Quantitative Analysis of Collective Migration by Single-Cell Tracking Aimed at Understanding Cancer Metastasis
Abstract
1. Introduction
2. Results
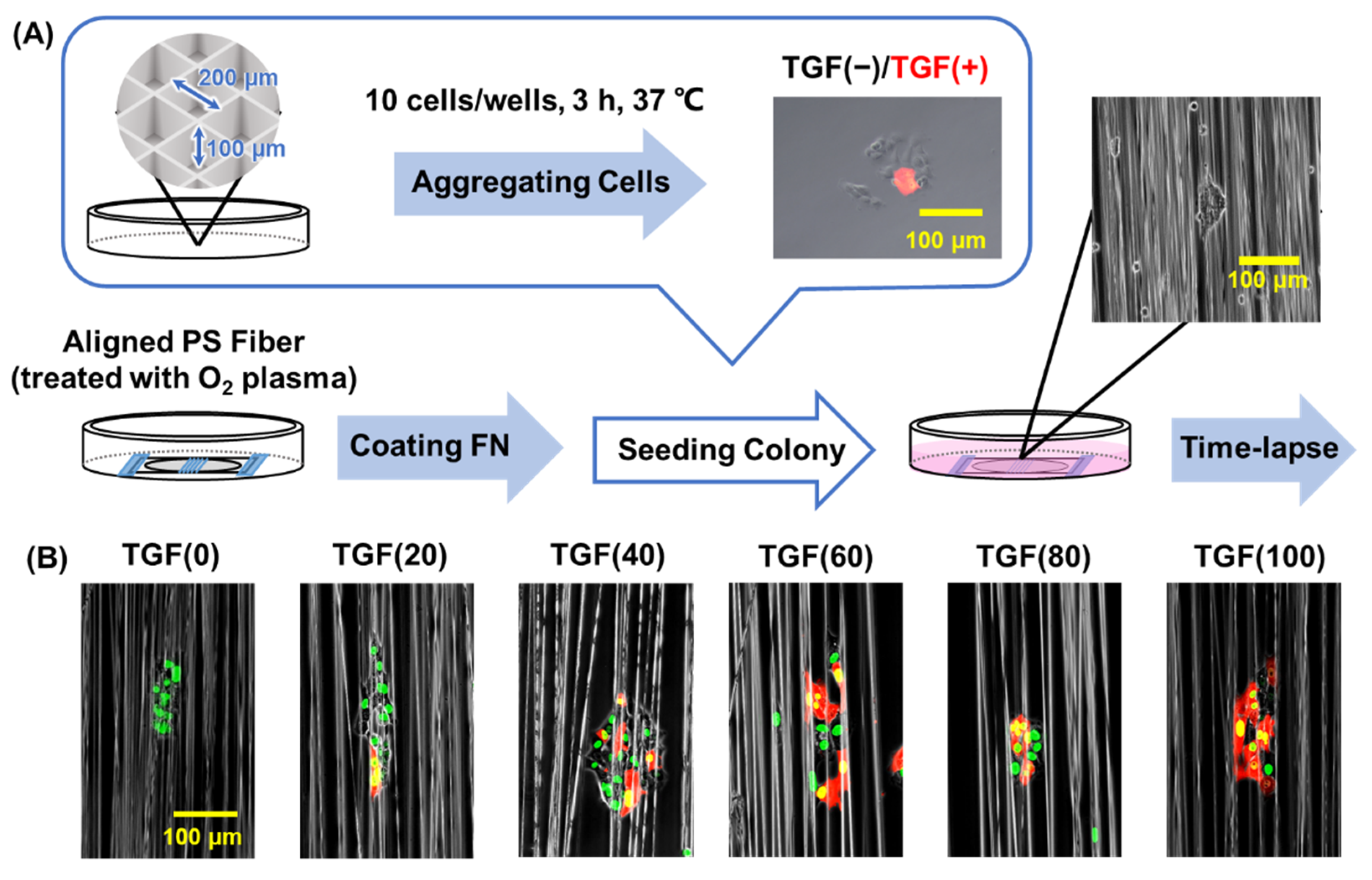
2.1. Cell Migration on Fiber
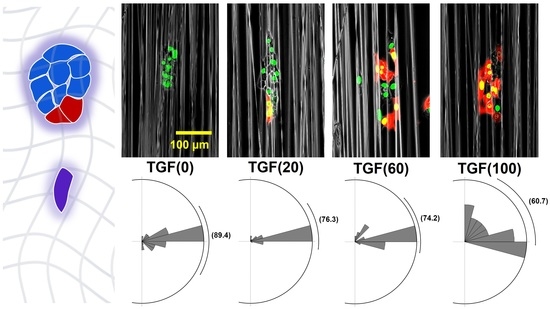
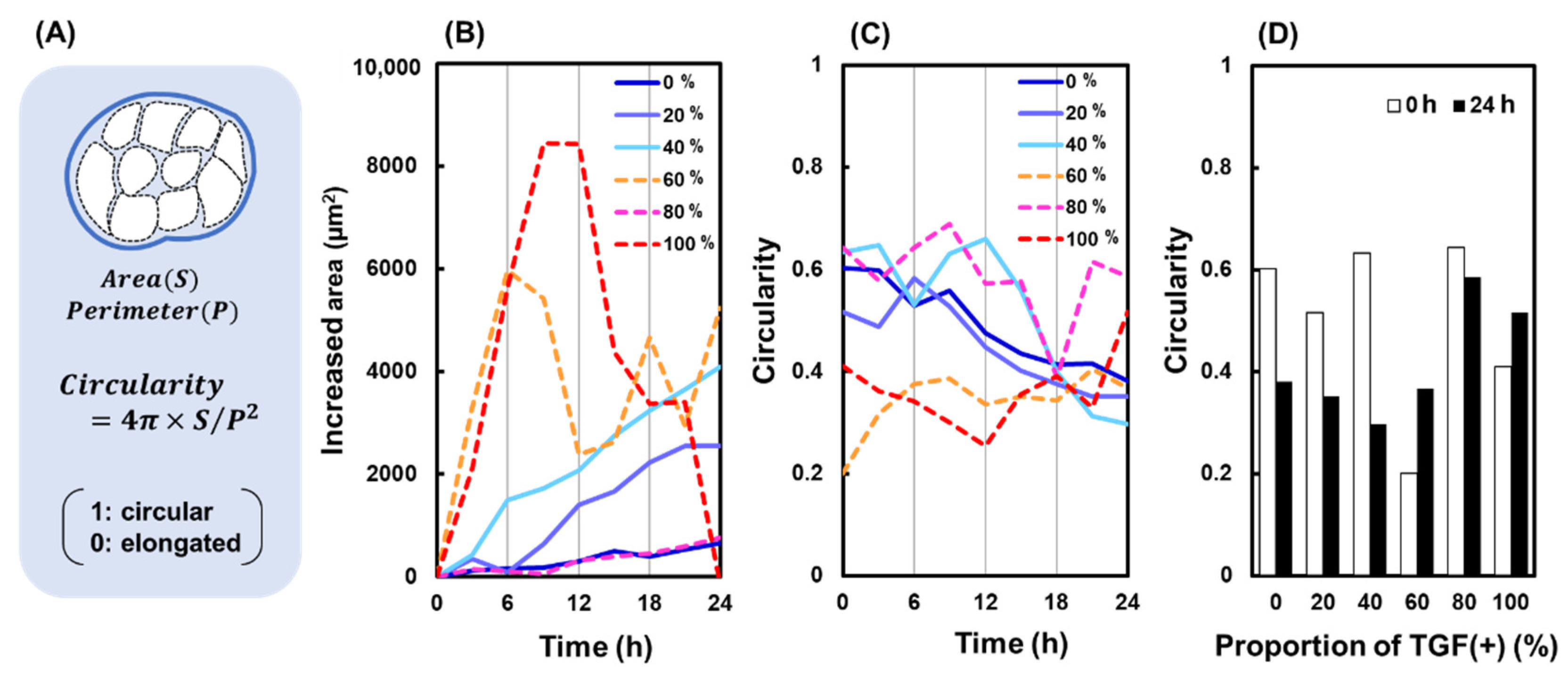
2.2. Colony Migration
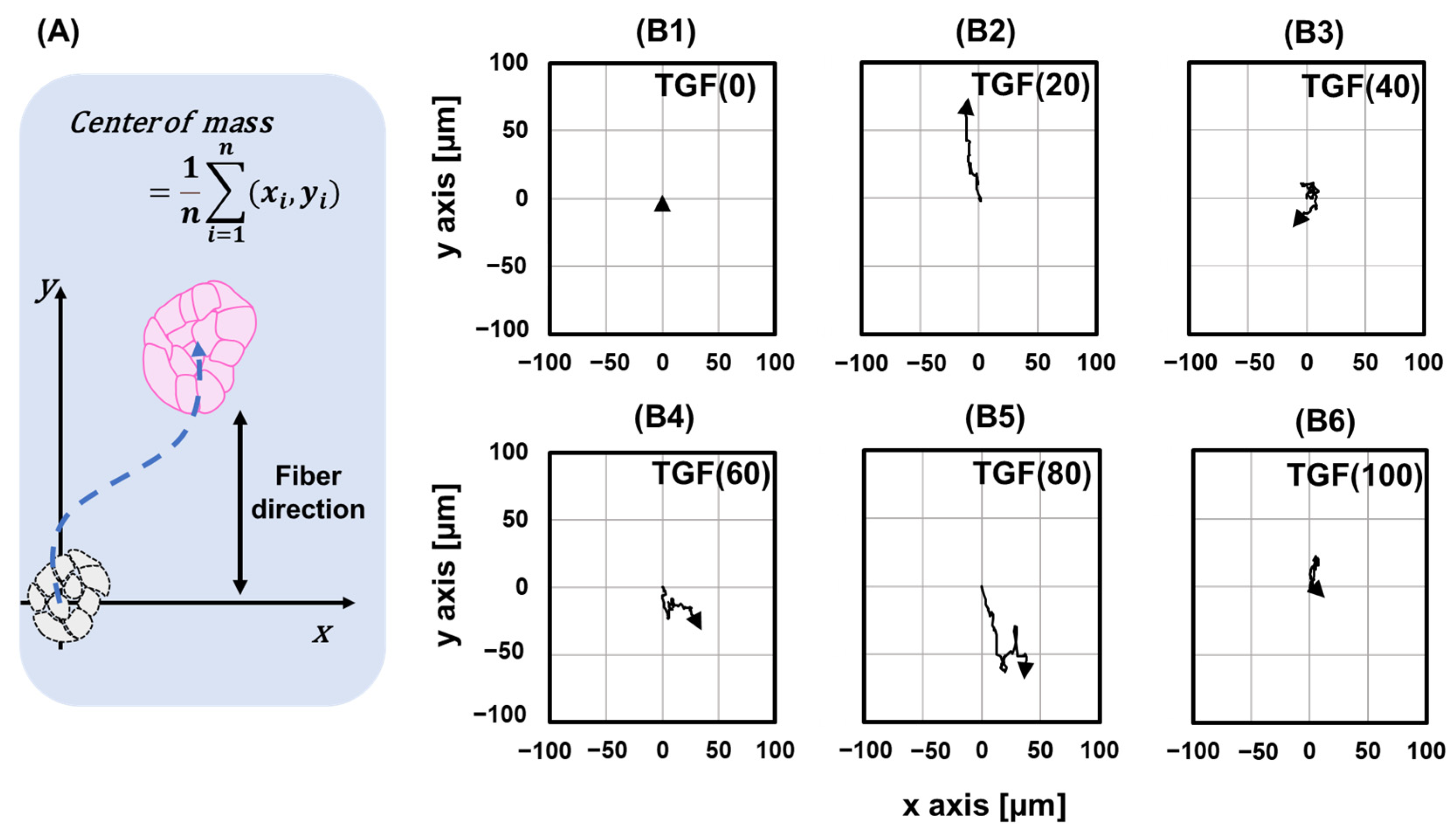
2.3. Migration of Single Cells
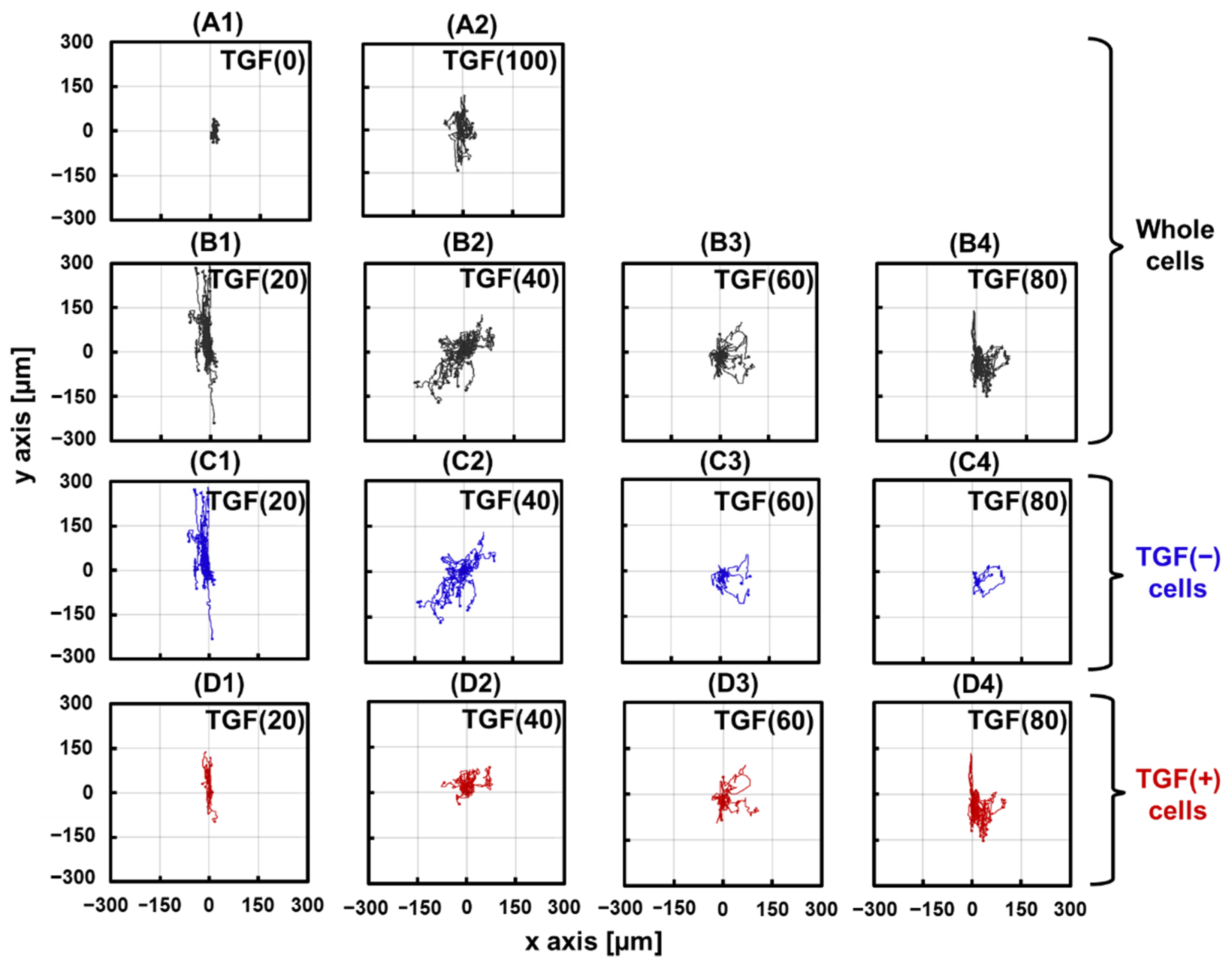
2.3.1. Changes in Cell Trajectories
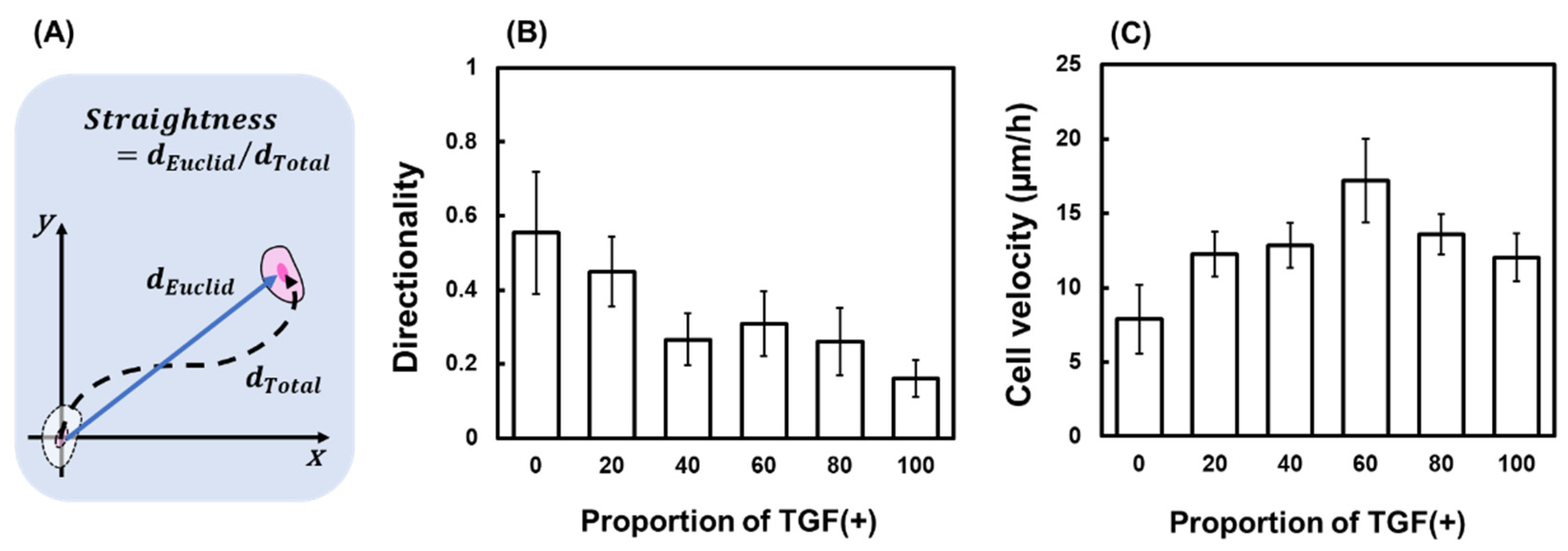
2.3.2. Changes in Straightness and Velocity
2.3.3. Changes in Direction Angle
3. Discussion
4. Materials and Methods
4.1. Electrospinning
4.2. Cell Culture
4.3. Morphological Analysis of Colonies
4.4. Migratory Analysis of Single Cell
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weiss, F.; Lauffenburger, D.; Friedl, P. Towards Targeting of Shared Mechanisms of Cancer Metastasis and Therapy Resistance. Nat. Rev. Cancer 2022, 22, 157–173. [Google Scholar] [CrossRef] [PubMed]
- Kölsch, V.; Charest, P.G.; Firtel, R.A. The Regulation of Cell Motility and Chemotaxis by Phospholipid Signaling. J. Cell Sci. 2008, 121, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Cooper, J.A. Actin, a Central Player in Cell Shape and Movement. Science 2009, 326, 1208–1212. [Google Scholar] [CrossRef]
- Mayor, R.; Etienne-Manneville, S. The Front and Rear of Collective Cell Migration. Nat. Rev. Mol. Cell Biol. 2016, 17, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Borghi, N.; Lowndes, M.; Maruthamuthu, V.; Gardel, M.L.; Nelson, W.J. Regulation of Cell Motile Behavior by Crosstalk between Cadherin- and Integrin-Mediated Adhesions. Proc. Natl. Acad. Sci. USA 2010, 107, 13324–13329. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The MiR-200 Family and MiR-205 Regulate Epithelial to Mesenchymal Transition by Targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A Reciprocal Repression between ZEB1 and Members of the MiR-200 Family Promotes EMT and Invasion in Cancer Cells. EMBO Rep. 2008, 9, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lamouille, S.; Derynck, R. TGF-β-Induced Epithelial to Mesenchymal Transition. Cell Res. 2009, 19, 156–172. [Google Scholar] [CrossRef] [PubMed]
- Plou, J.; Juste-Lanas, Y.; Olivares, V.; del Amo, C.; Borau, C.; García-Aznar, J.M. From Individual to Collective 3D Cancer Dissemination: Roles of Collagen Concentration and TGF-β. Sci. Rep. 2018, 8, 12723. [Google Scholar] [CrossRef] [PubMed]
- Zavadil, J.; Böttinger, E.P. TGF-β and Epithelial-to-Mesenchymal Transitions. Oncogene 2005, 24, 5764–5774. [Google Scholar] [CrossRef]
- Zhang, J.; Tian, X.J.; Zhang, H.; Teng, Y.; Li, R.; Bai, F.; Elankumaran, S.; Xing, J. TGF-β-Induced Epithelial-to-Mesenchymal Transition Proceeds through Stepwise Activation of Multiple Feedback Loops. Sci. Signal. 2014, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the Tumour Transition States Occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef]
- Huang, R.Y.J.; Wong, M.K.; Tan, T.Z.; Kuay, K.T.; Ng, A.H.C.; Chung, V.Y.; Chu, Y.S.; Matsumura, N.; Lai, H.C.; Lee, Y.F.; et al. An EMT Spectrum Defines an Anoikis-Resistant and Spheroidogenic Intermediate Mesenchymal State That Is Sensitive to e-Cadherin Restoration by a Src-Kinase Inhibitor, Saracatinib (AZD0530). Cell Death Dis. 2013, 4, e915. [Google Scholar] [CrossRef]
- Bocci, F.; Jolly, M.K.; Tripathi, S.C.; Aguilar, M.; Hanash, S.M.; Levine, H.; Onuchic, J.N. Numb Prevents a Complete Epithelial-Mesenchymal Transition by Modulating Notch Signaling. J. R. Soc. Interface 2017, 14, 20170512. [Google Scholar] [CrossRef] [PubMed]
- Fontana, F.; Marzagalli, M.; Sommariva, M.; Gagliano, N.; Limonta, P. In Vitro 3D Cultures to Model the Tumor Microenvironment. Cancers (Basel) 2021, 13, 2970. [Google Scholar] [CrossRef] [PubMed]
- Gamboa Castro, M.; Leggett, S.E.; Wong, I.Y. Clustering and Jamming in Epithelial-Mesenchymal Co-Cultures. Soft Matter 2016, 12, 8327–8337. [Google Scholar] [CrossRef]
- Sokullu, E.; Cücük, Z.L.; Sarabi, M.R.; Birtek, M.T.; Bagheri, H.S.; Tasoglu, S. Microfluidic Invasion Chemotaxis Platform for 3D Neurovascular Co-Culture. Fluids 2022, 7, 238. [Google Scholar] [CrossRef]
- Yu-Ju Wu, C.; Chen, C.H.; Lin, C.Y.; Feng, L.Y.; Lin, Y.C.; Wei, K.C.; Huang, C.Y.; Fang, J.Y.; Chen, P.Y. CCL5 of Glioma-Associated Microglia/Macrophages Regulates Glioma Migration and Invasion via Calcium-Dependent Matrix Metalloproteinase 2. Neuro-Oncol. 2020, 22, 253–266. [Google Scholar] [CrossRef]
- Vishwakarma, M.; Di Russo, J.; Probst, D.; Schwarz, U.S.; Das, T.; Spatz, J.P. Mechanical Interactions among Followers Determine the Emergence of Leaders in Migrating Epithelial Cell Collectives. Nat. Commun. 2018, 9, 3469. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Riahi, R.; Torab, P.; Zhang, D.D.; Wong, P.K. Collective Cell Migration in 3D Epithelial Wound Healing. ACS Nano 2019, 13, 1204–1212. [Google Scholar] [CrossRef]
- Bandzerewicz, A.; Gadomska-Gajadhur, A. Into the Tissues: Extracellular Matrix and Its Artificial Substitutes: Cell Signalling Mechanisms. Cells 2022, 11, 914. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.M.; Sixt, M. Mechanisms of 3D Cell Migration. Nat. Rev. Mol. Cell Biol. 2019, 20, 738–752. [Google Scholar] [CrossRef]
- Braghirolli, D.I.; Steffens, D.; Pranke, P. Electrospinning for Regenerative Medicine: A Review of the Main Topics. Drug Discov. Today 2014, 19, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Carbone, E.J.; Lo, K.W.H.; Laurencin, C.T. Electrospinning of Polymer Nanofibers for Tissue Regeneration. Prog. Polym. Sci. 2015, 46, 1–24. [Google Scholar] [CrossRef]
- Cheung, K.J.; Gabrielson, E.; Werb, Z.; Ewald, A.J. Collective Invasion in Breast Cancer Requires a Conserved Basal Epithelial Program. Cell 2013, 155, 1639–1651. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Kajita, M.K.; Deguchi, K.; Suye, S.; Fujita, S. Time-Series Clustering of Single-Cell Trajectories in Collective Cell Migration. Cancers 2022, 14, 4587. [Google Scholar] [CrossRef]
- Huang, W.-Y.; Suye, S.; Fujita, S. Cell Trapping via Migratory Inhibition within Density-Tuned Electrospun Nanofibers. ACS Appl. Bio Mater. 2021, 4, 7456–7466. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xin, Z.; Deguchi, K.; Suye, S.-i.; Fujita, S. Quantitative Analysis of Collective Migration by Single-Cell Tracking Aimed at Understanding Cancer Metastasis. Int. J. Mol. Sci. 2022, 23, 12372. https://doi.org/10.3390/ijms232012372
Xin Z, Deguchi K, Suye S-i, Fujita S. Quantitative Analysis of Collective Migration by Single-Cell Tracking Aimed at Understanding Cancer Metastasis. International Journal of Molecular Sciences. 2022; 23(20):12372. https://doi.org/10.3390/ijms232012372
Chicago/Turabian StyleXin, Zhuohan, Keiko Deguchi, Shin-ichiro Suye, and Satoshi Fujita. 2022. "Quantitative Analysis of Collective Migration by Single-Cell Tracking Aimed at Understanding Cancer Metastasis" International Journal of Molecular Sciences 23, no. 20: 12372. https://doi.org/10.3390/ijms232012372
APA StyleXin, Z., Deguchi, K., Suye, S.-i., & Fujita, S. (2022). Quantitative Analysis of Collective Migration by Single-Cell Tracking Aimed at Understanding Cancer Metastasis. International Journal of Molecular Sciences, 23(20), 12372. https://doi.org/10.3390/ijms232012372