Emerging Roles of RNA-Binding Proteins in Inner Ear Hair Cell Development and Regeneration
Abstract
1. Introduction
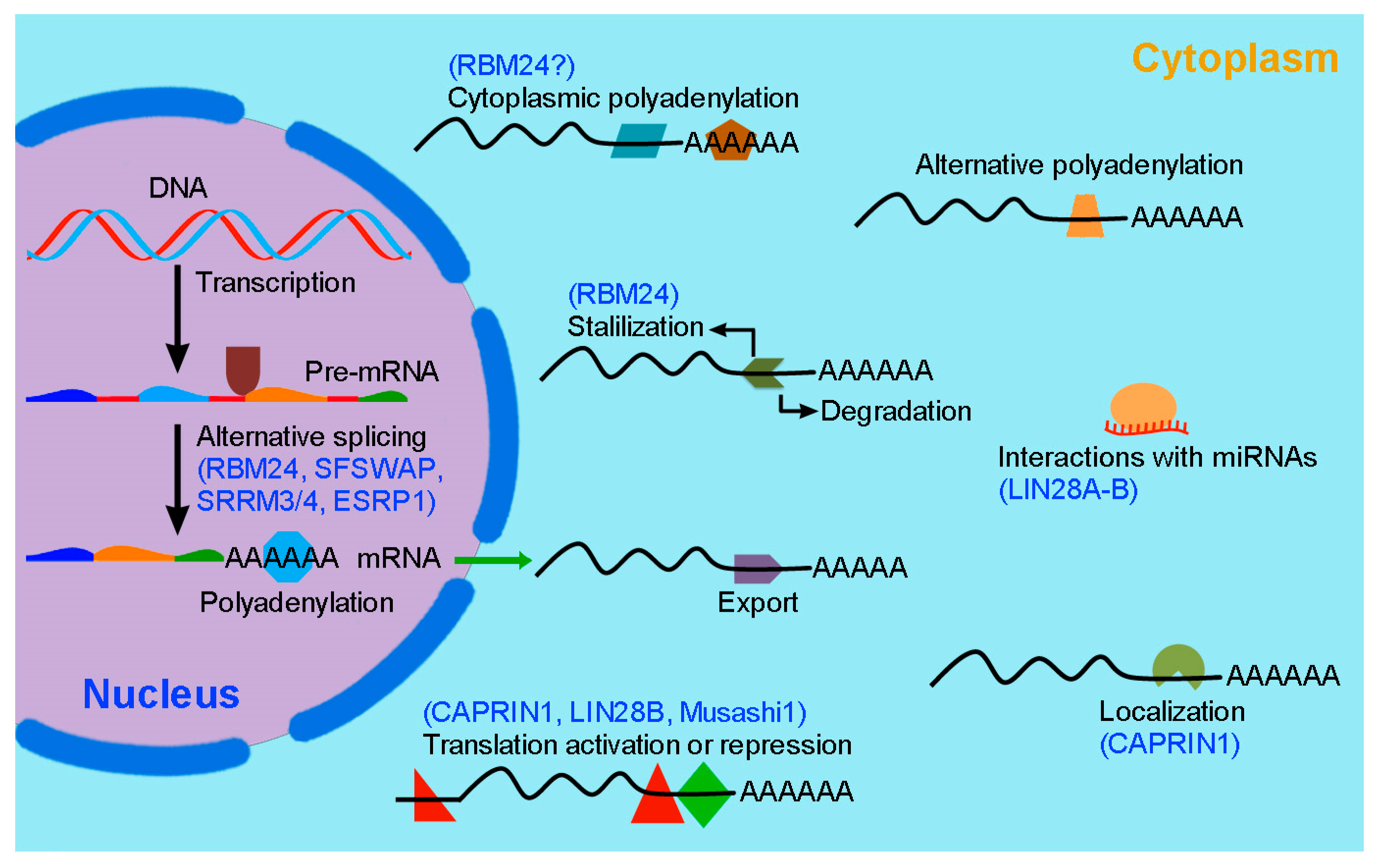
2. RBP-Mediated Post-Transcriptional Regulation in Inner Ear Hair Cell Development and Regeneration
2.1. LIN28 in Cochlear Hair Cell Development and Regeneration
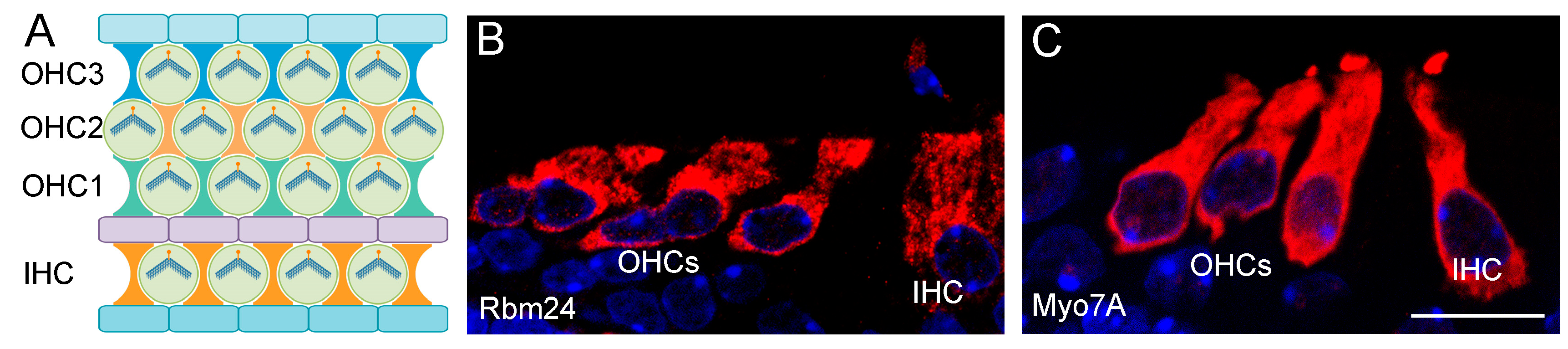
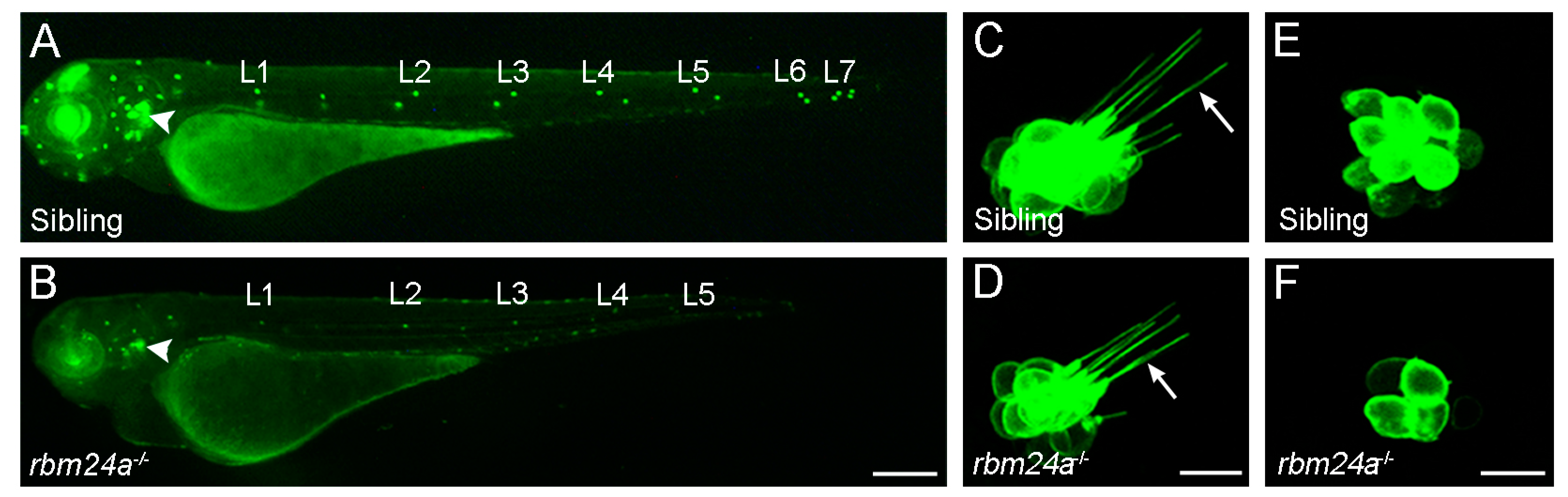
2.2. RBM24 Regulates mRNA Stability and Pre-mRNA Splicing in Hair Cells
2.3. SFSWAP Functions in Growth and Patterning of Inner Ear Sensory Organs
2.4. “Noise/Damage-Related” RBPs in Hearing Loss
2.5. Post-Transcriptional Inactivation of REST Transcriptional Repressor by SRRM4-Regulated Exon Inclusion
2.6. Mutations of the ESRP1 Gene in Humans Cause Alternative Splicing Defects and Hearing Loss
2.7. Possible Role of Musashi1 in the Maintenance of Stem Cell Fate for Hair Cell Regeneration
2.8. Other RBPs Implicated in Inner Ear Development and Hair Cell Regeneration
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Groves, A.K.; Fekete, D.M. Shaping sound in space: The regulation of inner ear patterning. Development 2012, 139, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Basch, M.L.; Brown, R.M.; Jen, H.I.; Groves, A.K. Where hearing starts: The development of the mammalian cochlea. J. Anat. 2016, 228, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Driver, E.C.; Kelley, M.W. Development of the cochlea. Development 2020, 147, dev162263. [Google Scholar] [CrossRef] [PubMed]
- Deans, M.R. Conserved and divergent principles of planar polarity revealed by hair cell development and function. Front. Neurosci. 2021, 15, 742391. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.L.; Shin, J.B. Mechanisms of hair cell damage and repair. Trends Neurosci. 2019, 42, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.S.; Tucci, D.L.; Merson, M.H.; O’Donoghue, G.M. Global hearing health care: New findings and perspectives. Lancet 2017, 390, 2503–2515. [Google Scholar] [CrossRef]
- Elliott, K.L.; Pavlínková, G.; Chizhikov, V.V.; Yamoah, E.N.; Fritzsch, B. Development in the mammalian auditory system depends on transcription factors. Int. J. Mol. Sci. 2021, 22, 4189. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.K.; Kelley, M.W. Molecular mechanisms of inner ear development. Cold Spring Harb. Perspect. Biol. 2012, 4, a008409. [Google Scholar] [CrossRef]
- Atkinson, P.J.; Huarcaya Najarro, E.; Sayyid, Z.N.; Cheng, A.G. Sensory hair cell development and regeneration: Similarities and differences. Development 2015, 142, 1561–1571. [Google Scholar] [CrossRef]
- Shi, D.L.; Grifone, R. RNA-binding proteins in the post-transcriptional control of skeletal muscle development, regeneration and disease. Front. Cell Dev. Biol. 2021, 9, 738978. [Google Scholar] [CrossRef] [PubMed]
- Brinegar, A.E.; Cooper, T.A. Roles for RNA-binding proteins in development and disease. Brain Res. 2016, 1647, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rohacek, A.M.; Bebee, T.W.; Tilton, R.K.; Radens, C.M.; McDermott-Roe, C.; Peart, N.; Kaur, M.; Zaykaner, M.; Cieply, B.; Musunuru, K.; et al. ESRP1 mutations cause hearing loss due to defects in alternative splicing that disrupt cochlear development. Dev. Cell 2017, 43, 318–331.e5. [Google Scholar] [CrossRef]
- Ye, Z.; Su, Z.; Xie, S.; Liu, Y.; Wang, Y.; Xu, X.; Zheng, Y.; Zhao, M.; Jiang, L. Yap-lin28a axis targets let7-Wnt pathway to restore progenitors for initiating regeneration. Elife 2020, 9, e55771. [Google Scholar] [CrossRef] [PubMed]
- Golden, E.J.; Benito-Gonzalez, A.; Doetzlhofer, A. The RNA-binding protein LIN28B regulates developmental timing in the mammalian cochlea. Proc. Natl. Acad. Sci. USA 2015, 112, E3864–E3873. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Doetzlhofer, A. LIN28B/let-7 control the ability of neonatal murine auditory supporting cells to generate hair cells through mTOR signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 22225–22236. [Google Scholar] [CrossRef]
- Li, X.J.; Morgan, C.; Goff, L.A.; Doetzlhofer, A. Follistatin promotes LIN28B-mediated supporting cell reprogramming and hair cell regeneration in the murine cochlea. Sci. Adv. 2022, 8, eabj7651. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, J.J.; Shi, D.L. Loss of Rbm24a causes defective hair cell development in the zebrafish inner ear and neuromasts. J. Genet. Genom. 2020, 47, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Yao, X.; Wang, C.; Chen, F.; Liu, D.; Shao, M.; Xu, Z. Rbm24a is necessary for hair cell development through regulating mRNA stability in zebrafish. Front. Cell Dev. Biol. 2020, 8, 604026. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Du, H.; Ren, R.; Wang, Y.; Xu, Z. Alternative splicing of Cdh23 exon 68 is regulated by RBM24, RBM38, and PTBP1. Neural Plast. 2020, 2020, 8898811. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Yuan, H.; Zhang, M.; Wang, C.; Cai, X.; Liu, J.; Xu, X.Q. Rbm24 regulates inner-ear-specific alternative splicing and is essential for maintaining auditory and motor coordination. RNA Biol. 2021, 18, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, C.; He, S.; Liu, Z. Mosaic CRISPR-stop enables rapid phenotyping of nonsense mutations in essential genes. Development 2021, 148, dev196899. [Google Scholar] [CrossRef] [PubMed]
- Moayedi, Y.; Basch, M.L.; Pacheco, N.L.; Gao, S.S.; Wang, R.; Harrison, W.; Xiao, N.; Oghalai, J.S.; Overbeek, P.A.; Mardon, G.; et al. The candidate splicing factor Sfswap regulates growth and patterning of inner ear sensory organs. PLoS Genet. 2014, 10, e1004055. [Google Scholar] [CrossRef][Green Version]
- Gao, S.S.; Wang, R.; Raphael, P.D.; Moayedi, Y.; Groves, A.K.; Zuo, J.; Applegate, B.E.; Oghalai, J.S. Vibration of the organ of Corti within the cochlear apex in mice. J. Neurophysiol. 2014, 112, 1192–1204. [Google Scholar] [CrossRef] [PubMed]
- Panganiban, C.H.; Barth, J.L.; Darbelli, L.; Xing, Y.; Zhang, J.; Li, H.; Noble, K.V.; Liu, T.; Brown, L.N.; Schulte, B.A.; et al. Noise-induced dysregulation of Quaking RNA binding proteins contributes to auditory nerve demyelination and hearing loss. J. Neurosci. 2018, 38, 2551–2568. [Google Scholar] [CrossRef]
- Towers, E.R.; Kelly, J.J.; Sud, R.; Gale, J.E.; Dawson, S.J. Caprin-1 is a target of the deafness gene Pou4f3 and is recruited to stress granules in cochlear hair cells in response to ototoxic damage. J. Cell Sci. 2011, 124, 1145–1155. [Google Scholar] [CrossRef]
- Gonçalves, A.C.; Towers, E.R.; Haq, N.; Porco, J.A., Jr.; Pelletier, J.; Dawson, S.J.; Gale, J.E. Drug-induced stress granule formation protects sensory hair cells in mouse cochlear explants during ototoxicity. Sci. Rep. 2019, 9, 12501. [Google Scholar] [CrossRef]
- Nolan, L.S.; Chen, J.; Gonçalves, A.C.; Bullen, A.; Towers, E.R.; Steel, K.P.; Dawson, S.J.; Gale, J.E. Targeted deletion of the RNA-binding protein Caprin1 leads to progressive hearing loss and impairs recovery from noise exposure in mice. Sci. Rep. 2022, 12, 2444. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Jahan, I.; Bonde, G.; Sun, X.; Hildebrand, M.S.; Engelhardt, J.F.; Smith, R.J.; Cornell, R.A.; Fritzsch, B.; Bánfi, B. A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx waltzer mouse. PLoS Genet. 2012, 8, e1002966. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Kelly, M.C.; Rehman, A.U.; Boger, E.T.; Morell, R.J.; Kelley, M.W.; Friedman, T.B.; Bánfi, B. Defects in the alternative splicing-dependent regulation of REST cause deafness. Cell 2018, 174, 536–548.e21. [Google Scholar] [CrossRef]
- Nakano, Y.; Wiechert, S.; Fritzsch, B.; Bánfi, B. Inhibition of a transcriptional repressor rescues hearing in a splicing factor-deficient mouse. Life Sci. Alliance 2020, 3, e202000841. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, H.; Yaoi, T.; Suzuki, T.; Okano, H.; Hisa, Y.; Fushiki, S. Spatiotemporal patterns of Musashi1 expression during inner ear development. Neuroreport 2004, 15, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Murata, J.; Murayama, A.; Horii, A.; Doi, K.; Harada, T.; Okano, H.; Kubo, T. Expression of Musashi1, a neural RNA-binding protein, in the cochlea of young adult mice. Neurosci. Lett. 2004, 354, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Savary, E.; Hugnot, J.P.; Chassigneux, Y.; Travo, C.; Duperray, C.; Van De Water, T.; Zine, A. Distinct population of hair cell progenitors can be isolated from the postnatal mouse cochlea using side population analysis. Stem Cells 2007, 25, 332–339. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wakasaki, T.; Niiro, H.; Jabbarzadeh-Tabrizi, S.; Ohashi, M.; Kimitsuki, T.; Nakagawa, T.; Komune, S.; Akashi, K. Musashi-1 is the candidate of the regulator of hair cell progenitors during inner ear regeneration. BMC Neurosci. 2017, 18, 64. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M.; Fujimoto, C.; Iwasaki, S.; Kashio, A.; Kikkawa, Y.S.; Kondo, K.; Okano, H.; Yamasoba, T. Alteration of Musashi1 intra-cellular distribution during regeneration following gentamicin-induced hair cell loss in the guinea pig crista ampullaris. Front. Cell. Neurosci. 2019, 13, 481. [Google Scholar] [CrossRef]
- Li, Y.; Xiang, J.; Duan, C. Insulin-like growth factor-binding protein-3 plays an important role in regulating pharyngeal skeleton and inner ear formation and differentiation. J. Biol. Chem. 2005, 280, 3613–3620. [Google Scholar] [CrossRef]
- Pei, W.; Xu, L.; Huang, S.C.; Pettie, K.; Idol, J.; Rissone, A.; Jimenez, E.; Sinclair, J.W.; Slevin, C.; Varshney, G.K.; et al. Guided genetic screen to identify genes essential in the regeneration of hair cells and other tissues. NPJ Regen. Med. 2018, 3, 11. [Google Scholar] [CrossRef]
- Pei, W.; Xu, L.; Chen, Z.; Slevin, C.C.; Pettie, K.P.; Wincovitch, S.; NISC Comparative Sequencing Program; Burgess, S.M. A subset of SMN complex members have a specific role in tissue regeneration via ERBB pathway-mediated proliferation. NPJ Regen. Med. 2020, 5, 6. [Google Scholar] [CrossRef]
- Maharana, S.K.; Saint-Jeannet, J.P. Molecular mechanisms of hearing loss in Nager syndrome. Dev. Biol. 2021, 476, 200–208. [Google Scholar] [CrossRef]
- Tsialikas, J.; Romer-Seibert, J. LIN28: Roles and regulation in development and beyond. Development 2015, 142, 2397–2404. [Google Scholar] [CrossRef]
- Zhu, H.; Shyh-Chang, N.; Segrè, A.V.; Shinoda, G.; Shah, S.P.; Einhorn, W.S.; Takeuchi, A.; Engreitz, J.M.; Hagan, J.P.; Kharas, M.G.; et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011, 147, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Grifone, R.; Shao, M.; Saquet, A.; Shi, D.L. RNA-binding protein Rbm24 as a multifaceted post-transcriptional regulator of embryonic lineage differentiation and cellular homeostasis. Cells 2020, 9, 1891. [Google Scholar] [CrossRef] [PubMed]
- Grifone, R.; Xie, X.; Bourgeois, A.; Saquet, A.; Duprez, D.; Shi, D.L. The RNA-binding protein Rbm24 is transiently expressed in myoblasts and is required for myogenic differentiation during vertebrate development. Mech. Dev. 2014, 134, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Grifone, R.; Saquet, A.; Xu, Z.; Shi, D.L. Expression patterns of Rbm24 in lens, nasal epithelium, and inner ear during mouse embryonic development. Dev. Dyn. 2018, 247, 1160–1169. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Lu, T.; Zhang, C.; Zhang, Y.Z.; Kong, S.H.; Shi, D.L. Rbm24 controls poly(A) tail length and translation efficiency of crystallin mRNAs in the lens via cytoplasmic polyadenylation. Proc. Natl. Acad. Sci. USA 2020, 117, 7245–7254. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Jen, H.I.; Kang, H.; Klisch, T.J.; Zoghbi, H.Y.; Groves, A.K. Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J. Neurosci. 2015, 35, 5870–5883. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.L. Planar cell polarity regulators in asymmetric organogenesis during development and disease. J. Genet. Genom. 2022. [Google Scholar] [CrossRef]
- Lush, M.E.; Diaz, D.C.; Koenecke, N.; Baek, S.; Boldt, H.; Peter, M.K.S.; Gaitan-Escudero, T.; Romero-Carvajal, A.; Busch-Nentwich, E.M.; Perera, A.G.; et al. scRNA-Seq reveals distinct stem cell populations that drive hair cell regeneration after loss of Fgf and Notch signaling. ELife 2019, 8, 44431. [Google Scholar] [CrossRef]
- Ghilardi, A.; Diana, A.; Bacchetta, R.; Santo, N.; Ascagni, M.; Prosperi, L.; Del Giacco, L. Inner ear and muscle developmental defects in Smpx-deficient zebrafish embryos. Int. J. Mol. Sci. 2021, 22, 6497. [Google Scholar] [CrossRef]
- Richardson, G.P.; Petit, C. Hair-bundle links: Genetics as the gateway to function. Cold Spring Harb. Perspect. Med. 2019, 9, a033142. [Google Scholar] [CrossRef]
- Bolz, H.; von Brederlow, B.; Ramírez, A.; Bryda, E.C.; Kutsche, K.; Nothwang, H.G.; Seeliger, M.; del C-Salcedó Cabrera, M.; Vila, M.C.; Molina, O.P.; et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat. Genet. 2001, 27, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Bork, J.M.; Peters, L.M.; Riazuddin, S.; Bernstein, S.L.; Ahmed, Z.M.; Ness, S.L.; Polomeno, R.; Ramesh, A.; Schloss, M.; Srisailpathy, C.R.; et al. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am. J. Hum. Genet. 2001, 68, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Di Palma, F.; Holme, R.H.; Bryda, E.C.; Belyantseva, I.A.; Pellegrino, R.; Kachar, B.; Steel, K.P.; Noben-Trauth, K. Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for Usher syndrome type 1D. Nat. Genet. 2001, 27, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Noben-Trauth, K.; Zheng, Q.Y.; Johnson, K.R. Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat. Genet. 2003, 35, 21–23. [Google Scholar] [CrossRef] [PubMed]
- Siemens, J.; Kazmierczak, P.; Reynolds, A.; Sticker, M.; Littlewood-Evans, A.; Müller, U. The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proc. Natl. Acad. Sci. USA 2002, 99, 14946–14951. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Hung, L.H.; Licht, T.; Kostin, S.; Looso, M.; Khrameeva, E.; Bindereif, A.; Schneider, A.; Braun, T. RBM24 is a major regulator of muscle-specific alternative splicing. Dev. Cell 2014, 31, 87–99. [Google Scholar] [CrossRef]
- Grifone, R.; Saquet, A.; Desgres, M.; Sangiorgi, C.; Gargano, C.; Li, Z.; Coletti, D.; Shi, D.L. Rbm24 displays dynamic functions required for myogenic differentiation during muscle regeneration. Sci. Rep. 2021, 11, 9423. [Google Scholar] [CrossRef]
- Sarkissian, M.; Winne, A.; Lafyatis, R. The mammalian homolog of suppressor-of-white-apricot regulates alternative mRNA splicing of CD45 exon 4 and fibronectin IIICS. J. Biol. Chem. 1996, 271, 31106–31114. [Google Scholar] [CrossRef]
- Darbelli, L.; Richard, S. Emerging functions of the Quaking RNA-binding proteins and link to human diseases. Wiley Interdiscip. Rev. RNA 2016, 7, 399–412. [Google Scholar] [CrossRef]
- Neumann, D.P.; Goodall, G.J.; Gregory, P.A. The Quaking RNA-binding proteins as regulators of cell differentiation. Wiley Interdiscip. Rev. RNA 2022, e1724. [Google Scholar] [CrossRef]
- Grill, B.; Wilson, G.M.; Zhang, K.X.; Wang, B.; Doyonnas, R.; Quadroni, M.; Schrader, J.W. Activation/division of lymphocytes results in increased levels of cytoplasmic activation/proliferation-associated protein-1: Prototype of a new family of proteins. J. Immunol. 2004, 172, 2389–2400. [Google Scholar] [CrossRef] [PubMed]
- Calarco, J.A.; Superina, S.; O’Hanlon, D.; Gabut, M.; Raj, B.; Pan, Q.; Skalska, U.; Clarke, L.; Gelinas, D.; van der Kooy, D.; et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell 2009, 138, 898–910. [Google Scholar] [CrossRef] [PubMed]
- Whitlon, D.S.; Gabel, C.; Zhang, X. Cochlear inner hair cells exist transiently in the fetal Bronx Waltzer (bv/bv) mouse. J. Comp. Neurol. 1996, 364, 515–522. [Google Scholar] [CrossRef]
- Sobkowicz, H.M.; Inagaki, M.; August, B.K.; Slapnick, S.M. Abortive synaptogenesis as a factor in the inner hair cell degeneration in the Bronx Waltzer (bv) mutant mouse. J. Neurocytol. 1999, 28, 17–38. [Google Scholar] [CrossRef]
- Cheong, M.A.; Steel, K.P. Early development and degeneration of vestibular hair cells in bronx waltzer mutant mice. Hear Res. 2002, 164, 179–189. [Google Scholar] [CrossRef]
- Hwang, J.Y.; Zukin, R.S. REST, a master transcriptional regulator in neurodegenerative disease. Curr. Opin. Neurobiol. 2018, 48, 193–200. [Google Scholar] [CrossRef]
- Raj, B.; Irimia, M.; Braunschweig, U.; Sterne-Weiler, T.; O’Hanlon, D.; Lin, Z.Y.; Chen, G.I.; Easton, L.E.; Ule, J.; Gingras, A.C.; et al. A global regulatory mechanism for activating an exon network required for neurogenesis. Mol. Cell 2014, 56, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Warzecha, C.C.; Carstens, R.P. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT). Semin. Cancer Biol. 2012, 22, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Bebee, T.W.; Park, J.W.; Sheridan, K.I.; Warzecha, C.C.; Cieply, B.W.; Rohacek, A.M.; Xing, Y.; Carstens, R.P. The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. Elife 2015, 4, e08954. [Google Scholar] [CrossRef] [PubMed]
- Burguera, D.; Marquez, Y.; Racioppi, C.; Permanyer, J.; Torres-Méndez, A.; Esposito, R.; Albuixech-Crespo, B.; Fanlo, L.; D’Agostino, Y.; Gohr, A.; et al. Evolutionary recruitment of flexible Esrp-dependent splicing programs into diverse embryonic morphogenetic processes. Nat. Commun. 2017, 8, 1799. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Hwang, J.Y.; Jung, S.; Park, H.; Bok, J.; Park, J.W. Position specific alternative splicing and gene expression profiles along the tonotopic axis of chick cochlea. Front. Mol. Biosci. 2021, 8, 726976. [Google Scholar] [CrossRef]
- Okano, H.; Kawahara, H.; Toriya, M.; Nakao, K.; Shibata, S.; Imai, T. Function of RNA-binding protein Musashi-1 in stem cells. Exp. Cell Res. 2005, 306, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Nicolson, T. The genetics of hair-cell function in zebrafish. J. Neurogenet. 2017, 31, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Kniss, J.S.; Jiang, L.; Piotrowski, T. Insights into sensory hair cell regeneration from the zebrafish lateral line. Curr. Opin. Genet. Dev. 2016, 40, 32–40. [Google Scholar] [CrossRef]
- Okano, T.; Kelley, M.W. Expression of insulin-like growth factor binding proteins during mouse cochlear development. Dev. Dyn. 2013, 242, 1210–1221. [Google Scholar] [CrossRef] [PubMed]
- Lush, M.E.; Piotrowski, T. Sensory hair cell regeneration in the zebrafish lateral line. Dev. Dyn. 2014, 243, 1187–1202. [Google Scholar] [CrossRef] [PubMed]
- Verma, B.; Akinyi, M.V.; Norppa, A.J.; Frilander, M.J. Minor spliceosome and disease. Semin. Cell Dev. Biol. 2018, 79, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Markmiller, S.; Cloonan, N.; Lardelli, R.M.; Doggett, K.; Keightley, M.C.; Boglev, Y.; Trotter, A.J.; Ng, A.Y.; Wilkins, S.J.; Verkade, H.; et al. Minor class splicing shapes the zebrafish transcriptome during development. Proc. Natl. Acad. Sci. USA 2014, 111, 3062–3067. [Google Scholar] [CrossRef] [PubMed]
- Li, D.K.; Tisdale, S.; Lotti, F.; Pellizzoni, L. SMN control of RNP assembly: From post-transcriptional gene regulation to motor neuron disease. Semin. Cell Dev. Biol. 2014, 32, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Orvis, J.; Gottfried, B.; Kancherla, J.; Adkins, R.S.; Song, Y.; Dror, A.A.; Olley, D.; Rose, K.; Chrysostomou, E.; Kelly, M.C.; et al. gEAR: Gene expression analysis resource portal for community-driven, multi-omic data exploration. Nat. Methods 2021, 18, 843–844. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, M.C.; Alam, S.S.; Kumar, S.; Jerome-Majewska, L.A. Spliceosomopathies and neurocristopathies: Two sides of the same coin? Dev. Dyn. 2020, 249, 924–945. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.; Li, S. SF3b4: A versatile player in eukaryotic cells. Front. Cell Dev. Biol. 2020, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Mittal, R.; Liu, G.; Polineni, S.P.; Bencie, N.; Yan, D.; Liu, X.Z. Role of microRNAs in inner ear development and hearing loss. Gene 2019, 686, 49–55. [Google Scholar] [CrossRef]
- Rehman, A.U.; Bird, J.E.; Faridi, R.; Shahzad, M.; Shah, S.; Lee, K.; Khan, S.N.; Imtiaz, A.; Ahmed, Z.M.; Riazuddin, S.; et al. Mutational spectrum of MYO15A and the molecular mechanisms of DFNB3 human deafness. Hum. Mutat. 2016, 37, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Brigande, J.V.; Heller, S. Quo vadis, hair cell regeneration? Nat. Neurosci. 2009, 12, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Qiang, R.; Dong, Y.; Zhang, Y.; Chen, Y.; Zhou, H.; Gao, X.; Chai, R. Hair cell regeneration from inner ear progenitors in the mammalian cochlea. Am. J. Stem. Cells 2020, 9, 25–35. [Google Scholar]
| RBPs | Functions | References |
|---|---|---|
| LIN28A | Upregulated after severe injury-induced progenitor regeneration in zebrafish neuromasts; required for recovery of progenitor cells and initiation of regeneration | [13] |
| LIN28B | Involved in hair cell production; antagonizes let-7 miRNA and increases Akt-mTORC1 activity; required for hair cell regeneration through mitotic and non-mitotic mechanisms | [14,15,16] |
| RBM24 | Possible transcriptional target of Atoh1 in mice; maintains the stability of smpx, gsdmeb, and otofa mRNAs in zebrafish; promotes inner ear-specific inclusion of exon 68 in Cdh23 mRNA; required for hair cell morphogenesis and survival | [17,18,19,20,21] |
| SFSWAP | Genetic interaction with Jagged1 in inner ear patterning; required for hearing and balance | [22,23] |
| QKI | Early target of noise-induced hearing loss; required for myelination of spiral ganglion neurons and auditory nerves | [24] |
| CAPRIN1 | Transcriptional target of Pou4f3; formation of stress granules in cochlear hair cells following ototoxin-induced damage; required for synapse formation between inner hair cells and spiral ganglion neurons as well as for maintenance of the auditory function | [25,26,27] |
| SRRM3, SRRM4 | Post-transcriptional inactivation of Rest protein in the inner ear by regulating the inclusion of exon 4 in Rest mRNA; required for the formation of cochlear and vestibular hair cells | [28,29,30] |
| ESRP1 | Regulation of alternative splicing; mutations are linked to SNHL in humans (OMIM#618013); required for Fgfr2 alternative splicing during inner ear morphogenesis and cochlear hair cell differentiation in mice | [12] |
| Musashi | Possible function in maintaining stem cell fate and in asymmetric division during hair cell regeneration | [31,32,33,34,35] |
| IGFBP3 | Required for inner ear growth, hair cell differentiation, and semicircular canal formation in zebrafish | [36] |
| RNPC3 | A component of the U12-dependent minor spliceosome; required for neuromast hair cell regeneration in zebrafish | [37] |
| SMN1, GEMIN3, GEMIN5 | Components of the SMN complex, which induces cell proliferation by inhibiting the ErbB pathway; required for hair cell regeneration following ototoxin-induced damage in zebrafish | [37,38] |
| SF3B4 | A component of the U2-dependent major spliceosome; mutations linked to hearing loss in humans; required for otic gene expression and otic development in Xenopus | [39] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, D.-L.; Cheng, X.-N.; Saquet, A.; Grifone, R. Emerging Roles of RNA-Binding Proteins in Inner Ear Hair Cell Development and Regeneration. Int. J. Mol. Sci. 2022, 23, 12393. https://doi.org/10.3390/ijms232012393
Shi D-L, Cheng X-N, Saquet A, Grifone R. Emerging Roles of RNA-Binding Proteins in Inner Ear Hair Cell Development and Regeneration. International Journal of Molecular Sciences. 2022; 23(20):12393. https://doi.org/10.3390/ijms232012393
Chicago/Turabian StyleShi, De-Li, Xiao-Ning Cheng, Audrey Saquet, and Raphaëlle Grifone. 2022. "Emerging Roles of RNA-Binding Proteins in Inner Ear Hair Cell Development and Regeneration" International Journal of Molecular Sciences 23, no. 20: 12393. https://doi.org/10.3390/ijms232012393
APA StyleShi, D.-L., Cheng, X.-N., Saquet, A., & Grifone, R. (2022). Emerging Roles of RNA-Binding Proteins in Inner Ear Hair Cell Development and Regeneration. International Journal of Molecular Sciences, 23(20), 12393. https://doi.org/10.3390/ijms232012393







