBN 978-3-319-45801-4. [Google Scholar]
- Masson, A.O.; Krawetz, R.J. Understanding Cartilage Protection in OA and Injury: A Spectrum of Possibilities. BMC Musculoskelet Disord 2020, 21, 432. [Google Scholar] [CrossRef] [PubMed]
- Velot, É.; Madry, H.; Venkatesan, J.K.; Bianchi, A.; Cucchiarini, M. Is Extracellular Vesicle-Based Therapy the next Answer for Cartilage Regeneration? Front. Bioeng. Biotechnol. 2021, 9, 259. [Google Scholar] [CrossRef] [PubMed]
- Asen, A.; Goebel, L.; Rey-Rico, A.; Sohier, J.; Zurakowski, D.; Cucchiarini, M.; Madry, H. Sustained Spatiotemporal Release of TGF-β1 Confers Enhanced Very Early Chondrogenic Differentiation during Osteochondral Repair in Specific Topographic Patterns. FASEB J. 2018, 32, 5298–5311. [Google Scholar] [CrossRef]
- Bianchi, A.; Velot, É.; Kempf, H.; Elkhoury, K.; Sanchez-Gonzalez, L.; Linder, M.; Kahn, C.; Arab-Tehrany, E. Nanoliposomes from Agro-Resources as Promising Delivery Systems for Chondrocytes. Int. J. Mol. Sci. 2020, 21, 3436. [Google Scholar] [CrossRef] [PubMed]
- Velot, É.; Elkhoury, K.; Kahn, C.; Kempf, H.; Linder, M.; Arab-Tehrany, E.; Bianchi, A. Efficient TGF-Β1 Delivery to Articular Chondrocytes In Vitro Using Agro-Based Liposomes. Int. J. Mol. Sci. 2022, 23, 2864. [Google Scholar] [CrossRef]
- He, K.; Huang, X.; Shan, R.; Yang, X.; Song, R.; Xie, F.; Huang, G. Intra-Articular Injection of Lornoxicam and MicroRNA-140 Co-Loaded Cationic Liposomes Enhanced the Therapeutic Treatment of Experimental Osteoarthritis. AAPS PharmSciTech 2022, 23, 9. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; He, T.; Bajpayee, A.G. Recent Advances in Targeted Drug Delivery for Treatment of Osteoarthritis. Curr. Opin. Rheumatol. 2021, 33, 94–109. [Google Scholar] [CrossRef]
- Younas, A.; Gu, H.; Zhao, Y.; Zhang, N. Novel Approaches of the Nanotechnology-Based Drug Delivery Systems for Knee Joint Injuries: A Review. Int. J. Pharm. 2021, 608, 121051. [Google Scholar] [CrossRef]
- Arab Tehrany, E.; Kahn, C.J.F.; Baravian, C.; Maherani, B.; Belhaj, N.; Wang, X.; Linder, M. Elaboration and Characterization of Nanoliposome Made of Soya; Rapeseed and Salmon Lecithins: Application to Cell Culture. Colloids Surf. B Biointerfaces 2012, 95, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Penhoat, A.; Guillot, N.; Meugnier, E.; Chanon, S.; Loizon, E.; Caillet, F.; Monnoye, M.; Vidal, H.; Gérard, P.; et al. Rapeseed and Soy Lecithin As Food Additives Vectors of α-Linolenic Acid: Impacts on High-Fat Induced Adiposity, Inflammation and Gut Microbiota in Mice. Curr. Dev. Nutr. 2021, 5, 364. [Google Scholar] [CrossRef]
- Calder, P.C. Omega-3 Polyunsaturated Fatty Acids and Inflammatory Processes: Nutrition or Pharmacology?: Omega-3 Fatty Acids and Inflammation. Br. J. Clin. Pharmacol. 2013, 75, 645–662. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, N.; Yang, A.; Huang, J.; Ren, X.; Xian, M.; Zou, H. Hop Bitter Acids: Resources, Biosynthesis, and Applications. Appl. Microbiol. Biotechnol. 2021, 105, 4343–4356. [Google Scholar] [CrossRef]
- Macchioni, V.; Picchi, V.; Carbone, K. Hop Leaves as an Alternative Source of Health-Active Compounds: Effect of Genotype and Drying Conditions. Plants 2021, 11, 99. [Google Scholar] [CrossRef]
- Brown, K.S.; Jamieson, P.; Wu, W.; Vaswani, A.; Alcazar Magana, A.; Choi, J.; Mattio, L.M.; Cheong, P.H.-Y.; Nelson, D.; Reardon, P.N.; et al. Computation-Assisted Identification of Bioactive Compounds in Botanical Extracts: A Case Study of Anti-Inflammatory Natural Products from Hops. Antioxidants 2022, 11, 1400. [Google Scholar] [CrossRef] [PubMed]
- Stracke, D.; Schulz, T.; Prehm, P. Inhibitors of Hyaluronan Export from Hops Prevent Osteoarthritic Reactions. Mol. Nutr. Food Res. 2011, 55, 485–494. [Google Scholar] [CrossRef]
- Ponticelli, M.; Russo, D.; Faraone, I.; Sinisgalli, C.; Labanca, F.; Lela, L.; Milella, L. The Promising Ability of Humulus lupulus L. Iso-α-Acids vs. Diabetes, Inflammation, and Metabolic Syndrome: A Systematic Review. Molecules 2021, 26, 954. [Google Scholar] [CrossRef]
- Jung, F.; Staltner, R.; Tahir, A.; Baumann, A.; Burger, K.; Halilbasic, E.; Hellerbrand, C.; Bergheim, I. Oral Intake of Xanthohumol Attenuates Lipoteichoic Acid-Induced Inflammatory Response in Human PBMCs. Eur. J. Nutr. 2022, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-N.; Sun, L.-C.; Chu, Y.-L.; Yu, R.-C.; Hsieh, C.-W.; Hsu, H.-Y.; Hsu, F.-C.; Cheng, K.-C. Bioactive Compounds with Anti-Oxidative and Anti-Inflammatory Activities of Hop Extracts. Food Chem. 2020, 330, 127244. [Google Scholar] [CrossRef] [PubMed]
- Hougee, S.; Faber, J.; Sanders, A.; Berg, W.B.; Garssen, J.; Smit, H.F.; Hoijer, M.A. Selective Inhibition of COX-2 by a Standardized CO2 Extract of Humulus lupulus in vitro and its Activity in a Mouse Model of Zymosan-Induced Arthritis. Planta Med. 2006, 72, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Knez Hrnčič, M.; Španinger, E.; Košir, I.; Knez, Ž.; Bren, U. Hop Compounds: Extraction Techniques, Chemical Analyses, Antioxidative, Antimicrobial, and Anticarcinogenic Effects. Nutrients 2019, 11, 257. [Google Scholar] [CrossRef]
- Dwivedi, G.; Flaman, L.; Alaybeyoglu, B.; Struglics, A.; Frank, E.H.; Chubinskya, S.; Trippel, S.B.; Rosen, V.; Cirit, M.; Grodzinsky, A.J. Inflammatory Cytokines and Mechanical Injury Induce Post-Traumatic Osteoarthritis-like Changes in a Human Cartilage-Bone-Synovium Microphysiological System. Arthritis Res. Ther. 2022, 24, 198. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, A.; Moulin, D.; Sebillaud, S.; Koufany, M.; Galteau, M.-M.; Netter, P.; Terlain, B.; Jouzeau, J.-Y. Contrasting Effects of Peroxisome-Proliferator-Activated Receptor (PPAR)Gamma Agonists on Membrane-Associated Prostaglandin E2 Synthase-1 in IL-1beta-Stimulated Rat Chondrocytes: Evidence for PPARgamma-Independent Inhibition by 15-Deoxy-Delta12,14prostaglandin J2. Arthritis Res. Ther. 2005, 7, R1325–R1337. [Google Scholar] [CrossRef]
- Kapoor, M.; Martel-Pelletier, J.; Lajeunesse, D.; Pelletier, J.-P.; Fahmi, H. Role of Proinflammatory Cytokines in the Pathophysiology of Osteoarthritis. Nat. Rev. Rheumatol. 2011, 7, 33–42. [Google Scholar] [CrossRef]
- Amin, A.R.; Dave, M.; Attur, M.; Abramson, S.B. COX-2, NO, and Cartilage Damage and Repair. Curr. Rheumatol. Rep. 2000, 2, 447–453. [Google Scholar] [CrossRef]
- Abramson, S.B.; Attur, M.; Amin, A.R.; Clancy, R. Nitric Oxide and Inflammatory Mediators in the Perpetuation of Osteoarthritis. Curr. Rheumatol. Rep. 2001, 3, 535–541. [Google Scholar] [CrossRef]
- Zanoli, P.; Zavatti, M. Pharmacognostic and Pharmacological Profile of Humulus lupulus L. J. Ethnopharmacol. 2008, 116, 383–396. [Google Scholar] [CrossRef]
- De Keukeleire, J.; Ooms, G.; Heyerick, A.; Roldan-Ruiz, I.; Van Bockstaele, E.; De Keukeleire, D. Formation and Accumulation of Alpha-Acids, Beta-Acids, Desmethylxanthohumol, and Xanthohumol during Flowering of Hops (Humulus lupulus L.). J. Agric. Food Chem. 2003, 51, 4436–4441. [Google Scholar] [CrossRef] [PubMed]
- Sarraf, C.; Desjardins, Y.; Leonhart, S.; Gosselin, A.; Gosselin, G. Agronomic and nutraceutical potential of hops (Humulus lupulus L.) grown in québec, Canada. Acta Hortic. 2013, 1010, 155–161. [Google Scholar] [CrossRef]
- Mongelli, A.; Rodolfi, M.; Ganino, T.; Marieschi, M.; Caligiani, A.; Dall’Asta, C.; Bruni, R. Are Humulus lupulus L. Ecotypes and Cultivars Suitable for the Cultivation of Aromatic Hop in Italy? A Phytochemical Approach. Ind. Crops Prod. 2016, 83, 693–700. [Google Scholar] [CrossRef]
- Patzak, J.; Krofta, K.; Henychová, A.; Nesvadba, V. Number and Size of Lupulin Glands, Glandular Trichomes of Hop (Humulus lupulus L.), Play a Key Role in Contents of Bitter Acids and Polyphenols in Hop Cone. Int. J. Food Sci. Technol. 2015, 50, 1864–1872. [Google Scholar] [CrossRef]
- Taniguchi, Y.; Matsukura, Y.; Ozaki, H.; Nishimura, K.; Shindo, K. Identification and Quantification of the Oxidation Products Derived from α-Acids and β-Acids during Storage of Hops (Humulus lupulus L.). J. Agric. Food Chem. 2013, 61, 3121–3130. [Google Scholar] [CrossRef]
- Gülz, P.-G.; Müller, E.; Herrmann, T.; Lösel, P. Epicuticular Leaf Waxes of the Hop (Humulus lupulus). Chemical Composition and Surface Structures. Z. Für Nat. C 1993, 48, 689–696. [Google Scholar] [CrossRef]
- Ceh, B.; Kac, M.; Košir, I.; Abram, V. Relationships between Xanthohumol and Polyphenol Content in Hop Leaves and Hop Cones with Regard to Water Supply and Cultivar. Int. J. Mol. Sci. 2007, 8, 989–1000. [Google Scholar] [CrossRef]
- Kang, E.-J.; Kim, H.J.; Choi, J.H.; Noh, J.-R.; Kim, J.-H.; Lee, I.B.; Choi, Y.-K.; Choi, D.-H.; An, J.; Oh, W.K.; et al. Humulus Japonicus Extract Ameliorates Collagen-induced Arthritis in Mice through Regulation of Overall Articular Inflammation. Int. J. Mol. Med. 2020, 45, 417–428. [Google Scholar] [CrossRef]
- Caban, M.; Chojnacka, K.; Owczarek, K.; Laskowska, J.; Fichna, J.; Podsedek, A.; Sosnowska, D.; Lewandowska, U. Spent Hops (Humulus lupulus L.) Extract as Modulator of the Inflammatory Response in Lipopolysaccharide Stimulated RAW 264.7 Macrophages. J. Physiol. Pharmacol. 2020, 71, 67–78. [Google Scholar] [CrossRef]
- Sommella, E.; Verna, G.; Liso, M.; Salviati, E.; Esposito, T.; Carbone, D.; Pecoraro, C.; Chieppa, M.; Campiglia, P. Hop-Derived Fraction Rich in Beta Acids and Prenylflavonoids Regulates the Inflammatory Response in Dendritic Cells Differently from Quercetin: Unveiling Metabolic Changes by Mass Spectrometry-Based Metabolomics. Food Funct. 2021, 12, 12800–12811. [Google Scholar] [CrossRef]
- Park, T.-S.; Ryu, Y.-K.; Park, H.-Y.; Kim, J.Y.; Go, J.; Noh, J.-R.; Kim, Y.-H.; Hwang, J.H.; Choi, D.-H.; Oh, W.-K.; et al. Humulus Japonicus Inhibits the Progression of Alzheimer’s Disease in a APP/PS1 Transgenic Mouse Model. Int. J. Mol. Med. 2017, 39, 21–30. [Google Scholar] [CrossRef]
- Lee, I.-S.; Lim, J.; Gal, J.; Kang, J.C.; Kim, H.J.; Kang, B.Y.; Choi, H.J. Anti-Inflammatory Activity of Xanthohumol Involves Heme Oxygenase-1 Induction via NRF2-ARE Signaling in Microglial BV2 Cells. Neurochem. Int. 2011, 58, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Van Cleemput, M.; Cattoor, K.; De Bosscher, K.; Haegeman, G.; De Keukeleire, D.; Heyerick, A. Hop (Humulus lupulus )-Derived Bitter Acids as Multipotent Bioactive Compounds. J. Nat. Prod. 2009, 72, 1220–1230. [Google Scholar] [CrossRef]
- Aigner, T.; McKenna, L. Molecular Pathology and Pathobiology of Osteoarthritic Cartilage. Cell. Mol. Life Sci. 2002, 59, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, K.; Yamaguchi, A.; Arita, J.; Sakurai, S.; Ikeda, A.; Takido, M. Inhibitory Effect of Edible Plant Extracts on 12-O-Tetradecanoylphorbol–13-Acetate-Induced Ear Oedema in Mice. Phytother. Res. 1993, 7, 185–189. [Google Scholar] [CrossRef]
- Salviati, E.; Ciaglia, E.; Sommella, E.; Montella, F.; Bertamino, A.; Ostacolo, C.; Parrino, B.; Rubino, R.; Vecchione, C.; Puca, A.; et al. Immunomodulatory Activity of Humulus lupulus Bitter Acids Fraction: Enhancement of Natural Killer Cells Function by NKp44 Activating Receptor Stimulation. J. Funct. Foods 2019, 61, 103469. [Google Scholar] [CrossRef]
- Lupinacci, E.; Meijerink, J.; Vincken, J.-P.; Gabriele, B.; Gruppen, H.; Witkamp, R.F. Xanthohumol from Hop (Humulus lupulus L.) Is an Efficient Inhibitor of Monocyte Chemoattractant Protein-1 and Tumor Necrosis Factor-α Release in LPS-Stimulated RAW 264.7 Mouse Macrophages and U937 Human Monocytes. J. Agric. Food Chem. 2009, 57, 7274–7281. [Google Scholar] [CrossRef]
- Fukuda, T.; Ohya, R.; Kobayashi, K.; Ano, Y. Matured Hop Bitter Acids in Beer Improve Lipopolysaccharide-Induced Depression-Like Behavior. Front. Neurosci. 2019, 13, 41. [Google Scholar] [CrossRef]
- Ao, X.; Yan, J.; Liu, S.; Chen, S.; Zou, L.; Yang, Y.; He, L.; Li, S.; Liu, A.; Zhao, K. Extraction, Isolation and Identification of Four Phenolic Compounds from Pleioblastus Amarus Shoots and Their Antioxidant and Anti-Inflammatory Properties in Vitro. Food Chem. 2022, 374, 131743. [Google Scholar] [CrossRef]
- Rahmati, M.; Mobasheri, A.; Mozafari, M. Inflammatory Mediators in Osteoarthritis: A Critical Review of the State-of-the-Art, Current Prospects, and Future Challenges. Bone 2016, 85, 81–90. [Google Scholar] [CrossRef]
- Nakata, K.; Hanai, T.; Take, Y.; Osada, T.; Tsuchiya, T.; Shima, D.; Fujimoto, Y. Disease-Modifying Effects of COX-2 Selective Inhibitors and Non-Selective NSAIDs in Osteoarthritis: A Systematic Review. Osteoarthr. Cartil. 2018, 26, 1263–1273. [Google Scholar] [CrossRef]
- Timur, U.T.; Caron, M.M.J.; Jeuken, R.M.; Bastiaansen-Jenniskens, Y.M.; Welting, T.J.M.; van Rhijn, L.W.; van Osch, G.J.V.M.; Emans, P.J. Chondroprotective Actions of Selective COX-2 Inhibitors In Vivo: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 6962. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, J.A.; Collins, J.A.; Loeser, R.F. Reactive Oxygen Species, Aging and Articular Cartilage Homeostasis. Free Radic. Biol. Med. 2019, 132, 73–82. [Google Scholar] [CrossRef]
- Ahmad, N.; Ansari, M.Y.; Haqqi, T.M. Role of INOS in Osteoarthritis: Pathological and Therapeutic Aspects. J. Cell. Physiol. 2020, 235, 6366–6376. [Google Scholar] [CrossRef] [PubMed]
- Roy, H.S.; Singh, R.; Ghosh, D. Recent Advances in Nanotherapeutic Strategies That Target Nitric Oxide Pathway for Preventing Cartilage Degeneration. Nitric Oxide 2021, 109–110, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Castro, C.M.; Corciulo, C.; Solesio, M.E.; Liang, F.; Pavlov, E.V.; Cronstein, B.N. Adenosine A2A Receptor (A2AR) Stimulation Enhances Mitochondrial Metabolism and Mitigates Reactive Oxygen Species-Mediated Mitochondrial Injury. FASEB J. 2020, 34, 5027–5045. [Google Scholar] [CrossRef]
- Jiang, C.; Luo, P.; Li, X.; Liu, P.; Li, Y.; Xu, J. Nrf2/ARE Is a Key Pathway for Curcumin-Mediated Protection of TMJ Chondrocytes from Oxidative Stress and Inflammation. Cell Stress Chaperones 2020, 25, 395–406. [Google Scholar] [CrossRef]
- van Beuningen, H.M.; Arntz, O.J.; van den Berg, W.B. In Vivo Effects of Interleukin-1 on Articular Cartilage. Prolongation of Proteoglycan Metabolic Disturbances in Old Mice. Arthritis Rheumatol. 1991, 34, 606–615. [Google Scholar] [CrossRef]
- van der Kraan, P.M.; Blaney Davidson, E.N.; Blom, A.; van den Berg, W.B. TGF-Beta Signaling in Chondrocyte Terminal Differentiation and Osteoarthritis. Osteoarthr. Cartil. 2009, 17, 1539–1545. [Google Scholar] [CrossRef]
- Guingamp, C.; Gegout-Pottie, P.; Philippe, L.; Terlain, B.; Netter, P.; Gillet, P. Mono-Iodoacetate-Induced Experimental Osteoarthritis: A Dose-Response Study of Loss of Mobility, Morphology, and Biochemistry. Arthritis Care Res. 1997, 40, 1670–1679. [Google Scholar] [CrossRef]
- Ma, H.-L.; Blanchet, T.J.; Peluso, D.; Hopkins, B.; Morris, E.A.; Glasson, S.S. Osteoarthritis Severity Is Sex Dependent in a Surgical Mouse Model. Osteoarthr. Cartil. 2007, 15, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Bianchi, A.; Presle, N.; Moulin, D.; Koufany, M.; Guillaume, C.; Kempf, H.; Pizard, A. Eplerenone Treatment Alleviates the Development of Joint Lesions in a New Rat Model of Spontaneous Metabolic-Associated Osteoarthritis. Ann. Rheum. Dis. 2018, 77, 315–316. [Google Scholar] [CrossRef]
- Goodman, S.M.; Springer, B.; Guyatt, G.; Abdel, M.P.; Dasa, V.; George, M.; Gewurz-Singer, O.; Giles, J.T.; Johnson, B.; Lee, S.; et al. 2017 American College of Rheumatology/American Association of Hip and Knee Surgeons Guideline for the Perioperative Management of Antirheumatic Medication in Patients With Rheumatic Diseases Undergoing Elective Total Hip or Total Knee Arthroplasty. Arthritis Rheumatol. 2017, 69, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Mankin, H.J.; Dorfman, H.; Lippiello, L.; Zarins, A. Biochemical and Metabolic Abnormalities in Articular Cartilage from Osteo-Arthritic Human Hips. II. Correlation of Morphology with Biochemical and Metabolic Data. J. Bone Jt. Surg. 1971, 53, 523–537. [Google Scholar] [CrossRef]
- Bianchi, A.; Guibert, M.; Cailotto, F.; Gasser, A.; Presle, N.; Mainard, D.; Netter, P.; Kempf, H.; Jouzeau, J.-Y.; Reboul, P. Fibroblast Growth Factor 23 Drives MMP13 Expression in Human Osteoarthritic Chondrocytes in a Klotho-Independent Manner. Osteoarthr. Cartil. 2016, 24, 1961–1969. [Google Scholar] [CrossRef]


| Time (min) | A (%) | B (%) |
|---|---|---|
| 0 | 90 | 10 |
| 2 | 90 | 10 |
| 4 | 80 | 20 |
| 5 | 75 | 25 |
| 6.9 | 69 | 31 |
| 9 | 69 | 31 |
| 15 | 50 | 50 |
| 17 | 38 | 62 |
| 25 | 22.5 | 77.5 |
| 32 | 20 | 80 |
| 35 | 17 | 83 |
| 35.5 | 0 | 100 |
| 39 | 0 | 100 |
| 40 | 90 | 10 |
| 45 | 90 | 10 |
| Genes | Sequences 5′-3′ |
|---|---|
| COX-2 | Fwd: GCT-GGA-ACA-TGG-AAT-TAC-CCA |
| Rev: CTT-TCT-GTA-CTG-CGG-GTG-GAA | |
| mPGES-1 | Fwd: TGG-TCA-TCA-AGA-TGT-ACG-TGG-T |
| Rev: GGG-TCG-CTC-CTG-CAA-TAC-T | |
| iNOS | Fwd: TGC-AAT-GAA-TGG-GGA-AAA-AG |
| Rev: ATT-CTG-CTG-CTT-GCT-GAG-GT | |
| RP29 | Fwd: CTC-TAA-CCG-CCA-CGG-TCT-GA |
| Rev: ACT-AGC-ATG-ATT-GGT-ATC-AC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velot, É.; Ducrocq, F.; Girardeau, L.; Hehn, A.; Piutti, S.; Kahn, C.; Linder, M.; Bianchi, A.; Arab-Tehrany, E. Hop Extract Anti-Inflammatory Effect on Human Chondrocytes Is Potentiated When Encapsulated in Rapeseed Lecithin Nanoliposomes. Int. J. Mol. Sci. 2022, 23, 12423. https://doi.org/10.3390/ijms232012423
Velot É, Ducrocq F, Girardeau L, Hehn A, Piutti S, Kahn C, Linder M, Bianchi A, Arab-Tehrany E. Hop Extract Anti-Inflammatory Effect on Human Chondrocytes Is Potentiated When Encapsulated in Rapeseed Lecithin Nanoliposomes. International Journal of Molecular Sciences. 2022; 23(20):12423. https://doi.org/10.3390/ijms232012423
Chicago/Turabian StyleVelot, Émilie, Florent Ducrocq, Loïc Girardeau, Alain Hehn, Séverine Piutti, Cyril Kahn, Michel Linder, Arnaud Bianchi, and Elmira Arab-Tehrany. 2022. "Hop Extract Anti-Inflammatory Effect on Human Chondrocytes Is Potentiated When Encapsulated in Rapeseed Lecithin Nanoliposomes" International Journal of Molecular Sciences 23, no. 20: 12423. https://doi.org/10.3390/ijms232012423
APA StyleVelot, É., Ducrocq, F., Girardeau, L., Hehn, A., Piutti, S., Kahn, C., Linder, M., Bianchi, A., & Arab-Tehrany, E. (2022). Hop Extract Anti-Inflammatory Effect on Human Chondrocytes Is Potentiated When Encapsulated in Rapeseed Lecithin Nanoliposomes. International Journal of Molecular Sciences, 23(20), 12423. https://doi.org/10.3390/ijms232012423








